Abstract
We have determined the entire DNA sequence of plasmid pKpQIL, the blaKPC-3-carrying plasmid harbored by the carbapenem-resistant Klebsiella pneumoniae clone sequence type 258 (ST 258) in Israel. pKpQIL is a 113,637-bp, self-transmissible plasmid that belongs to the incompatibility group IncFII. It consists of a large backbone of a pKPN4-like plasmid and carries the blaKPC-3-containing Tn4401a transposon of a pNYC-like plasmid.
Bacterial plasmids are extrachromosomal genetic elements that play a crucial role in the acquisition and dissemination of antibiotic resistance determinants through inter- and intraspecies horizontal gene transfer. Resistance to carbapenems in Enterobacteriaceae does not occur naturally but rather arises by the acquisition of various β-lactamases encoded by transferrable plasmids (15).
During the last decade, carbapenem-resistant KPC-producing Enterobacteriaceae strains, particularly Klebsiella pneumoniae, have emerged and spread globally (14). These KPC-producing strains harbor plasmids encoding KPC-type carbapenemases. Several KPC-encoding plasmids from K. pneumoniae were partially or fully sequenced, including blaKPC-2- and blaKPC-3-carrying plasmids from the United States (6) and blaKPC-2-carrying plasmids from Greece, Colombia (12), and China (19). These plasmids differed in size and harbored the transposon Tn4401, proven to be involved with the dissemination of blaKPC (12, 14). This element has been found entirely or in part in all of the blaKPC-encoding plasmid sequences to date and has been found in one of two isoforms, a or b, that differ by a 100-bp deletion upstream of the blaKPC gene (6). A variant of this transposon, harboring ISKpn8, was reported from China (19).
In 2006, an extremely drug-resistant KPC-3-producing K. pneumoniae strain emerged in Israel (8), causing a nationwide clonal outbreak. This strain is genetically related to K. pneumoniae outbreak isolates from various regions in the United States (13), identified as sequence type 258 (ST 258) (7). Soon thereafter, this sequence type was found to have spread further geographically to China and several countries within the Middle East, Europe, and South America (1, 4, 11, 20). The K. pneumoniae ST 258 strain isolated in Israel is susceptible only to gentamicin and colistin (8), thereby limiting therapeutic options and thus posing a clinical and public health threat (18). Clinical isolates of K. pneumoniae ST 258 studied during the last 4 years in Israel have been found to harbor an identical blaKPC-3-encoding plasmid of about 105 kb (13), designated pKpQIL (9). This plasmid was shown to be self-conjugative and exclusively responsible for carbapenem resistance in these isolates. pKpQIL contains the Tn4401 element and TEM-1 gene (9). In this paper, we report the complete sequence and analysis of pKpQIL.
Plasmid pKpQIL was purified from a representative clinical isolate of carbapenem-resistant KPC-3-producing K. pneumoniae ST 258, isolate 557. This isolate harbored two plasmids: pKpQIL and an additional 240-kb plasmid (9). Plasmid DNA was purified using a NucleoBond PC 100 plasmid midi kit (Macherey-Nagel GmbH, Düren, Germany) and transformed into Escherichia coli GeneHogs (Invitrogen Corp., Dorset, United Kingdom). Transformants harboring the blaKPC gene were identified by PCR and selected as described previously (8). pKpQIL was isolated and subjected to a complete DNA sequence analysis using “454 massively parallel DNA sequencing” (Dyn G. S. Ltd., Caesarea, Israel). The resulting sequences were assembled to eight contigs using the 454 Newbler assembler software (10). Sequence gaps on the plasmid were closed by PCR and sequencing. GeneMark (http://exon.biology.gatech.edu/gmhmm2_prok.cgi) was used to predict the putative open reading frames (ORFs). Overlapping and closely clustered ORFs were manually inspected. The G+C content of the plasmid was identified using the GC calculator (http://www.sciencebuddies.org/science-fair-projects/project_ideas/Genom_GC_Calculator.shtml). The nucleotide acid and deduced protein sequences were analyzed at the NCBI website (http://www.ncbi.nlm.nih.gov/).
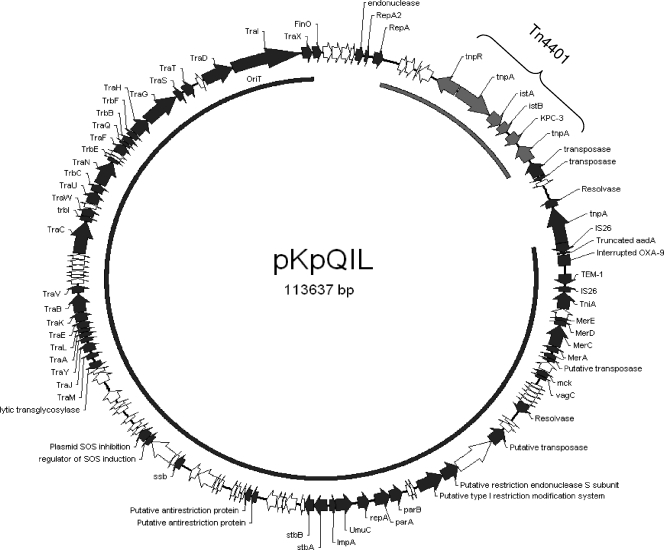
The entire DNA sequence of pKpQIL is composed of 113,637 bp forming a circular plasmid (Fig. 1) with a G+C content of 53.9%. One hundred thirty-six open reading frames were identified. The products of 67 ORFs showed a high similarity to proteins of known functions. Sequence analysis of pKpQIL showed a high similarity with two K. pneumoniae plasmids. Seventy-five percent of the sequence was highly similar to that of plasmid pKPN4 (GenBank accession no. CP000649), and 12% of pKpQIL was highly similar to the partially sequenced plasmid pNYC (GenBank accession no. EU176011).
FIG. 1.
Genetic map of the full-length, 113-kb pKpQIL plasmid. The arrows indicate gene location and orientation. Predicted ORFs with known functions are indicated by black arrows. ORFs of hypothetical proteins are indicated by white arrows. Boundaries of Tn4401 are marked with a bracket. The plasmid map was drawn using Redasoft visual cloning software. The black arc inside the circle shows the region in common with plasmid pKPN4 of K. pneumoniae MGH 78578, and the gray arc indicates the region in common with plasmid pNYC of K. pneumoniae strain YC.
The region in common between pKpQIL and pKPN4 consists of 86,610 bp (positions 26360 to 112970 on pKpQIL). This region shows 99% identity with plasmid pKPN4 at both the sequence and the gene organization level. This plasmid is one of five plasmids harbored by multidrug-resistant K. pneumoniae MGH 78578 (ATCC 700721) that was isolated from the sputum of a 66-year-old male pneumonia patient hospitalized in Massachusetts General Hospital in 1994 (http://www.expasy.ch/sprot/hamap/KLEP7.html). The common coding region of these two plasmids consists of genes responsible for plasmid maintenance, transmission, and antibiotic resistance (Fig. 1).
The genes identified on pKpQIL that are responsible for plasmid maintenance and stability, as in other enteric plasmids (2), include plasmid segregation genes (parA and parB), type I restriction modification systems, and mediators of plasmid stability (stbA and stbB). The tra gene region, involved in plasmid transfer via conjugation, makes up approximately 35 kb (30% of the plasmid length) of both plasmids, supporting our previous findings that pKpQIL is self-transmissible (9). An additional common region between pKpQIL and pKPN4 carries antibiotic resistance genes, including the aadA gene, blaOXA-9, and blaTEM-1 and is similarly positioned in the transposon Tn1331, as reported previously (17). Sequence analysis of this region showed two main modifications. The first is the truncation of aadA being a result of the IS26 insertion at position 682 of the aadA gene. A second is a nonsense mutation (A→G) at position 350 of the blaOXA-9 gene, leading to inactivation of this gene. The presence of blaOXA-9 and blaTEM-1 together with blaKPC-3 was previously reported in other K. pneumoniae isolates (5, 16, 21).
From positions 4829 to 18898 (14,070 bp), pKpQIL shows 99% identity with the plasmid pNYC of the K. pneumoniae strain YC (12). This region contains the 10-kb blaKPC-3-carrying Tn4401 transposon responsible for carbapenem resistance that has been previously characterized (6, 12). The Tn4401 transposon of pKpQIL has a 100-bp deletion and is therefore isoform a, as in agreement with previous findings (7).
The remaining part of pKpQIL (overall, 12,954 bp) includes 4,828 bp upstream of Tn4401 and 8,126 bp downstream of Tn4401. The 4,828 bp show a 94% homology to the plasmid pKP91 of K. pneumoniae 342 (GenBank accession number NC_011281). This region includes four hypothetical proteins, an endonuclease, and the origin of replication of pKpQIL-RepA and -RepA2 (Fig. 1). RepAs of pKpQIL show 96 to 97% homology to RepAs of pKP91. The 8,126-bp region, located downstream of Tn4401, encodes for two transposases, a hypothetical protein, a putative resolvase, and IS26 (Fig. 1).
pKpQIL was found to belong to the IncFII-like incompatibility group, based on the RepA sequence (3). RepA showed 89% amino acid identity with the IncFII RepA from Salmonella enterica plasmid pSLT (GenBank accession number AE006471). Previously reported IncFII plasmids were also identified to carry antibiotic resistance genes, including CTX-M- and SHV-type extended-spectrum β-lactamase (ESBL)-encoding genes, plasmid-mediated quinolone resistance genes, or ampC genes (2). pKpQIL is the first blaKPC-carrying plasmid found to belong to the IncFII group. PCR-based replicon typing (PBRT) applied by Carattoli et al. to select blaKPC-2-carrying plasmids from different outbreak strains resulted in negative results for all the replicons (2). Other blaKPC--carrying plasmids analyzed in the same study belonged to other replicon types, such as plasmid 9 of K. pneumoniae from the United States (GenBank accession number FJ223607), found to belong to the IncN-like type (2), and plasmid pBC633 of K. pneumoniae strain KN633 from Colombia (GenBank accession number EU176012), which had a ColE-like replication origin (2).
pKpQIL was found to carry two replicons. In addition to the IncFII-like replicon mentioned previously, another origin of replication (RepA, position 53179 to 54075) exists on the common part of pKpQIL and pKPN4 and shows 100% homology to RepA of pKPN4. These findings correlate to other known IncF plasmids that often carry more than one replicon and are known as the low-copy-number plasmids described by Carattoli et al. (2). The IncF-like plasmids are known to be limited in their host range to the genera of Enterobacteriaceae (2).
The KPC-encoding plasmids from K. pneumoniae that have been fully sequenced are plasmid 9 (GenBank accession number FJ223607), plasmid 12 (GenBank accession number FJ223605), and plasmid 15S (GenBank accession number FJ223606). Other than the common Tn4401 region, necessary for the spread of the blaKPC gene (14), these KPC-encoding plasmids are different in their gene organization and in their antibiotic resistance features. The majority of these plasmids (pKP048 from China [19], pNGR from Greece, and pNYC from the United States [12]) were demonstrated to be self-conjugative, as was pKpQIL (9), while others, such as pBC633 (GenBank accession number EU176012), were not (12).
Previous findings indicate that all blaKPC-carrying plasmids investigated harbor the Tn4401 structure that is located on different plasmid backbones. The complete sequencing of pKpQIL presented in the current study suggests that it was formed as a result of the plasmid rearrangement of a pKPN4-like and pNYC-like plasmid. Additional studies on KPC-encoding plasmids harbored by K. pneumoniae ST 258 from other geographical areas and plasmid comparison studies are warranted in order to reveal the mechanisms driving the success of certain KPC-carrying plasmids compared to others.
Nucleotide sequence accession number.
The complete sequence of pKpQIL is available from the GenBank database under accession number GU595196.
Acknowledgments
This work was supported in part by the European Commission Research grant FP7: SATURN—Impact of Specific Antibiotic Therapies on the Prevalence of Human Host Resistant Bacteria, grant number 241796.
This work was performed in partial fulfillment of the requirements for a Ph.D. degree (Azita Leavitt, Sackler Faculty of Medicine, Tel Aviv University, Israel).
We thank Omri Wurtzel from the Weizmann Institute for his bioinformatic advice and Daphne Karfunkel for the critical reading of the manuscript.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Cai, J. C., H. W. Zhou, R. Zhang, and G. X. Chen. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli, A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 4.Cuzon, G., T. Naas, M. C. Demachy, and P. Nordmann. 2008. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in Klebsiella pneumoniae isolate from Greece. Antimicrob. Agents Chemother. 52:796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dortet, L., I. Radu, V. Gautier, F. Blot, E. Chachaty, and G. Arlet. 2008. Intercontinental travels of patients and dissemination of plasmid-mediated carbapenemase KPC-3 associated with OXA-9 and TEM-1. J. Antimicrob. Chemother. 61:455-457. [DOI] [PubMed] [Google Scholar]
- 6.Gootz, T. D., M. K. Lescoe, F. Dib-Hajj, B. A. Dougherty, H. Wen, D. L. Phyllis, and R. C. Huard. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitchel, B., J. K. Rasheed, J. B. Patel, A. Srinivasan, S. Navon-Venezia, Y. Carmeli, A. Brolund, and C. G. Giske. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavitt, A., S. Navon-Venezia, I. Chmelnitsky, M. J. Schwaber, and Y. Carmeli. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leavitt, A., I. Chmelnitsky, I. Ofek, Y. Carmeli, and S. Navon-Venezia. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65:243-248. [DOI] [PubMed] [Google Scholar]
- 10.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navon-Venezia, S., A. Leavitt, M. J. Schwaber, J. K. Rasheed, A. Srinivasan, J. B. Patel, Y. Carmeli, and Israeli KPC Kpn Study Group. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 15.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 16.Rice, L. B., L. L. Carias, R. A. Hutton, S. D. Rudin, A. Endimiani, and R. A. Bonomo. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumonia. Antimicrob. Agents Chemother. 52:3427-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwaber, M. J., and Y. Carmeli. 2008. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA 300:2911-2913. [DOI] [PubMed] [Google Scholar]
- 19.Shen, P., Z. Wei, Y. Jiang, X. Du, S. Ji, Y. Yu, and L. Li. 2009. Novel genetic environment of the carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodford, N., J. Zhang, M. Warner, M. E. Kaufmann, J. Matos, A. Macdonald, D. Brudney, D. Sompolinsky, S. Navon-Venezia, and D. M. Livermore. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62:1261-1264. [DOI] [PubMed] [Google Scholar]