Abstract
Rifampin has concentration-dependent activity against Mycobacterium tuberculosis. However, marked intersubject variation of rifampin concentrations occurs. In this study, we evaluated rifampin pharmacokinetics in relation to tuberculosis, geographic region, race, and single nucleotide polymorphisms of the human transporter genes SLCO1B1, SLCO1B3, and MDR1. Seventy-two adults with pulmonary tuberculosis from Africa, North America, and Spain were evaluated during multidrug intensive-phase therapy, and their results were compared to those from 16 healthy controls from North America. Rifampin pharmacokinetic values were similar between tuberculosis patients and controls (geometric mean [GM] area under the concentration-time curve from 0 to 24 h [AUC0-24] of 40.2 versus 40.9 μg·h/ml; P = 0.9). However, in multivariable analyses, the rifampin AUC0-24 was significantly affected by rifampin dosage (in mg/kg of body weight), polymorphisms in the SLCO1B1 gene, and the presence of tuberculosis by geographic region. The adjusted rifampin AUC0-24 was lowest in patients with tuberculosis from Africa compared to that in non-African patients or control subjects. The adjusted rifampin AUC0-24 was also 36% lower among participants with SLCO1B1 genotype c.463CA than that among participants with SLCO1B1 genotype c.463CC (adjusted GM, 29.8 versus 46.7 μg·h/ml; P = 0.001). Polymorphisms in the SLCO1B1 gene associated with lower rifampin exposure were more frequent among black subjects. In conclusion, marked intersubject variation of the rifampin AUC0-24 values was observed, but the mean values of the AUC0-24 did not significantly vary between patients with tuberculosis and healthy controls. Lower rifampin exposure was associated with the polymorphism of the SLCO1B1 c.463C>A gene. When adjusted for the patient mg/kg dosage and transporter gene polymorphisms, rifampin exposure was lower in patients with tuberculosis, which suggests that additional absorption or metabolic processes affect rifampin exposure with tuberculosis disease.
Rifamycins have consistently demonstrated concentration-dependent activity against Mycobacterium tuberculosis in both in vitro and in vivo models (14, 17, 18, 24). There is marked intersubject variation of rifamycin concentrations among patients on standard tuberculosis therapy, and the basis for variation is not well characterized. Advanced HIV infection has been associated with low rifampin concentrations, but marked differences in rifampin concentrations are also found among tuberculosis patients without HIV infection and between patient groups (7, 21, 27, 29). Low rifamycin exposures can have clinical significance. In a treatment trial of rifabutin in patients with HIV-associated tuberculosis, low rifabutin exposure (area under the concentration-time curve [AUC]) was associated with treatment failure or relapse with acquired rifamycin-resistant M. tuberculosis (33). In treatments with intermittent rifamycin-containing regimens (with lower total rifamycin exposure per week), increases in relapse rates were observed (5, 20).
Two recent Tuberculosis Trials Consortium (TBTC) clinical trials demonstrated a decreased initial response to therapy (by 2-month culture conversion) among patients enrolled at African sites rather than non-African sites (3, 9). Possible explanations for the decreased response include differences in severity of tuberculosis, morbidity due to HIV infection, differences in microbiologic techniques between sites, or the pharmacokinetics of antitubercular drugs, particularly rifampin.
Our primary study objectives were the comparison of rifampin pharmacokinetics among patients with tuberculosis and healthy controls and between regions (Africa versus non-Africa) and races (black versus all other races). Different human phenotypes may result from alternative forms of a gene at the same physical location (called alleles), and gene polymorphisms are reported to vary among races. Because organic anion transporter peptides (OATP) play key roles in transport and disposition of xenobiotics in the gastrointestinal tract and liver, the secondary objectives of the study were to determine the relationships between rifampin pharmacokinetics and human genetic polymorphisms of drug influx and efflux transporter genes (anion-transporting polypeptides [SLCO1B1 and SLCO1B3] and P-glycoprotein [MDR1]).
Rifampin pharmacokinetic data of healthy controls were previously described (32).
(Some of the pharmacokinetic and pharmacogenetic data for patients with tuberculosis were previously reported at the Second International Workshop on Clinical Pharmacology of Tuberculosis Drugs in 2009 [30] and as abstract A791 at the American Thoracic Society International Conference in 2008 [31].)
MATERIALS AND METHODS
Experimental design.
Patients were recruited as a convenience sample to this pharmacokinetic substudy from Tuberculosis Trials Consortium (TBTC) treatment trials 27 and 28 at research sites with capacity for intensive pharmacokinetic sampling. Both treatment trials enrolled adults with smear-positive pulmonary tuberculosis from Brazil, Canada, South Africa, Spain, Uganda, and the United States. Both trials were prospective, placebo-controlled, randomized phase II studies comparing different regimens during the intensive phase of tuberculosis treatment (the first 2 months). Study 27 had a factorial design that compared moxifloxacin and ethambutol (with isoniazid, rifampin, and pyrazinamide) and administration of drugs five versus three times per week (3). Study 28 compared moxifloxacin and isoniazid (with rifampin, pyrazinamide, and ethambutol) administered five times per week (9). The healthy controls were enrolled as a convenience sample from Texas (32). The primary endpoint of both treatment trials was sputum culture status after 2 months of treatment.
We enrolled 77 patients with tuberculosis from eight sites in Africa, Spain, and the United States in the pharmacokinetic study. However, samples from five patients from Africa were lost in a single shipment. Rifampin pharmacokinetic parameters in 72 patients with active tuberculosis and in 16 healthy controls were evaluated. For comparisons of black versus white subjects (see Table 6), one patient and one control subject of Asian race were excluded from analysis. Race and ethnicity were identified by self-report of the study participant.
The Institutional Review Boards of the Centers for Disease Control and Prevention and of participating TBTC sites approved the study; informed consent was obtained from all participants.
Rifampin tablets and sample collection.
For 80 of 86 (93%) cases in the analyses (35 from Uganda and 45 from the United States), the rifampin tablets for the pharmacokinetic study were good manufacturing practice (GMP)-grade drugs purchased from VersaPharm in the United States. The remaining rifampin tablets were obtained from Sandoz (for two patients from South Africa), Sanofi Aventis (for two from Spain), and UDL Laboratories (for two from the United States). Analyses by high-pressure liquid chromatography (HPLC) of sample rifampin tablets from Uganda demonstrated expected rifampin concentrations (C. Peloquin, unpublished data).
The pharmacokinetic sampling was performed between the 9th and 40th doses (administered 5 or 7 days per week and after at least three consecutive daily doses) of study treatment administered by directly observed therapy (DOT). Standard rifampin doses were used (600 mg for patients weighing >45 kg and 450 mg for patients weighing ≤45 kg). Pharmacokinetic sampling in normal volunteers was performed with the 10th consecutive daily dose of rifampin administered by DOT. All study drugs for pharmacokinetic sampling were administered in a fasting state. Blood samples were collected just before observed dosing of the study drug and then at 1, 2, 6, 8 to 10, 11 to 13, and 23 to 25 h thereafter.
Rifampin and human gene analyses.
Standard techniques were used to determine plasma drug concentrations of rifampin as previously described (22), with the following modifications. The HPLC procedures for analysis were performed with the following equipment: a Thermo Finnigan (San Jose, CA) model P4000 pump, a model AS 3000 fixed-volume autosampler, and a model UV2000 detector. The standard curves for the rifampin concentrations in plasma covered a range of 0.5 to 50 μg/ml. The absolute recovery of rifampin from serum was 95.5%. The within-day precision (coefficient of variation [CV]) of validation-quality control samples was 2.4 to 4.6%, and the overall validation precision was 6.3 to 7.1%.
Genomic DNA was extracted from patients' whole blood by using the QIAamp DNA blood minikit (catalog no. 51106) obtained from Qiagen (Gaithersburg, MD). TaqMan real-time PCR was used to detect single nucleotide polymorphisms (SNPs) of seven human transporter genes (MDR1 c.3435C>T, SLCO1B1 c.11187G>A, SLCO1B1 c.388A>G, SLCO1B1 c.463C>A, SLCO1B1 521T>C, SLCO1B1 c.1463G>C, and SLCO1B3 c.334T>G) (26). In brief, PCR amplification was carried out by using AmpliTaq Gold DNA polymerase obtained from Applied Biosystems (Foster City, CA). The reaction was carried out in a final volume of 25 μl containing 5 ng of sample DNA. Cycle parameters used were as follows: 95°C for 10 min and then 50 cycles of 92°C for 15 s and 60°C for 1 min. Real-time SNP detections were performed and analyzed on a Stratagene (La Jolla, CA) MX3000P PCR system. Detections were duplicated at separate times for reconfirmation.
Statistical and pharmacokinetic analyses.
Analyses of area under the concentration-time curve from 0 to 24 h (AUC0-24) values were performed using noncompartmental techniques (WinNonlin version 4; Pharsight Corporation, Mountain View, CA). Demographic data were reported where applicable in Table 1 as arithmetic means, and pharmacokinetic data in Tables 2, 3, and 4 were reported as both arithmetic means (± standard deviations [SD]) and geometric means. The statistical tests used transformed pharmacokinetic data to the log base e in Tables 2, 3, and 4, because in the analysis of covariance (ANCOVA) model (Table 5), transformed rifampin pharmacokinetic parameters better satisfied the assumptions of the model (see below). Data analyses were performed using SAS version 9.1 (SAS Institute Inc., Cary, NC) for determination of the false discovery rate and NCSS 2007 (Kaysville, UT) for other tests. For dichotomous data, differences between groups were determined using Pearson's chi-square statistic, and for nonparametric analyses, the Kruskal-Wallis test was used. Differences between groups were considered statistically significant at the level of P values of <0.05. The aim of the multivariate ANCOVA was to adjust for all factors that could affect rifampin blood concentrations (like rifampin dose and patient weight) and to delineate other factors associated with drug absorption, metabolism, or elimination. Patient covariates were explored with univariate analyses. Preselected terms were combined into a full multivariable model using backward elimination and forward addition. Two- and three-way interactions of terms in the final main effects model were checked for significance. Model terms with a P value of <0.05 were retained in the final model. The rifampin pharmacokinetic parameters were transformed to the log base e scale to better satisfy the assumptions of the analysis; residuals of the rifampin AUC and peak concentration transformed to the log base e were normally distributed by the Shapiro-Wilk test but were not with untransformed data. Hence, the log base e data were used for analysis. Each of the seven human transporter gene polymorphisms was treated as a main effect with two or three levels, depending on the gene. Tuberculosis disease by geographic region (present in Africans, present in non-Africans, or not present in non-Africans), HIV status (infected or not infected), race (black, white, or Asian), and sex were also treated as main effects, and age, rifampin dosage in mg/kg of body weight, and the number of rifampin doses taken at pharmacokinetic sampling were treated as covariates. Validity of the model was studied by skewness and kurtosis of residuals, consistent with a bell-shaped distribution. Homogeneity of variance for validity of the model was supported by plotting of residuals versus predicted values. When the null hypothesis of equal means was rejected, adjusted least-squares means were reported to ensure fair comparisons. Adjusted means in these results refers to the means adjusted for all model effects and unequal sample sizes. In order to account for analysis of multiple terms in the model (genes and other factors), the false discovery rate (25) was computed along with the individual significance of each term as it was eliminated from or retained in the model. In secondary analyses, the ANCOVA model was examined to also determine the effects of coadministration of moxifloxacin, isoniazid, or ethambutol on rifampin exposure of the covariate. Because of the study design, the individual effect of pyrazinamide on the rifampin concentration could not be evaluated.
TABLE 1.
Demographic and clinical data from the 72 study subjects with tuberculosis and 16 uninfected control subjects
| Characteristic | Values fore: |
P value | ||
|---|---|---|---|---|
| Patients with tuberculosis from: |
Healthy controls from the U.S. | |||
| Africa | U.S. and Spain | |||
| Demographic factors | ||||
| Race | <.001a | |||
| Black | 37 (100) | 5 (14) | 4 (25) | |
| White | 0 (0) | 29 (83) | 11 (69) | |
| Asian | 0 (0) | 1 (3) | 1 (6) | |
| Ethnicity, Hispanic | 0 (0) | 25 (71) | 8 (50) | <.001a |
| Clinical data | ||||
| Mean age in yr (SD)b | 30.1 (7.3) | 43.2 (15.0) | 40.1 (11.4) | <.001b |
| No. of males (%) | 30 (81) | 29 (83) | 12 (75) | 0.80a |
| Mean wt in kg (SD)b | 54.9 (8.2) | 62.6 (13.4) | 85.5 (15.9) | <.001b |
| No. of subjects with HIV infection | 1 (3) | 2 (6) | 0 | 0.54a |
| No. of initial AFB smears of sputa at treatment trialc | 0.01c | |||
| 1+ | 4 (11) | 7 (29) | ||
| 2+ | 1 (3) | 5 (21) | ||
| 3+ | 13 (35) | 3 (13) | ||
| 4+ | 19 (51) | 9 (38) | ||
| No. of subjects with cavitation on chest X-rayc | 0.60c | |||
| No cavities on chest X-ray | 10 (27) | 8 (24) | ||
| Cavities at <4 cm total | 18 (49) | 14 (41) | ||
| Cavities at ≥4 cm total | 9 (24) | 12 (35) | ||
| Mean dosage of rifampin in mg/kg (SD)b | 10.9 (1.3) | 9.8 (1.7) | 7.3 (1.3) | <.001b |
| Mean no. of rifampin doses at time of PK sampling (SD) | 27.5 (7.3) | 21.8 (10.0) | 10 (0) | <.001b |
| No. (%) of gene SNPsd | ||||
| SLCO1B1 c.463C>A | 0.45a | |||
| CC (73) | 30 (81) | 31 (89) | 12 (75) | |
| CA (15) | 7 (19) | 4 (11) | 4 (25) | |
| SLCO1B1 c.521T>C | 0.08a | |||
| TT (74) | 35 (95) | 25 (71) | 14 (88) | |
| TC (11) | 2 (5) | 8 (23) | 1 (6) | |
| CC (3) | 0 (0) | 2 (6) | 1 (6) | |
| SLCO1B1 c.1463G>C | 0.81a | |||
| GG (84) | 34 (95) | 34 (97) | 15 (94) | |
| GC (4) | 2 (5) | 1 (3) | 1 (6) | |
| SLCO1B1 c.388A>G | <.001a | |||
| AA (12) | 1 (3) | 10 (29) | 1 (6) | |
| AG (32) | 7 (19) | 15 (43) | 10 (63) | |
| GG (44) | 29 (78) | 10 (29) | 5 (31) | |
| SLCO1B1 c.11187G>A | 0.37a | |||
| GG (80) | 33 (89) | 31 (89) | 16 (100) | |
| GA (8) | 4 (11) | 4 (11) | ||
| SLCO1B3 334T>G | <.001a | |||
| TT (19) | 15 (41) | 2 (6) | 2 (13) | |
| TG (42) | 19 (51) | 14 (40) | 9 (56) | |
| GG (27) | 3 (8) | 19 (54) | 5 (31) | |
| MDR1 c.3435C>T | 0.004a | |||
| CC (57) | 32 (86) | 19 (54) | 6 (38) | |
| CT (26) | 5 (14) | 13 (37) | 8 (50) | |
| TT (5) | 0 (0) | 3 (9) | 2 (13) | |
P values obtained by chi-square testing.
Arithmetic mean, with P values obtained by ANOVA.
P values obtained by chi-square testing among patients with tuberculosis for AFB smear status and pulmonary cavitation.
Alleles are alternative forms of a gene at the same physical locus, and different alleles may result in different phenotypic traits. For example, for the human transporter gene SLCO1B1 c.463C>A, 71 study participants were found to have a genotype homozygous for the wild-type allele (CC), 15 were heterozygous for wild and alternative alleles (CA), and none were homozygous for the alternative alleles (CC). The SNP of the SLCO1B1 c.463C>A gene was significantly associated with a phenotypic change in rifampin exposure (P < 0.001). For each transporter gene polymorphism, the number of individuals with each genotype and the associated phenotype are described in Table 3.
The number of subjects from each group are as follows: 72 patients with tuberculosis (37 from Africa and 35 from the United States and Spain) and 16 healthy controls from the United States.
TABLE 2.
Rifampin pharmacokinetic parameters in univariate analyses among 72 patients with tuberculosis from Africa, the United States, and Spain and 16 control subjects from the United Statesc
| Pharmacokinetic parameterd | No. of subjects | Arithmetic mean (± SD) | Geometric mean |
P value |
|
|---|---|---|---|---|---|
| Healthy controls vs all TB patientsa | Healthy controls vs TB patients from Africa vs TB patients not from Africab | ||||
| AUC0-24 | |||||
| All subjects | 86 | 46.0 (23.9) | 40.3 | ||
| Healthy controls | 16 | 44.0 (18.6) | 40.9 | ||
| All subjects with TB | 70 | 46.4 (25.1) | 40.2 | 0.90 | |
| TB, Africa | 37 | 43.2 (22.3) | 37.8 | 0.58 | |
| TB, non-Africa | 33 | 50.1 (27.8) | 43.1 | ||
| AUC0-24/body wt in kg | |||||
| All subjects | 86 | 0.79 (0.49) | 0.65 | ||
| Healthy controls | 16 | 0.55 (0.32) | 0.49 | ||
| All subjects with TB | 70 | 0.84 (0.51) | 0.70 | 0.04 | |
| Subjects with TB, Africa | 37 | 0.82 (0.46) | 0.70 | 0.11 | |
| Subjects with TB, non-Africa | 33 | 0.86 (0.57) | 0.70 | ||
| Cmax | |||||
| All subjects | 88 | 8.4 (4.8) | 7.0 | ||
| Healthy controls | 16 | 9.9 (3.6) | 9.2 | ||
| All subjects with TB | 72 | 8.1 (5.0) | 6.6 | 0.06 | |
| Subjects with TB, Africa | 37 | 8.0 (5.0) | 6.6 | 0.19 | |
| Subjects with TB, non-Africa | 35 | 8.2 (5.0) | 6.6 | ||
| Cl/F | |||||
| All subjects | 86 | 17.0 (10.4) | 14.7 | ||
| Healthy controls | 16 | 15.7 (5.8) | 14.7 | 0.99 | |
| All subjects with TB | 70 | 17.3 (11.3) | 14.7 | ||
| Subjects with TB, Africa | 37 | 18.2 (12.4) | 15.5 | 0.67 | |
| Subjects with TB, non-Africa | 33 | 16.3 (9.9) | 13.8 | ||
| Vz/F | |||||
| All subjects | 86 | 57.2 (59.4) | 45.0 | ||
| Healthy controls | 16 | 41.6 (16.9) | 39.0 | 0.25 | |
| All subjects with TB | 70 | 60.8 (64.9) | 46.5 | 0.40 | |
| Subjects with TB, Africa | 37 | 56.1 (47.4) | 44.3 | ||
| Subjects with TB, non-Africa | 33 | 66.0 (80.7) | 49.1 | ||
| Half-life | |||||
| All subjects | 86 | 2.3 (1.1) | 2.1 | ||
| Healthy controls | 16 | 1.9 (0.3) | 1.8 | ||
| All subjects with TB | 70 | 2.4 (1.2) | 2.2 | 0.51 | |
| Subjects with TB, Africa | 37 | 2.1 (0.8) | 2.0 | 0.005 | |
| Subjects with TB, non-Africa | 33 | 2.7 (1.5) | 2.5 | ||
P values obtained by unpaired t testing for comparison of healthy controls to all patients with tuberculosis for a ln-transformed variable. TB, tuberculosis.
P values obtained by one-way ANOVA for comparison of healthy controls to patients with tuberculosis from and not from Africa for a ln-transformed variable.
Adequate pharmacokinetic data for calculation of AUC0-24 and half-life values in 70 of 72 patients with tuberculosis.
Cl/F, apparent oral clearance; Vz/F, apparent volume of distribution based on the terminal phase.
TABLE 3.
Effects of demographics, clinical factors, and transporter gene polymorphisms on rifampin AUC0-24 values
| Factor (no. of subjects) | Rifampin AUC0-24 |
P valuea | |
|---|---|---|---|
| Arithmetic mean (SD) | Geometric mean | ||
| Age, divided at median valuec | 0.73 | ||
| ≤33 yr (42) | 46.7 (22.7) | 41.1 | |
| >33 yr (44) | 45.3 (25.3) | 39.6 | |
| Sex | 0.44 | ||
| Male (70) | 43.7 (19.2) | 39.5 | |
| Female (16) | 56.2 (37.6) | 44.3 | |
| Groups | 0.59 | ||
| Healthy controls (16) | 44.0 (18.6) | 40.9 | |
| TB patients from Africa (37) | 43.2 (22.3) | 37.8 | |
| TB patients not from Africa (33) | 50.1 (27.8) | 43.1 | |
| Race | 0.21 | ||
| Black (46) | 42.1 (21.1) | 37.0 | |
| White (38) | 51.3 (26.8) | 45.2 | |
| Asian (2) | 34.3 | 34.1 | |
| HIV | 0.35 | ||
| Infected (3) | 34.3 (21.5) | 30.4 | |
| Not infected (82) | 46.6 (24.1) | 40.9 | |
| Diabetes mellitus | 0.82 | ||
| Present (8) | 48.4 (33.9) | 41.7 | |
| Not present (78) | 45.7 (23.0) | 40.2 | |
| Rifampin dosage in mg/kg divided at median valuec | 0.02 | ||
| <10.2 (44) | 39.5 (18.0) | 35.6 | |
| ≥10.2 (42) | 52.8 (27.5) | 46.0 | |
| Cavities on chest X-ray | 0.50b | ||
| None (16) | 44.2 (30.2) | 37.3 | |
| Cavities < 4 cm (32) | 46.0 (24.0) | 40.2 | |
| Cavities ≥ 4 cm (21) | 50.4 (22.8) | 45.0 | |
| Gene SNP | |||
| SLCO1B1 c.463C>A | <.001 | ||
| CC (71) | 49.4 (23.9) | 44.4 | |
| CA (15) | 29.9 (16.6) | 25.6 | |
| SLCO1B1 c.521T>C | 0.11 | ||
| TT (73) | 43.8 (22.6) | 38.5 | |
| TC (11) | 56.3 (30.5) | 50.2 | |
| CC (2) | 68.1 (15.6) | 67.2 | |
| SLCO1B1 c.1463G>C | 0.21 | ||
| GG (82) | 45.2 (23.6) | 39.7 | |
| GC (4) | 61.6 (28.7) | 55.7 | |
| SLCO1B1 c.388A>G | 0.29 | ||
| AA (11) | 50.1 (19.3) | 45.9 | |
| AG (32) | 49.1 (27.1) | 43.4 | |
| GG (43) | 42.7 (22.5) | 36.9 | |
| SLCO1B1 c.11187G>A | 0.39 | ||
| GG (78) | 44.8 (22.4) | 39.7 | |
| GA (8) | 57.4 (36.0) | 47.1 | |
| MDR1 c.3435C>T | 0.67 | ||
| CC (56) | 45.6 (25.0) | 39.5 | |
| CT (25) | 45.1 (21.4) | 40.7 | |
| TT (5) | 55.2 (27.2) | 49.2 | |
| SLCO1B3 c.334T>G | 0.99 | ||
| TT (19) | 44.5 (22.2) | 39.6 | |
| TG (42) | 46.4 (24.7) | 40.5 | |
| GG (25) | 46.4 (24.8) | 40.6 | |
P values obtained by ANOVA among all subjects for ln-transformed rifampin AUC0-24 values.
P values obtained by ANOVA among patients with tuberculosis for ln-transformed AUC0-24 values.
Low and high means shown for descriptive purposes. Pearson's correlation was nonsignificant for age (slope = 0.004, R-square = 0.01, P = 0.37) but significant for rifampin dosage (slope = 0.07, R-square = 0.06, P = 0.02).
TABLE 4.
Rifampin pharmacokinetic parameters among subjects with SLCO1B1 c.463C>A and c.521T>C SNPs in univariate analyses
| Parameter (no. of subjects)e | Arithmetic mean (± SD) | Geometric mean | RGM | P valueb |
|---|---|---|---|---|
| SLCO1B1 c.463C>Aa | ||||
| AUC0-24 | ||||
| All (86) | 46.0 (23.9) | 40.3 | ||
| CC (71) | 49.4 (24.0) | 44.4 | 0.58 | 0.0002 |
| CA (15) | 29.9 (16.6) | 25.6 | ||
| Cmax | ||||
| All (88) | 8.4 (4.8) | 7.0 | ||
| CC (73) | 8.8 (4.8) | 7.5 | 0.66 | 0.03 |
| CA (15) | 6.3 (4.5) | 5.0 | ||
| Clss/F | ||||
| All (86) | 17.0 (10.4) | 14.7 | ||
| CC (71) | 15.1 (7.9) | 13.5 | 1.63 | 0.0009 |
| CA (15) | 26.1 (15.7) | 22.0 | ||
| Vz/F | ||||
| All (86) | 57.2 (59.4) | 45.0 | ||
| CC (71) | 47.1 (30.0) | 41.0 | 1.71 | 0.0004 |
| CA (15) | 104.8 (118.1) | 70.1 | ||
| Half-life | ||||
| All (86) | 2.3 (1.1) | 2.1 | ||
| CC (71) | 2.2 (0.8) | 2.1 | 1.05 | 0.74 |
| CA (15) | 2.6 (2.1) | 2.2 | ||
| SLCO1B1 c.521T>Cc | ||||
| AUC0-24 | ||||
| TT (73) | 43.8 (22.6) | 38.5 | 1.37 | 0.11d |
| TC (11) | 56.3 (30.5) | 50.2 | ||
| CC (2) | 68.1 (15.6) | 67.2 | ||
| Cmax | ||||
| TT (74) | 8.4 (5.0) | 7.0 | 1.04 | 0.92d |
| TC (11) | 8.3 (3.3) | 7.4 | ||
| CC (3) | 9.5 (6.8) | 6.7 | ||
| Clss/F | ||||
| TT (73) | 17.8 (10.9) | 15.4 | 0.74 | 0.14d |
| TC (11) | 13.2 (5.6) | 11.9 | ||
| CC (2) | 9.1 (2.1) | 8.9 | ||
| Vz/F | ||||
| TT (73) | 57.6 (62.7) | 44.4 | 1.09 | 0.89d |
| TC (11) | 58.5 (39.2) | 50.9 | ||
| CC (2) | 37.0 (4.5) | 36.9 | ||
| Half-life | ||||
| TT (73) | 2.1 (1.0) | 2.0 | 1.47 | <0.001d |
| TC (11) | 3.3 (1.5) | 3.0 | ||
| CC (2) | 2.9 (0.3) | 2.9 |
The ratio of geometric means (RGM) for SLCO1B1 c.463C>A genotypes was calculated with CA/CC.
P values obtained by t testing of an unpaired ln-transformed variable for the SLCO1B1 c.463CC genotype versus the c.463CA genotype.
The RGM of SLCO1B1 c.521 was calculated with (TC + CC)/TT.
P values obtained by ANCOVA of a ln-transformed variable.
Clss/F, apparent oral plasma clearance at steady state; Vz/F, apparent volume of distribution based on the terminal phase.
TABLE 5.
Analysis by multivariate ANCOVA of the effects on rifampin pharmacokinetic parameters by transporter gene polymorphism, demographic and clinical factors, and rifampin dosage
| Main effects | Adjusted mean rifampin AUC0-24 (μg·h/ml) | Adjusted mean of rifampin peak concn | ANCOVA P valuea | False discovery rate P value |
|---|---|---|---|---|
| Rifampin AUC0-24 (n = 86) | ||||
| Rifampin dosage vs ln AUC0-24, slope = 1.041b | 0.002 | 0.02 | ||
| SLCO1B1 c.463C>A | ||||
| CC (n = 71) | 46.7 | 0.001 | 0.02 | |
| CA (n = 15) | 29.8 | |||
| Groupsd | 0.02d | |||
| (1) Healthy controls (n = 16) | 49.7 | (1) vs (2) = .007 | (1) vs (2) = .03 | |
| (2) Subjects with TB, Africa (n = 37) | 29.0 | (2) vs (3) = 0.07 | (2) vs (3) = 0.23 | |
| (3) Subjects with TB, non-Africa (n = 33) | 36.0 | (3) vs (1) = 0.06 | (3) vs (1) = 0.23 | |
| Rifampin Cmax (n = 88) | ||||
| Rifampin dosage vs ln Cmax, slope = 1.53c | 0.0003 | 0.002 | ||
| Groupsd | 0.0006d | |||
| (1) Healthy controls (n = 16) | 14.5 | (1) vs (2) = 0.0002 | (1) vs (2) = 0.002 | |
| (2) Subjects with TB, Africa (n = 37) | 5.5 | (2) vs (3) = 0.24 | (2) vs (3) = 0.56 | |
| (3) Subjects with TB, non-Africa (n = 35) | 6.5 | (3) vs (1) = 0.0004 | (3) vs (1) = 0.002 |
Main effects retained in the ANCOVA model, with P values of <0.05.
ln AUC0-24 = 1.041 times (ln rifampin dosage in mg/kg).
ln Cmax = 1.53 times (ln rifampin dosage in mg/kg).
Comparisons in ANCOVA using post hoc least significant difference (LSD) method.
RESULTS
We evaluated rifampin pharmacokinetics in 72 patients with tuberculosis and in 16 healthy controls. Patients with active tuberculosis, particularly those from Africa, weighed less than controls, were sampled for pharmacokinetics after administration of more rifampin doses (Table 1) and, based on the standard rifampin dose algorithm, received a higher rifampin dosage (10.9, 9.8, and 7.3 mg/kg for African patients, non-African patients, and healthy controls, respectively). African patients were younger, and all were of the black race, compared to 14% of non-African tuberculosis patients and 25% of the healthy controls (P < 0.001). Patients from Africa had higher baseline acid-fast bacillus (AFB)-grade smear counts than non-African patients, but other clinical features of patients with tuberculosis were similar by region of enrollment. Differences were found between regions in the distribution of polymorphisms of SLCO1B1 c.388A>G, SLCO1B3 c.334T>G, and MDR1 c.3435C>T (Table 1).
Effects of tuberculosis, geographic region, and demographic factors on rifampin pharmacokinetic parameters.
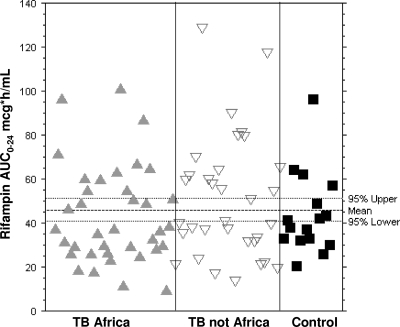
There were no significant differences in rifampin pharmacokinetic parameters by univariate analysis between patients with tuberculosis and healthy controls (Table 2). Normalized rifampin AUC0-24 values (AUC0-24 divided by body weight) were significantly decreased in healthy controls because of greater body weight in the control group. When subjects were separated into three groups—patients with tuberculosis from Africa, patients with tuberculosis from sites outside Africa, and healthy controls—there were no significant differences in AUC0-24, peak concentration, or apparent clearance, although rifampin half-life was significantly longer in non-African patients with tuberculosis than in others (Table 2). Of note, there were large intersubject differences (greater than a 10-fold range) in rifampin AUC0-24 values among study participants (Fig. 1). More patients with tuberculosis irrespective of geographic origin had low peak concentrations of rifampin (defined as <8 μg/ml) compared to those of healthy controls (27) (proportions of low maximum concentrations of drug in serum [Cmax] in tuberculosis patients from Africa, in patients not from Africa, and in healthy controls, 52%, 41%, and 7%, respectively; P = 0.01, chi-square test).
FIG. 1.
Rifampin mean exposures (AUC0-24) were similar among patients with tuberculosis from Africa (▴), patients with tuberculosis from the United States and Spain (▿), and healthy controls (▪), but there was a more than 10-fold range in rifampin AUC values among subjects. The arithmetic means and 95% confidence intervals for all cases are indicated in the scatter plot.
In univariate analysis of demographic and clinical factors, subjects receiving a lower rifampin dosage (median value < 10.2 mg/kg) had a lower geometric mean (GM) AUC0-24 value for rifampin (P = 0.02) (Table 3). Rifampin exposure was lower among black versus white patients, although the difference was not significant (GM AUC0-24 values of 37.0 μg·h/ml versus 45.2 μg·h/ml; ratio of geometric mean [RGM] AUC0-24, 0.82; P = 0.08, unpaired t test).
Effect of human transporter gene polymorphisms on rifampin pharmacokinetic parameters.
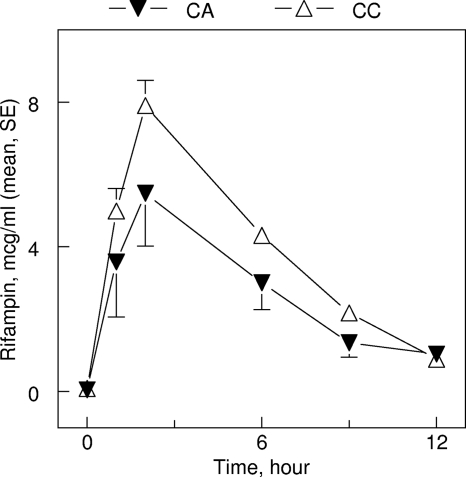
A single nucleotide polymorphism (SNP) of SLCO1B1 gene c.463C>A significantly affected rifampin pharmacokinetic parameters in univariate analysis (Tables 3 and 4). Patients with the SLCO1B1 c.463C>A polymorphism had 42% lower rifampin exposure (25.6 versus 44.4 μg·h/ml; P = 0.0002, unpaired t test), 34% lower peak concentration levels (P = 0.03), and 63% greater apparent oral clearance (P = 0.0009) (Fig. 2; Table 4). Differences in rifampin plasma concentrations by the c.463C>A polymorphism were most notable in the first 2 h after the drug administration (Fig. 2). SNP of the SLCO1B1 c.521T>C gene did not significantly affect rifampin pharmacokinetic parameters in univariate analysis, i.e., nonsignificantly greater rifampin exposure (AUC0-24 values of 67.2, 50.2, and 38.5 μg·h/ml for the c.521CC, -TC, and -CC genotypes, respectively; P = 0.11) and decreased apparent oral clearance (P = 0.14) were observed. SNPs of other transporter genes were not associated with altered rifampin exposure (Table 3).
FIG. 2.
Plasma rifampin concentrations (in μg/ml) versus time (in hours) in patients grouped by SLCO1B1 c.463CA polymorphisms (▾) versus SLCO1B1 c.463CC genotypes (▵) (arithmetic means and standard errors [SE] are shown with vertical bars).
Multivariate analysis of factors associated with rifampin pharmacokinetics.
In multivariate analyses, the rifampin AUC0-24 was significantly affected by rifampin dosage (in mg/kg), SLCO1B1 c.463C>A polymorphism, and presence of tuberculosis by the region of enrollment (Table 5). The adjusted mean rifampin AUC0-24 was correlated directly with rifampin dosage, decreasing with the SLCO1B1 c.463CA versus -CC genotype (29.8 versus 46.7 μg·h/ml, respectively; P = 0.001, ANCOVA) and also showing the lowest values in the group of patients with tuberculosis from Africa compared to those of non-African patients with tuberculosis or healthy controls (29.0, 36.0, and 49.7 μg·h/ml, respectively; P = 0.02, ANCOVA) (post hoc comparison of African patients with tuberculosis and healthy controls, P value of 0.007 by the least-significant-difference test). The rifampin AUC0-24 was not significantly associated with the number of rifampin doses at the time of pharmacokinetic sampling or with the use of moxifloxacin, isoniazid, or ethambutol.
Distribution of SLCO1B1 single nucleotide polymorphisms between races.
The SLCO1B1 c.463CA and -CC genotypes were detected in 15 and 71 subjects, respectively. Subjects of black race were 2.4-fold more likely than whites (24 versus 10%; P = 0.09, chi-square test) to have the SLCO1B1 c.463CA genotype associated with lower rifampin exposure (Tables 3 and 6). There was also a disproportionate distribution of the SLCO1B1 c.521T>C genotypes among races, i.e., subjects of black race were 1.4-fold more likely than whites (96 versus 70%, respectively; P = 0.005) (Table 6) to have the c.521TT polymorphism associated with a shorter rifampin half-life.
TABLE 6.
Distribution of SLCO1B1 c.463C>A and c.521T>C SNPs between black and white races
| SLCO1B1 genotype | Allele | No. (%) of subjects with the indicated race |
P valuea | |
|---|---|---|---|---|
| Black | White | |||
| c.463C>A | CC | 35 (76) | 36 (90) | 0.09 |
| CA | 11 (24) | 4 (10) | ||
| Total | 46 (100) | 40 (100) | ||
| c.521T>C | TT | 44 (96) | 28 (70) | 0.005 |
| CT or CC | 2 (4) | 12 (30) | ||
| Total | 46 (100) | 40 (100) | ||
P values obtained by chi-square testing.
DISCUSSION
We evaluated the possible reasons for the large intersubject differences of rifampin concentrations among persons receiving standard treatment dosages. The geometric mean rifampin AUC0-24 and peak concentration were not significantly different between patients with tuberculosis and healthy controls or between African and non-African patients with tuberculosis. However, there were substantial intersubject differences of rifampin exposures (greater than a 10-fold range of rifampin AUC0-24 values) (Fig. 1). We found that a SNP in the SLCO1B1 gene was associated with rifampin exposure.
In multivariate ANCOVAs that were adjusted for rifampin dosage (in mg/kg), the adjusted mean rifampin AUC0-24 was significantly lower in patients with tuberculosis from Africa than in healthy controls from North America. The similarity of rifampin (arithmetic and geometric) mean AUC0-24 values among African and non-African patients and controls in univariate analyses in part was due to lower body weight in African versus non-African patients and controls and, as a result, higher rifampin dosage in mg/kg in Africans patients (10.9, 9.8, and 7.3 mg/kg, respectively). Previous studies have shown regional differences in rifampin preparations (2, 6, 10, 12, 29). van Crevel et al. attributed low rifampin concentrations in 70% of patients in Indonesia to the decreased bioavailability of local drug preparations (29). To ensure a uniformly bioavailable formulation in this pharmacokinetic study, 93% of patients (from the United States and Africa) used rifampin manufactured by a single U.S. manufacturer. Further, potency of the study rifampin used in the trial was independently confirmed by testing of 12 sample tablets obtained from the drug supply used at the Uganda trial site.
The rifampin AUC0-24 was significantly affected by the SNP of SLCO1B1 c.463C>A. Peptide products of the SLCO family of genes, organic anion transporter peptides (OATP), mediate transport of xenobiotics in the gastrointestinal tract and liver (11, 13). Chronic rifampin use induces OATP and P-glycoprotein (MDR1) via activation of the nuclear pregnane X receptor (28). Further, rifampin is a potent inhibitor of OATP1B1 (19). Thus, there potentially are alternate ways for SNP of OATP1B1 c.463 to affect rifampin pharmacokinetic parameters. In this study, the adjusted mean rifampin AUC0-24 was 36% lower in 15 people with the SLCO1B1 c.463CA genotype than in 71 people with the c.463CC genotype (P = 0.001). Effects on rifampin absorption and/or clearance by the c.463CA polymorphism are suggested by the decreased GM rifampin exposure and peak concentration. Opposing effects of increased apparent oral clearance and volume of distribution resulted in no change in half-life. Subjects of black race (most were enrolled from African sites) were more likely to have the SLCO1B1 c.463CA polymorphism associated with lower rifampin exposure. In a previous study, the SLCO c.521T>C polymorphism did not significantly affect the pharmacokinetics of single-dose rifampin in Asian subjects (15). With multidose therapy, we showed that the SLCO c.521TC and -TT genotypes were associated with longer rifampin half-life in univariate but not multivariate analyses.
In this study, plasma rifampin concentrations were measured, but concentrations in lung or other body compartments were not. Conte et al. compared rifampin concentrations in lung epithelial lining fluid, plasma, and alveolar cells (8). Considerable interpatient variability was found, and rifampin was not detected in epithelial lining fluid samples of 20% of subjects, despite detection in plasma samples (8).
In multivariate ANCOVA, significant effects on the rifampin AUC0-24 were demonstrated by tuberculosis disease by geographic region after adjustments for subject rifampin dosage and SLCO polymorphisms. This suggests that additional processes may affect rifampin exposure. For example, data from a prior study suggested a decrease in absorptive capacity with tuberculosis because patients with low rifampin blood concentrations also demonstrated decreased excretion of orally administered mannitol and lactulose (23). Differences in rifampin exposures also could be caused by different rates of metabolism. Jamis-Dow and colleagues demonstrated in human liver microsomes that rifampin is metabolized to 25-desacetyl rifampin by B-esterases (16). However, the effect of B-esterase gene polymorphisms on rifampin pharmacokinetic parameters has not been evaluated.
The substantial intersubject differences in rifampin exposure (Fig. 1) are noteworthy. The important metric for comparing pharmacokinetics among populations, however, may not be the mean (or median) value; it may be the proportion of participants with a value below some threshold rifampin concentration. In this regard, it is notable that a number of patients with active tuberculosis had low rifampin concentrations. There is currently great interest in higher dosages and more frequent dosing of rifamycins to increase the potency of tuberculosis treatment regimens and allow treatment shortening. Improved therapeutic efficacy may result from increased rifamycin dosage not only by increased mean (and median) rifamycin concentrations but also by greater concentrations above a minimum threshold among patients with the lowest exposures.
In TBTC studies 27 and 28, from which the patients in this pharmacokinetic substudy were recruited, patients who enrolled at two African sites had much lower 2-month sputum culture conversion than did patients who enrolled at sites in North America, Spain, and Brazil. For example, the proportion of patients with negative cultures after 2 months of treatment in study 27 was 60% (108/179) among Africans patients, compared to 84% (91/108) among patients recruited elsewhere (P < 0.0001). However, the difference in 2-month culture conversions between African and non-African patients was not simply explained by a difference in the unadjusted mean rifampin AUC or peak concentration. More likely, the lower response to intensive-phase therapy among patients enrolled at African sites is multifactorial.
Our study has several limitations. First, patients with tuberculosis were recruited as a convenience sample into the pharmacokinetic substudy during two clinical trials. However, differences in the individual drugs in these regimens did not appear to significantly influence rifampin exposure or other factors in multivariate analyses. Second, we did not evaluate rifampin pharmacokinetics in tissue compartments (e.g., alveolar lining fluid) that may be important in its action (8). Because drug transporters mediate uptake in the intestine, liver, and brain, further investigation is needed to assess the role of SNPs of drug transporters (SLCO1B1, SLCO1B3, and MDR1) in other tissue compartments. Third, the sample size (and power) of the study was not sufficient to evaluate subgroup differences (e.g., only three patients with HIV coinfection were sampled). Fourth, study subjects were sampled after a greater mean number of daily rifampin doses were administered to patients with tuberculosis than to controls. However, steady-state conditions are reported to be achieved after the sixth daily dose of rifampin (1, 4), and in this study, all study subjects were sampled after at least nine rifampin doses and after at least three consecutive daily doses. Also, in multivariate analyses, the number of rifampin doses received at the time of pharmacokinetic sampling was not significantly associated with rifampin exposure or peak concentration. Fifth, although we did not adjust for the multiple comparisons made in the analysis, the effect of multiple comparisons was assessed using the false discovery rate. Finally, residual confounding by measured or unmeasured variables is possible.
The strengths of this pharmacokinetic study are the comparison of healthy controls to well-characterized patients who were receiving directly supervised therapy during carefully conducted clinical trials, the enrollment of patients with tuberculosis from three continents, and the use of pharmacokinetic sampling at 7 time points to capture complete pharmacokinetic data for study subjects.
In summary, marked intersubject variations of rifampin AUC0-24 values was observed, but mean AUC0-24 values did not significantly vary between patients with tuberculosis and healthy controls. Lower rifampin exposure was associated with the polymorphism of the SLCO1B1 c.463C>A gene. When adjusted for rifampin dosage and transporter gene polymorphisms, rifampin exposure was significantly lower with tuberculosis disease in Africans, which suggests that additional absorption or metabolic processes affect rifampin exposure.
Acknowledgments
This work was supported by the Centers for Disease Control and Prevention, the Global Alliance for TB Drug Development, and the Veterans Administration. Assistance in evaluation of patients was provided by the Frederic C. Bartter General Clinical Research Center at the VAMC San Antonio (supported by NIH grant MO1-RR-01346). This study was sponsored by the Tuberculosis Trials Consortium.
We are grateful to Elsa Villarino and Kenneth Castro for their support and leadership within the CDC, to Stefan Goldberg and Jose Maria Miro Meda for their assistance with the study, to Lorna Bozeman for help with study logistics, and to Timothy Sterling, Chad Heilig, and Martino Laurenzi for review of the manuscript.
Footnotes
Published ahead of print on 26 July 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Borin, M. T., J. T. Chambers, B. J. Carel, S. Gagnon, and W. W. Freimuth. 1997. Pharmacokinetic study of the interaction between rifampin and delavirdine mesylate. Clin. Pharmacol. Ther. 61:544-553. [DOI] [PubMed] [Google Scholar]
- 2.Buniva, G., V. Pagani, and A. Carozzi. 1983. Bioavailability of rifampicin capsules. Int. J. Clin. Pharmacol. Ther. Toxicol. 21:404-409. [PubMed] [Google Scholar]
- 3.Burman, W. J., S. Goldberg, J. L. Johnson, G. Muzanye, M. Engle, A. W. Mosher, S. Choudhri, C. L. Daley, S. S. Munsiff, Z. Zhao, A. Vernon, and R. E. Chaisson. 2006. Moxifloxacin versus ethambutol in the first 2 months of treatment for pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 174:331-338. [DOI] [PubMed] [Google Scholar]
- 4.Burman, B., K. Gallicano, and C. Peloquin. 2001. Comparative pharmacokinetics and pharmacodynamics of the rifamycin antibacterials. Clin. Pharmacokinet. 40:327-341. [DOI] [PubMed] [Google Scholar]
- 5.Chang, K. C., C. C. Leung, W. W. Yew, S. L. Chan, and C. M. Tam. 2006. Dosing schedules of 6-month regimens and relapse for pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 174:1153-1158. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee, P. 2001. India's trade in fake drugs—bringing the counterfeiters to book. Lancet 357:1776. [DOI] [PubMed] [Google Scholar]
- 7.Choudhri, S. H., M. Hawken, S. Gathua, G. O. Minyiri, W. Watkins, J. Sahai, D. S. Sitar, F. Y. Aoki, and R. Long. 1997. Pharmacokinetics of antimycobacterial drugs in patients with tuberculosis, AIDS and diarrhea. Clin. Infect. Dis. 25:104-111. [DOI] [PubMed] [Google Scholar]
- 8.Conte, E. Jr., J. A. Golden, J. E. Kipps. E. T. Lin, and E. Zurlinden. 2004. Effect of sex and AIDS status on the plasma and intrapulmonary pharmacokinetics of rifampicin. Clin. Pharmacokinet. 43:395-404. [DOI] [PubMed] [Google Scholar]
- 9.Dorman, S. E., J. L. Johnson, S. Goldberg, G. Muzanye, N. Padayatchi, L. Bozeman, C. M. Heilig, J. Bernardo, S. Choudhri, J. H. Grosset, E. Guy, P. Guyadeen, M. C. Leus, G. Maltas, D. Menzies, E. L. Nuermberger, M. Villarino, A. Vernon, R. E. Chaisson, and the Tuberculosis Trials Consortium. 2009. Substitution of moxifloxacin for isoniazid during intensive phase treatment of pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 180:273-280. [DOI] [PubMed] [Google Scholar]
- 10.Ellard, G. A., and P. B. Fourie. 1999. Rifampicin bioavailability: a review of its pharmacology and the chemotherapeutic necessity for ensuring optimal absorption. Int. J. Tuber. Lung Dis. 3(Suppl. 3):S301-S308. [PubMed] [Google Scholar]
- 11.Englund, G., F. Rorsman, A. Ronnblom, U. Karlbom, U. L. Lazorova, J. Gråsjö, A. Kindmark, and P. Artursson. 2006. Regional levels of drug transporters along the human intestinal tract: co-expression of ABC and SLC transporters and comparison with Caco-2 cells. Eur. J. Pharm. Sci. 29:269-277. [DOI] [PubMed] [Google Scholar]
- 12.Fox, W. 1990. Drug combinations and the bioavailability of rifampicin. Tubercle 71:241-245. [DOI] [PubMed] [Google Scholar]
- 13.Glaeser, H., D. G. Bailey, G. K. Dresser, J. C. Gregor, U. I. Schwarz, J. S. McGrath, E. Jolicoeur, W. Lee, B. F. Leake, R. G. Tirona, and R. B. Kim. 2007. Intestinal drug transporter expression and the impact of grapefruit juice in humans. Clin. Pharm. Ther. 81:362-370. [DOI] [PubMed] [Google Scholar]
- 14.Grosset, J. H., I. M. Rosenthal, M. Zhang, and E. L. Nuermberger. 2008. Dose-ranging comparison of rifampin and rifapentine in combination with isoniazid and pyrazinamide in the mouse model of tuberculosis, abstr. A-1825. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 15.He, Y. J., W. Zhang, Y. Chen, D. Guo, J. H. Tu, L. Y. Xu, Z. R. Tan, B. L. Chen, Z. Li, G. Zhou, B. N. Yu, J. Kirchheiner, and H. H. Zhou. 2009. Rifampicin alters atorvastatin plasma concentration on the basis of SLCO1B1 521T>C polymorphism. Clin. Chim. Acta 405:49-52. [DOI] [PubMed] [Google Scholar]
- 16.Jamis-Dow, C. A., A. G. Katki, J. M. Collins, and R. W. Klecker. 1997. Rifampin and rifabutin and their metabolism by human liver esterases. Xenobiotica 27:1015-1024. [DOI] [PubMed] [Google Scholar]
- 17.Jayaram, R., S. Gaonkar, P. Kaur, B. L. Suresh, B. N. Mahesh, R. Jayashree, V. Nandi, S. Bharat, R. K. Shandil, E. Kantharaj, and V. Balasubramanian. 2003. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob. Agents Chemother. 47:2118-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji, B., C. Truffot-Pernot, C. Lacroix, M. C. Raviglione, R. J. O'Brien, P. Olliaro, G. Roscigno, and J. H. Grosset. 1993. Effectiveness of rifampin, rifabutin, and rifapentine for preventive therapy of tuberculosis in mice. Am. Rev. Respir. Dis. 148:1541-1546. [DOI] [PubMed] [Google Scholar]
- 19.Lau, Y. Y., Y. Huang, L. Frassetto, and L. Z. Benet. 2007. Effect of OATP1B transporter inhibition on the pharmacokinetics of atorvastatin in healthy volunteers. Clin. Pharm. Ther. 81:194-204. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., S. S. Munsiff, C. R. Driver, and J. Sackoff. 2005. Relapse and acquired rifampin resistance in HIV-infected patients with tuberculosis treated with rifampin-or rifabutin-based regimens in New York City, 1997-2000. Clin. Infect. Dis. 41:83-91. [DOI] [PubMed] [Google Scholar]
- 21.McIlleron, H., P. Wash, A. Burger, J. Norman, P. I. Folb, and P. Smith. 2006. Determinants of rifampin, isoniazid, pyrazinamide and ethambutol pharmacokinetics in a cohort of tuberculosis patients. Antimicrob. Agents Chemother. 50:1170-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peloquin, C. A., R. Namdar, M. D. Singleton, and D. E. Nix. 1999. Pharmacokinetics of rifampin under fasting conditions, with food, and with antacids. Chest 115:12-18. [DOI] [PubMed] [Google Scholar]
- 23.Pinheiro, V. G. F., L. M. A. Ramos, E. C. Barroso, L. M. A. Ramos, H. S. A. Monteiro, O. Y. Bushen, M. C. Façanha, C. A. Peloquin, R. L. Guerrant, and A. A. M. Lima. 2006. Intestinal permeability and malabsorption of rifampin and isoniazid in active pulmonary tuberculosis. Braz. J. Infect. Dis. 10:374-379. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal, I. M., K. Williams, S. Tyagi, C. A. Peloquin, A. A. Vernon, W. R. Bishai, J. H. Grosset, and E. L. Nuermberger. 2006. Potent twice-weekly rifapentine-containing regimens in murine tuberculosis. Am. J. Respir. Crit. Care Med. 174:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosner, B. 2006. Fundamentals of biostatistics, 6th ed. Duxbury Press, Belmont, CA.
- 26.Song, P., S. Li, B. Meibohm, A. O. Gaber, M. R. Honaker, M. Kotb, and C. R. Yates. 2002. Detection of MDR1 single nucleotide polymorphisms C3435T and G2677T using real-time polymerase chain reaction: MDR1 single nucleotide polymorphism genotyping assay. AAPS PharmSci. 4:E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tappero, J. W., W. Z. Bradford, T. B. Agerton, P. Hopewell, A. L. Reingold, S. Lockman, A. Oyewo, E. A. Talbot, T. A. Kenyon, T. L. Moeti, H. J. Moffat, and C. A. Peloquin. 2005. Serum concentrations of antimycobacterial drugs in patients with pulmonary tuberculosis in Botswana. Clin. Infect. Dis. 41:461-469. [DOI] [PubMed] [Google Scholar]
- 28.Tirona, R. G., B. F. Leake, A. W. Wolkoff, and R. B. Kim. 2003. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J. Pharm. Exp. Ther. 304:223-228. [DOI] [PubMed] [Google Scholar]
- 29.van Crevel, R., B. Alisjahbana, W. C. M. de Lange, F. Borst, H. Danusantoso, J. W. M. Meer, D. van der, Burger, and R. H. H. Nelwan. 2002. Low plasma concentrations of rifampicin in tuberculosis patients in Indonesia. Int. J. Tuber. Lung Dis. 6:497-502. [DOI] [PubMed] [Google Scholar]
- 30.Weiner, M., C. Peloquin, W. Burman, C. C. Luo, M. Engle, T. Prihoda, W. Mac Kenzie, E. Bliven, J. Johnson, and A. Vernon. 2009. The effects of tuberculosis infection, race and human gene SLCO1B1 polymorphisms on rifampin concentrations. Abstr. Virology-Education Second International Workshop Clin. Pharmacol. Tuberc. Drugs, San Francisco, CA, 11 September 2009.
- 31.Weiner, M., C. Peloquin, J. Johnson, W. Burman, T. Prihoda, N. Padayatchi, S. Goldberg, M. Engle, G. Muzanye, and the Tuberculosis Trials Consortium. 2008. Pharmacokinetics of rifampin and moxifloxacin in patients with pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 177(Suppl.):A791. [Google Scholar]
- 32.Weiner, M., W. Burman, C. C. Luo, C. A. Peloquin, M. Engle, S. Goldberg, V. Agarwal, and A. Vernon. 2007. Effects of rifampin and multidrug resistance gene polymorphism on concentrations of moxifloxacin. Antimicrob. Agents Chemother. 51:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner, M., D. Benator, W. Burman, C. A. Peloquin, A. Khan, A. Vernon, B. Jones, C. Silva-Trigo, Z. Zhao, T. Hodge and Tuberculosis Trials Consortium. 2005. Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin. Infect. Dis. 40:1481-1491. [DOI] [PubMed] [Google Scholar]