Abstract
We had shown that virus resistance to ADS-J1 was associated with amino acid changes in the envelope glycoprotein, mostly located in the gp120 coding region. Time-of-addition and endocytic virus transfer assays clearly demonstrated that ADS-J1 behaved as a gp120 inhibitor. ADS-J1-resistant virus was cross-resistant to the polyanion dextran sulfate, and recombination of gp120 recovered only the ADS-J1-resistant phenotype. In summary, ADS-J1 blocks an early step of virus entry that appears to be driven by gp120 alone.
The essential steps of HIV-1 entry in the host cell offer several potential targets for the development of novel antiviral agents (19, 24, 33, 42). Agents that disrupt gp41-mediated membrane fusion, collectively called fusion inhibitors, were the first entry inhibitors to be approved for the treatment of HIV infection. Enfuvirtide (T20, Fuzeon) is a 36-amino-acid synthetic peptide with a sequence identical to a part of the C-terminal heptad repeat 2 (HR2) region of gp41 that binds to the N-terminal heptad repeat 1 (HR1) in an antiparallel manner, forming a coiled-coil structure during the prefusion step. Mutations in the highly conserved amino acid motif 36 to 45 in the HR1 domain confer resistance to T20 (35), providing strong evidence that HR1 is the site of interaction of T20. However, mutations in other regions of HIV-1 envelope (Env) have been also associated with T20 resistance (26, 27).
Several low-molecular-weight (SMW) compounds have been identified as blockers of the initial steps of virus entry, including CCR5 coreceptor (33, 42). However, the identification of SMW compounds targeting gp41 has been elusive. A polyanionic compound, ADS-J1, was previously identified in silico as a potential candidate and shown to bind to gp41 peptides and interfere with the formation of the gp41 coiled-coil domain in an in vitro enzyme-linked immunosorbent assay (ELISA) model of HR1-HR2 interaction (16, 30, 31). Conversely, we had shown that ADS-J1 blocked the binding of HIV particles to lymphoid MT-4 cells and inhibited HIV replication at a time/site of interaction similar to those of the polyanion dextran sulfate (DS), a well-described, nonspecific inhibitor of virus entry (3). Moreover, at least four HIV-1 strains resistant to ADS-J1 were generated. The resistance to ADS-J1 was associated with gp120 based on the fact that the majority of the mutations were located in the gp120 coding sequence, mainly in the V3 loop region. Although three of the resistant strains contained mutations in gp41, one of them, HIV-1 ARA45C, did not (3). In addition, molecular modeling suggested that the gp120 V3 loop was the preferential binding site for ADS-J1 onto HIV-1, and mutations induced by the inhibitor significantly changed the stereoelectronic properties of the gp120 surface, justifying a marked drop in the affinity of ADS-J1 toward an ADS-J1-resistant HIV-1 strain (36). At that time, we considered conclusive the evidence of the mode of action of ADS-J1.
More recently, Wang et al. (43) suggested that ADS-J1 could bind directly to a trimeric peptide containing the gp41 pocket region (IQN17) in a surface plasmon resonance (SPR) assay and indicated that ADS-J1 can be used as a lead compound for the design of novel HIV-1 fusion inhibitors (44). Therefore, we thought it relevant to provide further evidence of the mode of action of ADS-J1.
ADS-J1-resistant HIV is cross-resistant to agents targeting gp120.
We evaluated the activity of ADS-J1 against a panel of HIV strains resistant to entry inhibitors, targeting either gp120 or gp41 (Table 1), which have been described elsewhere (3, 4, 12, 13, 21, 23, 34). Anti-HIV activity and cytotoxicity measurements in MT-4 cells were done as described elsewhere (6, 39, 40). The AR177-resistant strain (21) was selected to generate ADS-J1 resistance in order to bypass the activity of ADS-J1 as a gp120-CD4 interaction inhibitor and to evaluate its properties as a gp41 fusion inhibitor (3). The AR177-resistant virus is hypersensitive to T20 (Tables 1 and 2) due to a change of an aspartic acid to glycine at gp41 position 34 that occurs after virus culture in the absence of selective pressure.
TABLE 1.
Anti-HIV activity of selected compounds against virus strains made resistant to entry inhibitors
| Compound | EC50 (μM)a |
EC50 (mM) (fold resistance)b |
||||||
|---|---|---|---|---|---|---|---|---|
| HIV-1 NL4-3 | AR177-resc | ADS-J1-res Ara49 | ADS-J1-res Ara45C | BMS-res | AMD3100-res | T20/C34-res | SFV-res | |
| ADS-J1 | 0.1 ± 0.04 | 2.04 ± 0.24 | 14.6 ± 2.4 (146) | 12.3 ± 1.4 (123) | 0.2 ± 0.002 (2) | 1.7 ± 0.4 (17) | 0.4 ± 0.16 (4) | 0.08 ± 0.01(1) |
| BMS-155 | 0.02 ± 0.004 | 0.005 ± 0.004 | 0.1 ± 0.04 (5) | 0.05 ± 0.01 (3) | >2.7 ± n.a. (>350) | 0.03 ± 0.008 (2) | 0.031 ± 0.004 (2) | 0.007 ± 0.0007 (0) |
| AMD3100 | 0.002 ± 0.002 | 0.01 ± 0.006 | 0.003 ± 0.0006 (2) | 0.006 ± 0.0005 (3) | 0.004 ± 0.002 (2) | 0.7 ± (350) | 0.003 ± 0.002 (2) | 0.002 ± 0.0008 (1) |
| Dextran sulfate | 0.05 ± 0.01 | 0.37 ± 0.26 | 10.07 ± 7.12 (200) | 17.2 ± 1.74 (344) | 0.005 ± 0.002 (0) | >25 ± n.a. (>500) | 0.05 ± 0.03 (1) | 0.005 ± 0.003 (0) |
| C34 | 0.0002 ± 0.0003 | 0.0005 ± 0.0003 | 0.0007 ± 0.0006 (4) | 0.001 ± 0.001 (5) | 0.001 ± 0.0003 (5) | 0.005 ± 0.0008 (25) | 0.01 ± 0.003 (60) | 0.15 ± 0.03 (750) |
| T20 | 0.09 ± 0.04 | 0.004 ± 0.004 | 0.5 ± 0.06 (5) | 0.5 ± 0.49 (5) | 0.2 ± 0.10 (3) | 0.06 ± 0.01 (1) | >2.5 ± n.a. (>20) | 0.9 ± 0.01 (10) |
| Sifuvirtide | 0.001 ± 0.00005 | 0.0004 ± 0.0005 | 0.002 ± 0.003 (2) | 0.002 ± 0.002 (2) | 0.002 ± 0.001 (2) | 0.008 ± 0.001 (4) | 0.01 ± 0.01 (10) | 1.03 ± 0.04 (>1,000) |
| AZT | 0.007 ± 0.004 | 0.007 ± 0.008 | 0.015 ± 0.01 (2) | 0.01 ± 0.007 (1) | 0.01 ± 0.006 (1) | 0.004 ± 0.005 (1) | 0.007 ± 0.001 (1) | 0.003 ± 0.0008 (0) |
EC50: 50% effective concentration, or the concentration needed to block replication of the wild-type NL4-3 HIV-1 in MT-4 cells.
Fold change in EC50 compared to that of the wild-type HIV-1 NL4-3 strain. Data represent the means and standard deviations of results of at least two independent evaluations done in triplicate. n.a., not available.
res, HIV-1 strain resistant to the corresponding drug.
TABLE 2.
Recombination of gp120 from ADS-J1-resistant virus restores ADS-J1 resistance
| Compound | EC50a (μΜ) |
EC50 (mM) (fold resistance)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Wt NL4-3 | AR177-resc | gp 120 Wt and gp41 Wt | gp120 ARA49 and gp41ARA49 | gp120 ARA45C and gp41 ARA45C | gp120 ARA49 and gp41 Wt | gp120 ARA45C and gp41 Wt | gp120 Wt and gp41 ARA49 | gp120 Wt and gp41 ARA45C | |
| ADS-J1 | 0.1 ± 0.04 | 2.04 ± 0.24 | 2.35 ± 0.2 (1) | 10.88 ± 0.3 (6) | 11.1 ± 0.26 (6) | 10.73 ± 0.3 (6) | 12.23 ± 0.11 (6) | 1.9 ± 0.1 (1) | 2.01 ± 0.04 (1) |
| BMS-155 | 0.007 ± 0.004 | 0.005 ± 0.004 | 0.02 ± 0.001 (4) | 0.01 ± 0.003 (2) | 0.008 ± 0.001 (2) | 0.008 ± 0.003 (2) | 0.02 ± 0.01 (4) | 0.008 ± 0.003 (2) | 0.02 ± 0.007 (4) |
| AMD3100 | 0.002 ± 0.002 | 0.01 ± 0.006 | 0.02 ± 0.004 (2) | 0.004 ± 0.001 (0) | 0.003 ± 0.0005 (0) | 0.004 ± 0.0006 (0) | 0.003 ± 0.001 (0) | 0.017 ± 0.004 (2) | 0.02 ± 0.003 (2) |
| Dextran sulfate | 0.05 ± 0.01 | 0.3 ± 0.26 | 0.08 ± 0.005 (0) | 2.2 ± 0.6 (7) | 1.66 ± 0.18 (6) | 1.93 ± 0.86 (6) | 2.5 (8) | 0.08 ± 0.001 (0) | 0.077 ± 0.03 (0) |
| C34 | 0.0003 ± 0.0003 | 0.0003 ± 0.0003 | 0.0007 ± 0.0003 (2) | 0.0004 ± 0.0005 (1) | 0.0001 ± 0.0004 (0) | 0.0003 ± 0.0004 (1) | 0.0004 ± 0.0002 (0) | 0.0008 ± 0.0002 (3) | 0.001 ± 0.0004 (4) |
| T20 | 0.09 ± 0.04 | 0.004 ± 0.004 | 0.07 ± 0.06 (19) | 0.35 ± 0.36 (89) | 0.03 ± 0.02 (8) | 0.085 ± 0.08 (21) | 0.09 ± 0.02 (22) | 0.4 ± 0.21 (99) | 0.5 ± 0.43 (144) |
| AZT | 0.006 ± 0.004 | 0.008 ± 0.008 | 0.005 ± 0.0002 (1) | 0.003 ± 0.002 (0) | 0.002 ± 0.0007 (0) | 0.003 ± 0.002 (0) | 0.004 ± 0.0005 (1) | 0.01 ± 0.007 (1) | 0.005 ± 0.0005 (1) |
EC50: 50% effective concentration, or the concentration needed to block replication of the virus in MT-4 cells.
Fold change in EC50 compared to that of the parental AR177-resistant strain used to generate the ADS-J1-resistant strains. Data represent the means and standard deviations of results of at least two independent evaluations done in triplicate.
res, HIV-1 strain resistant to the corresponding drug.
ADS-J1 blocked the replication of viruses resistant to the gp41 fusion inhibitors C34, T20 and sifuvirtide (SFV) at concentrations similar to that of the wild-type NL4-3 strain. Although ADS-J1 was similarly active against a BMS-155-resistant virus, it was 17-fold less potent when tested against the AMD3100-resistant strain. AMD3100-resistant HIV has been shown to be cross-resistant to ADS-J1 and to DS (22, 23), suggesting that mutations that confer resistance to AMD3100 affect the sensitivity to other agents targeting gp120. The ADS-J1-resistant strains Ara49 and Ara45C were clearly cross-resistant to the gp120 blocking agent DS (23) (200- and >300-fold change compared to the wild-type NL4-3 virus), while T20- and C34-resistant virus remained sensitive to ADS-J1. This result suggests that ADS-J1 targets gp120 instead of gp41.
Recombination of ADS-J1-resistant gp120 into wild-type HxB2 confers resistance to ADS-J1.
To identify the gene responsible for the resistance to ADS-J1, gp120 and/or gp41 from ADS-J1-resistant virus and the virus passaged in parallel but without any selective pressure (Arwt45B2 virus) were recombined into the pJJ5-Δenv HXB2 backbone (17) as described before (5) (Fig. 1). HXB2-env clones were transfected into MT-4/CCR5+ cells with the Amaxa Nucleofector system (Lonza, Madrid, Spain). Viral stocks were generated, and proviral DNA was extracted to confirm chimeric env sequences. As shown in Table 2, the recombination of the full envelope containing the gp120 and gp41 sequence of the ADS-J1-resistant strains conferred resistance to ADS-J1 (109-fold resistant compared to wild-type NL4-3, similar to results shown in Table 1). Recombination of gp120 from the ADS-J1-resistant strains and wild-type gp41 was sufficient to confer resistance to ADS-J1. Recombination of the gp41 coding sequence of the ADS-J1 virus alone did not induce any change or modify the sensitivity to the fusion inhibitors tested (T20 and C34), confirming that resistance depends on the gp120 coding sequence and did not affect gp41.
FIG. 1.
gp120 and gp41 sequences from the HIV-1 NL4-3 and Arwt45B2 and the recombinant strains generated. Arwt45B2 is the virus obtained after passages in parallel to ARA49 and ARA45C in the absence of ADS-J1. Arwt45B2 was used as the complementary moiety to the ADSJ-1-resistant portion in the generation of the recombinant strains. The numbering corresponds to the HXB2 HIV-1 strain. Variable loops of gp120 are indicated by boxes.
ADS-J1 interferes in gp120 but not gp41 function in a time-of-drug-addition assay.
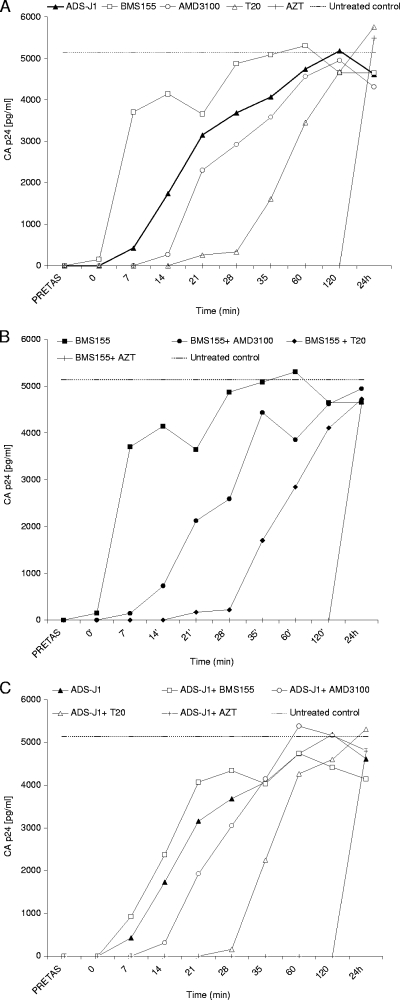
The time/site-of-drug-addition (TOA) experiments allow the determination of the last step blocked by an anti-HIV drug (3, 15). The time delay before the addition of a drug is an estimate of its mode of action. Compounds with dual mechanism (e.g., inhibition of entry and reverse transcription [RT]) would be interpreted as inhibitors of the last step. To identify the time/site of interaction of ADS-J1, drugs acting at different steps of virus entry, as well as combinations of these compounds with BMS-155 or ADS-J1, were added at various times postinfection. BMS-155 lost its activity if added later than 7 min postinfection. The addition of the CXCR4 antagonist AMD3100 could be delayed up to 14 min, while the addition of T20 could be delayed up to 35 min. AZT remained completely active when added up to 2 h postinfection (Fig. 2A). When ADS-J1 was tested alone, its activity was lost at minute 14 postinfection (Fig. 2A). To demonstrate that the TOA assay shows the effect on the latest step inhibited, combinations of anti-HIV agents were tested. The addition of BMS-155 (Fig. 2B) or ADS-J1 (Fig. 2C) to AMD3100, T20, or AZT recapitulated the activity of AMD3100, T20, or AZT alone, indicating that ADS-J1 did not share a mode of action with AMD3100, T20, or AZT.
FIG. 2.
Effect of time of drug addition on the inhibition of HIV entry inhibitors. MT-4 cells infected with HIV-1 NL4-3 at a multiplicity of infection of 0.5 at 20°C for 1 h and 37°C thereafter were treated during virus infection and/or at various times postinfection. Treatment with test compounds alone (A) or in combination with BMS-155 (B) or ADS-J1 (C) was performed at replication-blocking concentrations (roughly 100-fold the 50% effective concentration [EC50]). Viral p24 levels in the culture medium were monitored at 30 h postinfection. The figure shows a representative result of three experiments.
ADS-J1 does not prevent virus interaction with CD4 in cell-to-cell HIV transmission.
A flow cytometry-based assay was used to simultaneously quantify HIV-1 envelope (Env)-mediated cell death, endocytic cell-to-cell viral transfer (11, 15, 28), cell death (8), and cell-to-cell fusion, allowing the rapid identification of the mode of action of active compounds (9, 10). In this assay, agents that block virus entry prevent Env-mediated cell death and agents with activity at any entry step after gp120-CD4 interaction increase HIV-1 endocytosis. Thus, overnight cocultures of MOLT-Uninfected or MOLT-NL-43 cells with nonstimulated CD4+ T cells were performed and intracellular p24 and cell death were evaluated. As expected, all HIV entry inhibitors, Leu3a (dilution, 1/100), AMD3100 (12.05 μM), C34 (1.18 μM), BMS-155 (5.33 μM), and ADS-J1 (4.25 μM), blocked HIV envelope-induced cell death at the concentrations tested (cell death for each of the compounds tested was roughly 4%, similar to that in the untreated coculture) (9). HIV-1 transfer from the infected MOLT-NL4-3 cells to CD4+ T cells was measured as the percentage of CD4+ T cells positively labeled with p24 antigen. The anti-CD4 monoclonal antibody (MAb) Leu3a (dilution, 1/100) blocked the transfer of p24 antigen up to 97% ± 6% compared to untreated samples. BMS-155 and ADS-J1 failed to block virus transfer at all the concentrations tested. Conversely, the gp41 fusion inhibitor C-34 and the CXCR4 coreceptor antagonist AMD3100 increased the amount of transferred NL4-3 antigen to CD4+ T cells (2-fold and 2.4-fold, respectively). These results suggest that ADS-J1, similarly to BMS-155, did not prevent virus-CD4 interaction; however, its mechanism of action is clearly distinct from those of agents affecting later steps in the HIV entry process.
The identification of the mechanism of action of antiviral compounds may be confounded by the experimental settings or the evaluation of agents in models that do not or only partially represent the mechanism used by viruses (14, 21). Development of drug resistance in cell culture is a valuable tool to unravel the mechanisms of action of anti-HIV agents (37, 38). The resistance to AMD3100 is mapped in or near the gp120 V3 loop region (18), the putative site of interaction with CXCR4; resistance to polyanions such as dextran sulfate, AR177, or negatively charged albumins was also located in the gp120 coding sequence (12, 21, 23). Selection of drug resistance to agents that block virus entry or fusion could be, however, confounded by the plasticity of the HIV-1 envelope that allows for the incorporation of mutations that do not necessarily hamper the replication capacity of the virus (37). Clearly, recombination of gp120 of the ADS-J1-resistant virus was sufficient to recover the resistant phenotype. Conversely, recombination of the ADS-J1-resistant gp41 did not alter the virus sensitivity to the drug, pointing to gp120 as the target gene for resistance to ADS-J1.
In our previous work, we showed that ADS-J1-resistant strains may contain mutations in both gp120 and gp41; however, we had also shown that one of the selected viruses, ARA45C, containing mutations in the gp120 V3 loop but without mutations in gp41, was resistant to ADS-J1, a result that was overlooked by Wang et al. when discussing the role of gp120 in ADS-J1 resistance (43). Distinct patterns of mutations may emerge when selecting for resistance to entry inhibitors (5, 7, 19, 20, 32, 33, 38, 41). Resistance often requires acquisition of multiple mutations that may induce further variation not necessarily representing a direct site of interaction of the drug but a compensatory mechanism. Therefore, mutations arise in the gp41 of CCR5-resistant agents (2) or the gp120 of gp41-targeting agents (27) or, as we reported, in gp41 of ADS-J1-resistant virus (3). However, taken together, our results strongly suggest that ADS-J1 is not a virus fusion inhibitor through interaction with gp41 but a gp-120 interacting compound.
It is puzzling that ADS-J1 remained relatively active against the polyanion AR177-resistant virus when first tested. However, we have shown that a DS-resistant virus, generated after passage of infected cells with DS at a molecular weight of 5,000, remained sensitive to DS molecules of higher molecular weight (>40,000) (23). The number and position of negative charges in polyanions may affect their anti-HIV potency, explaining the discrepant results with ADS-J1.
We clearly show that ADS-J1 lost its anti-HIV activity at a time/site before that corresponding to a fusion inhibitor. When the combination of T20 and ADS-J1 was evaluated, only the activity of T20 could be detected, confirming that ADS-J1 did not behave as a fusion inhibitor. In a TOA experiment, Wang and colleagues (43) compared the activity of ADS-J1 to that of the RTI AZT and, therefore, lacked the appropriate controls that could resolve the time of action of ADS-J1. To confirm their results, they used a so-called time-of-removal coculture assay in which they compared the activity of ADS-J1 alone or in combination with soluble CD4 (sCD4). However, they failed to include a relevant control, that is, the activity of sCD4 alone. sCD4 binds to gp120-expressing cells, independently blocking gp120 binding to cell surface CD4 but also all subsequent downstream effects of Env leading to virus entry (1, 25, 29). Therefore, the alleged increased potency of ADS-J1 in the presence of sCD4, interpreted as an increased exposure of the drug-binding site in gp41, may be the consequence of the additive effect of two active compounds: ADS-J1 and sCD4.
Since peptidic fusion inhibitors are not orally bioavailable and must be administered via injection, the development of small molecule inhibitors of gp41-mediated fusion remains a challenging and relevant objective in drug development (42). Unbiased identification of the mechanism of action of potential lead structures is a prerequisite for successful drug development. Here, we demonstrate that mutations in gp120 conferred resistance to ADS-J1 and that early gp120-dependent entry was the functional site of interaction of ADS-J1, therefore suggesting that this compound might not be considered a gp41 fusion inhibitor.
Acknowledgments
ADS-J1 was provided by Shibo Jiang, Lindsley F. Kimball Research Institute, New York, to generate ADS-J1-resistant HIV-1. We thank the National Institutes of Health (AIDS Research and Reference Reagent Program) and the EU Programme EVA Centralised Facility for AIDS Reagents, NIBSC, United Kingdom, for reagents.
This work was supported in part by the Spanish Ministerio de Ciencia e Innovación projects BFU2009-06958 and SAF2007-63622-C02. E.G.-O. holds a scholarship from the Catalan Agència de Gestió d'Ajuts Universitaris i de Recerca. E.B. and M.P. are research fellows from Fondo de Investigación Sanitaria (FIS).
We declare no conflict of interest.
Footnotes
Published ahead of print on 19 July 2010.
REFERENCES
- 1.Alexander, L., S. Zhang, B. McAuliffe, D. Connors, N. Zhou, T. Wang, M. Agler, J. Kadow, and P.-F. Lin. 2009. Inhibition of envelope-mediated CD4+-T-cell depletion by human immunodeficiency virus attachment inhibitors. Antimicrob. Agents Chemother. 53:4726-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anastassopoulou, C. G., T. J. Ketas, P. J. Klasse, and J. P. Moore. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armand-Ugon, M., I. Clotet-Codina, C. Tintori, F. Manetti, B. Clotet, M. Botta, and J. A. Este. 2005. The anti-HIV activity of ADS-J1 targets the HIV-1 gp120. Virology 343:141-149. [DOI] [PubMed] [Google Scholar]
- 4.Armand-Ugon, M., A. Gutierrez, B. Clotet, and J. A. Este. 2003. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59:137-142. [DOI] [PubMed] [Google Scholar]
- 5.Armand-Ugón, M., G. Moncunill, M. P. Mena, E. Gonzalez, E. Ballana, B. Clotet, and J. A. Esté. 2010. Different selection patterns of resistance and cross-resistance to HIV-1 agents targeting CCR5. J. Antimicrob. Chemother. 65:417-424. [DOI] [PubMed] [Google Scholar]
- 6.Ballana, E., E. Pauls, J. Senserrich, B. Clotet, F. Perron-Sierra, G. C. Tucker, and J. A. Este. 2009. Cell adhesion through alphaV-containing integrins is required for efficient HIV-1 infection in macrophages. Blood 113:1278-1286. [DOI] [PubMed] [Google Scholar]
- 7.Berro, R., R. W. Sanders, M. Lu, P. J. Klasse, and J. P. Moore. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco, J., J. Barretina, G. Henson, G. Bridger, E. De Clercq, B. Clotet, and J. A. Este. 2000. The CXCR4 antagonist AMD3100 efficiently inhibits cell-surface-expressed human immunodeficiency virus type 1 envelope-induced apoptosis. Antimicrob. Agents Chemother. 44:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco, J., I. Clotet-Codina, B. Bosch, M. Armand-Ugon, B. Clotet, and J. A. Este. 2005. Multiparametric assay to screen and dissect the mode of action of anti-human immunodeficiency virus envelope drugs. Antimicrob. Agents Chemother. 49:3926-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch, B., J. Blanco, I. Clotet-Codina, E. Pauls, M. Armand-Ugon, B. Grigorov, D. Muriaux, B. Clotet, J. Darlix, and J. A. Este. 2005. Inhibition of coreceptor independent cell-to-cell HIV-1 transmission by a CD4-IgG2 fusion protein. Antimicrob. Agents Chemother. 49:4296-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch, B., B. Grigorov, J. Senserrich, B. Clotet, J. L. Darlix, D. Muriaux, and J. A. Este. 2008. A clathrin-dynamin-dependent endocytic pathway for the uptake of HIV-1 by direct T cell-T cell transmission. Antiviral Res. 80:185-193. [DOI] [PubMed] [Google Scholar]
- 12.Cabrera, C., M. Witvrouw, A. Gutierrez, B. Clotet, M. E. Kuipers, P. J. Swart, D. K. Meijer, J. Desmyter, E. De Clercq, and J. A. Esté. 1999. Resistance of the human immunodeficiency virus to the inhibitory action of negatively charged albumins on virus binding to CD4. AIDS Res. Hum. Retroviruses 15:1535-1543. [DOI] [PubMed] [Google Scholar]
- 13.Caporuscio, F., A. Tafi, E. Gonzalez, F. Manetti, J. A. Este, and M. Botta. 2009. A dynamic target-based pharmacophoric model mapping the CD4 binding site on HIV-1 gp120 to identify new inhibitors of gp120-CD4 protein-protein interactions. Bioorg. Med. Chem. Lett. 19:6087-6091. [DOI] [PubMed] [Google Scholar]
- 14.Cherepanov, P., J. A. Este, R. F. Rando, J. O. Ojwang, G. Reekmans, R. Steinfeld, G. David, E. De Clercq, and Z. Debyser. 1997. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol. Pharmacol. 52:771-780. [DOI] [PubMed] [Google Scholar]
- 15.Clotet-Codina, I., B. Bosch, J. Senserrich, M. T. Fernandez-Figueras, R. Pena, E. Ballana, M. Bofill, B. Clotet, and J. A. Este. 2009. HIV endocytosis after dendritic cell to T cell viral transfer leads to productive virus infection. Antiviral Res. 83:94-98. [DOI] [PubMed] [Google Scholar]
- 16.Debnath, A. K., L. Radigan, and S. Jiang. 1999. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 42:3203-3209. [DOI] [PubMed] [Google Scholar]
- 17.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vreese, K., I. Van Nerum, K. Vermeire, J. Anne, and E. De Clercq. 1997. Sensitivity of human immunodeficiency virus to bicyclam derivatives is influenced by the three-dimensional structure of gp120. Antimicrob. Agents Chemother. 41:2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Este, J. A. 2003. Virus entry as a target for anti-HIV intervention. Curr. Med. Chem. 10:1617-1632. [DOI] [PubMed] [Google Scholar]
- 20.Este, J. A., C. Cabrera, J. Blanco, A. Gutierrez, G. Bridger, G. Henson, B. Clotet, D. Schols, and E. De Clercq. 1999. Shift of clinical human immunodeficiency virus type 1 isolates from X4 to R5 and prevention of emergence of the syncytium-inducing phenotype by blockade of CXCR4. J. Virol. 73:5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Este, J. A., C. Cabrera, D. Schols, P. Cherepanov, A. Gutierrez, M. Witvrouw, C. Pannecouque, Z. Debyser, R. F. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (Zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 22.Este, J. A., K. De Vreese, M. Witvrouw, J. C. Schmit, A. M. Vandamme, J. Anne, J. Desmyter, G. W. Henson, G. Bridger, and E. De Clercq. 1996. Antiviral activity of the bicyclam derivative JM3100 against drug-resistant strains of human immunodeficiency virus type 1. Antiviral Res. 29:297-307. [DOI] [PubMed] [Google Scholar]
- 23.Este, J. A., D. Schols, K. De Vreese, K. Van Laethem, A. M. Vandamme, J. Desmyter, and E. De Clercq. 1997. Development of resistance of human immunodeficiency virus type 1 to dextran sulfate associated with the emergence of specific mutations in the envelope gp120 glycoprotein. Mol. Pharmacol. 52:98-104. [DOI] [PubMed] [Google Scholar]
- 24.Este, J. A., and A. Telenti. 2007. HIV entry inhibitors. Lancet 370:81-88. [DOI] [PubMed] [Google Scholar]
- 25.Fisher, R. A., J. M. Bertonis, W. Meier, V. A. Johnson, D. S. Costopoulos, T. Liu, R. Tizard, B. D. Walker, M. S. Hirsch, R. T. Schooley, et al. 1988. HIV infection is blocked in vitro by recombinant soluble CD4. Nature 331:76-78. [DOI] [PubMed] [Google Scholar]
- 26.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 78:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann, F. G., L. Egerer, F. Brauer, C. Gerum, H. Schwalbe, U. Dietrich, and D. von Laer. 2009. Mutations in gp120 contribute to the resistance of human immunodeficiency virus type 1 to membrane-anchored C-peptide maC46. J. Virol. 83:4844-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubner, W., G. P. McNerney, P. Chen, B. M. Dale, R. E. Gordon, F. Y. Chuang, X. D. Li, D. M. Asmuth, T. Huser, and B. K. Chen. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussey, R. E., N. E. Richardson, M. Kowalski, N. R. Brown, H. C. Chang, R. F. Siliciano, T. Dorfman, B. Walker, J. Sodroski, and E. L. Reinherz. 1988. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature 331:78-81. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, S., and A. K. Debnath. 2000. Development of HIV entry inhibitors targeted to the coiled-coil regions of gp41. Biochem. Biophys. Res. Commun. 269:641-646. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, S., and A. K. Debnath. 2000. A salt bridge between an N-terminal coiled coil of gp41 and an antiviral agent targeted to the gp41 core is important for anti-HIV-1 activity. Biochem. Biophys. Res. Commun. 270:153-157. [DOI] [PubMed] [Google Scholar]
- 32.Kitrinos, K. M., H. Amrine-Madsen, D. M. Irlbeck, J. M. Word, and J. F. Demarest, on behalf of the CCR100136 Study Team. 2009. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope. Antimicrob. Agents Chemother. 53:1124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuritzkes, D. R. 2009. HIV-1 entry inhibitors: an overview. Curr. Opin. HIV AIDS 4:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madani, N., A. L. Perdigoto, K. Srinivasan, J. M. Cox, J. J. Chruma, J. LaLonde, M. Head, A. B. Smith III, and J. G. Sodroski. 2004. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J. Virol. 78:3742-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makinson, A., and J. Reynes. 2009. The fusion inhibitor enfuvirtide in recent antiretroviral strategies. Curr. Opin. HIV AIDS 4:150-158. [DOI] [PubMed] [Google Scholar]
- 36.Manetti, F., C. Tintori, M. Armand-Ugon, I. Clotet-Codina, S. Massa, R. Ragno, J. A. Este, and M. Botta. 2006. A combination of molecular dynamics and docking calculations to explore the binding mode of ADS-J1, a polyanionic compound endowed with anti-HIV-1 activity. J. Chem. Inf. Model. 46:1344-1351. [DOI] [PubMed] [Google Scholar]
- 37.Menéndez-Arias, L. 2010. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 85:210-231. [DOI] [PubMed] [Google Scholar]
- 38.Menendez-Arias, L., and J. A. Este. 2004. HIV-resistance to viral entry inhibitors. Curr. Pharm. Des. 10:1845-1860. [DOI] [PubMed] [Google Scholar]
- 39.Moncunill, G., M. Armand-Ugon, I. Clotet-Codina, E. Pauls, E. Ballana, A. Llano, B. Romagnoli, J. W. Vrijbloed, F. O. Gombert, B. Clotet, S. De Marco, and J. A. Este. 2008. Anti-HIV activity and resistance profile of the CXC chemokine receptor 4 antagonist POL3026. Mol. Pharmacol. 73:1264-1273. [DOI] [PubMed] [Google Scholar]
- 40.Moncunill, G., M. Armand-Ugon, E. Pauls, B. Clotet, and J. A. Este. 2008. HIV-1 escape to CCR5 coreceptor antagonism through selection of CXCR4-using variants in vitro. AIDS 22:23-31. [DOI] [PubMed] [Google Scholar]
- 41.Moore, J. P., and D. R. Kuritzkes. 2009. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr. Opin. HIV AIDS 4:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilton, J. C., and R. W. Doms. 2010. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res. 85:91-100. [DOI] [PubMed] [Google Scholar]
- 43.Wang, H., Z. Qi, A. Guo, Q. Mao, H. Lu, X. An, C. Xia, X. Li, A. K. Debnath, S. Wu, S. Liu, and S. Jiang. 2009. ADS-J1 inhibits HIV-1 entry by interacting with the gp41 pocket region and blocking fusion-active gp41 core formation. Antimicrob. Agents Chemother. 53:4987-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, Y., L. Lu, L. Xu, H. Yang, S. Jiang, and Y. H. Chen. 2010. Identification of a gp41 core-binding molecule with homologous sequence of human TNNI3K-like protein as a novel human immunodeficiency virus type 1 entry inhibitor. J. Virol. 84:9359-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]