Abstract
Of the 9 vancomycin-resistant Staphylococcus aureus (VRSA) cases reported to date in the literature, 7 occurred in Michigan. In 5 of the 7 Michigan VRSA cases, an Inc18-like vanA plasmid was identified in the VRSA isolate and/or an associated vancomycin-resistant Enterococcus (VRE) isolate from the same patient. This plasmid may play a critical role in the emergence of VRSA. We studied the geographical distribution of the plasmid by testing 1,641 VRE isolates from three separate collections by PCR for plasmid-specific genes traA, repR, and vanA. Isolates from one collection (phase 2) were recovered from surveillance cultures collected in 17 hospitals in 13 states. All VRE isolates from 2 Michigan institutions (n = 386) and between 60 and 70 VRE isolates (n = 883) from the other hospitals were tested. Fifteen VRE isolates (3.9%) from Michigan were positive for an Inc18-like vanA plasmid (9 E. faecalis [12.5%], 3 E. faecium [1.0%], 2 E. avium, and 1 E. raffinosus). Six VRE isolates (0.6%) from outside Michigan were positive (3 E. faecalis [2.7%] and 3 E. faecium [0.4%]). Of all E. faecalis isolates tested, 6.0% were positive for the plasmid, compared to 0.6% for E. faecium and 3.0% for other spp. Fourteen of the 15 plasmid-positive isolates from Michigan had the same Tn1546 insertion site location as the VRSA-associated Inc18-like plasmid, whereas 5 of 6 plasmid-positive isolates from outside Michigan differed in this characteristic. Most plasmid-positive E. faecalis isolates demonstrated diverse patterns by PFGE, with the exception of three pairs with indistinguishable patterns, suggesting that the plasmid is mobile in nature. Although VRE isolates with the VRSA-associated Inc18-like vanA plasmid were more common in Michigan, they remain rare. Periodic surveillance of VRE isolates for the plasmid may be useful in predicting the occurrence of VRSA.
Currently, vancomycin-resistant Staphylococcus aureus (VRSA) infections are rare. Thus far, nine cases have been documented in the United States (9, 20), and two have been reported in other countries, including Iran (1) and India (22, 27). Despite this rarity, 7 of the 9 U.S. VRSA cases occurred in the metropolitan Detroit, MI, area. All U.S. VRSA isolates demonstrated either unique pulsed-field gel electrophoresis (PFGE) patterns or unique plasmid restriction patterns (20, 33). This suggests that each VRSA isolate, including the 7 from Michigan, acquired resistance independently and was not the result of transmission of a common VRSA strain between patients.
VRSA isolates are thought to occur by in vivo transfer of a vanA plasmid from an Enterococcus isolate to an S. aureus isolate. For most of the VRSA cases, a vancomycin-resistant Enterococcus (VRE) isolate was either coisolated from the same body site as the VRSA isolate or was found to colonize the patient at another body site, such as the nares or rectum (Table 1 shows a summary of VRE isolates from VRSA patients). There is limited evidence that these VRE isolates are the vanA donors. In previous studies (29, 30, 33), we found that certain VRSA isolates carried vanA on a transposon, and in some cases, a plasmid was identified that was identical to the vanA plasmid or transposon in a VRE isolate from the same patient. In VRSA cases 3, 4, and 5, the VRE and VRSA isolates shared the same vanA plasmid. For all other VRSA cases, the VRSA isolates carried the same Tn1546-like element as their associated VRE isolate (10, 33). Hence, it was proposed that the VRSA phenotype occurred by conjugative transfer of a vanA plasmid from VRE to S. aureus (29, 33).
TABLE 1.
Inc18-like vanA plasmid characteristics of VRSA and VRSA-associated VRE isolates from nine VRSA cases in the United Statesc
| Case | State | Species | Site of isolation | Inc18-like vanA plasmid | Tn1546 arrangementa | Tn1546 insertion siteb | Reference or source |
|---|---|---|---|---|---|---|---|
| 1 | MI | VRSA | Foot wound | No | Wild-type sequence | − | 29, 33 |
| E. faecalis | Foot wound | Yes | Wild-type sequence | + | 10 | ||
| 2 | PA | VRSA | Foot | No | Insertions and deletions | − | 4 |
| E. faecium | Suspected contamination | NA | NA | ||||
| 3 | NY | VRSA | Urine | No | Insertions and deletions | − | 30 |
| E. faecium | Rectum | No | Insertions and deletions | − | Our unpublished data | ||
| 4 | MI | VRSA | Toe wound | Yes | Wild-type sequence | + | 33 |
| E. faecalis | Rectal swab | Yes | Wild-type sequence | + | 33 | ||
| 5 | MI | VRSA | Abdominal wound | Yes | Wild-type sequence | + | 33 |
| E. faecalis | Abdominal wound | Yes | Wild-type sequence | + | 33 | ||
| 6 | MI | VRSA | Wound | No | Wild-type sequence | − | 33 |
| E. faecalis/E. avium | Rectal | Yes | Wild-type sequence | + | 33 | ||
| 7 | MI | VRSA | Left elbow | Yes | NA | − | 33 |
| NA | NA | NA | NA | − | |||
| 8 | MI | VRSA | Foot | No | NA | − | Our unpublished data |
| NA | NA | NA | NA | − | |||
| 9 | MI | VRSA | Foot | No | NA | − | Our unpublished data |
| E. faecalis | NA | NA | NA | NA |
The wild-type arrangement of Tn1546 was considered to be the prototype (2).
The Tn1546 insertion site test result was defined as positive (+) if the junction sites were the same as those of plasmids pWZ909, pWZ7140, or pWZ1668; if not, the test result was defined as negative (−).
NA, not available.
Inc18 incompatibility plasmids are a family of broad-host-range conjugative plasmids that occur naturally in Enterococcus and Streptococcus spp. Plasmids pIP501 and pAMβ1 are two well-characterized examples in this group (15). These plasmids carry multiple antimicrobial-resistance genes, including genes that confer resistance to macrolides, lincosamides, and the streptogramin B (MLS) group, and they can be transferred to a wide variety of bacteria, including streptococci (26), lactococci (17), staphylococci (23), and enterococci (14). Specifically, Inc18-like vanA plasmids were identified in VRE isolates from 4 Michigan VRSA patients (cases 1, 4, 5, and 6) and in VRSA isolates from 3 patients (cases 4, 5, and 7) (10, 33). In all VRSA cases where an Enterococcus sp. with an Inc18-like vanA plasmid was found, the isolate was E. faecalis but each isolate demonstrated a different PFGE pattern, indicating that there was not a single enterococcus vanA donor in Michigan. These results suggest that Inc18-like vanA plasmids may be more likely than other vanA plasmids to transfer from an Enterococcus sp. to S. aureus. If VRE isolates with Inc18-like vanA plasmids are more common in Michigan than in other geographic areas, this may at least partially explain why VRSA isolates have occurred primarily in Michigan. In this study, we examined health care-associated VRE isolates from institutions in various geographical locations within the United States for the presence of an Inc18-like vanA plasmid in order to determine the occurrence of Inc18-like vanA plasmids in VRE and to examine the geographical distribution of these Inc18-positive VRE isolates.
MATERIALS AND METHODS
Bacterial isolate collection and species determination.
The numbers and sources of VRE clinical isolates examined in this study are listed in Table 2. This study was conducted in three phases.
TABLE 2.
Description of VRE isolates collected in this study
| Study phase | Source of isolates | No. of VRE isolates by species |
Total no. of isolates | ||
|---|---|---|---|---|---|
| E. faecalis | E. faecium | Other spp. | |||
| 1 | 4 laboratories, 1 in IL, 1 in NC, 2 in MI | 131 | 41 | 0 | 172 |
| 2 | 19 hospitals in 14 states, including 2 in MI | 184 | 995 | 90 | 1,269 |
| 3 | 19 laboratories in MI | 200 | 0 | 0 | 200 |
| Total | 42 facilities | 515 | 1,036 | 90 | 1,641 |
Phase 1 was a pilot study to compare the occurrences of VRE isolates with Inc18-like vanA plasmids in metropolitan Detroit, MI, and in two other areas: Chicago, IL, and Durham, NC. Isolates were requested from one health care institution in each of these three areas. Laboratories were asked to send approximately 30 VRE isolates, including approximately 20 E. faecalis and 10 E. faecium isolates that were recovered from either a clinical or surveillance culture during January 2002 to May 2006.
Phase 2 was an investigation of the occurrence of Inc18-like vanA plasmids in VRE isolates recovered from surveillance cultures collected in many different regions within the United States. A total of 1,269 VRE isolates were obtained from the “Strategies to Reduce Transmission of Antimicrobial Resistant Bacteria in Intensive Care Units” (STAR*ICU) trial, a cluster-randomized trial of infection control strategies to reduce the transmission of methicillin-resistant S. aureus (MRSA) and VRE in ICUs that was supported by the National Institute of Allergy and Infectious Diseases (http://clinicaltrials.gov/show/NCT00342745). The Institutional Review Boards (IRBs) at all sites waived the requirement for informed consent and Health Insurance Portability and Accountability Act (HIPAA) authorization based on the criteria of the Federal Code of Regulations, Title 45, Part 46.116 (d) and section 164.512 (i) of the HIPAA privacy rule. As part of this study, stool or perianal swabs to screen for colonization with VRE were collected on admission and weekly from patients in 19 ICUs located in 14 states, including 2 institutions in Michigan, between June 2005 and August 2006. Swabs were processed for culture at the National Institutes of Health Clinical Center Microbiology Laboratory. VRE isolates were confirmed to be vanA positive, but the isolates were not identified to the species level. For the current study, all VRE isolates from the two Michigan institutions and between 50 and 70 randomly chosen VRE isolates from other institutions were tested for the Inc18-like vanA plasmid. Isolates were chosen so that no more than one isolate per patient was sampled, and since the isolates were not identified to the species level, no preference for species was made.
Phase 3 was an analysis of vancomycin-resistant E. faecalis isolates from Michigan. The Michigan Department of Community Health collected a large number of VRE isolates from various parts of the state between 1995 and 2002 as part of a surveillance program. In an effort to enrich for isolates with the Inc18-like vanA plasmid, only E. faecalis isolates were tested for the plasmid (n = 200).
Species identification of VRE was confirmed at the CDC by PCR amplification of species-specific ligase sequences (8), standard biochemical assays for Enterococcus identification (25), or 16S rRNA gene sequence analysis (21).
Methods for detection and characterization of Inc18-like vanA plasmids.
Primer sequences for PCR assays are listed in Table 3. PCR assays were performed for the detection of vanA and the two Inc18-like, plasmid-specific genes traA and repR (8). Another PCR assay was developed to detect the Tn1546 insertion site in the Inc18-like vanA plasmid. Specifically, primer pairs ORF18F/ORF19R and ORF27F/ORF28R were designed for detection of the Tn1546 left- and right-side junctions, respectively. If needed, E. faecalis and E. faecium were identified by PCR detection of species-specific ligase (ddl) genes (primer pairs ddl F1 and R1 for E. faecalis and ddl F2 and R2 for E. faecium) (Table 3).
TABLE 3.
PCR primers used in this study
| Primer name | DNA sequence (5′→3′) | Fragment size (bp) |
|---|---|---|
| vanA F | CATGAATAGAATAAAAGTTGCTGCAATA | 1,032 |
| vanA R | CCCCTTTAACGCTAATACGATCAA | |
| traA F | TAATCGCAATGGCTTCTTATC | 475 |
| traA R | TCTGCCCAATCTTTACGAAT | |
| repR F | GCTTCATGACGGCTTGTTA | 565 |
| repR R | TTGGCTGCTTTGACAGATTTA | |
| ddl F1 | ATCAAGTACAGTTAGTCT | 941 |
| ddl R1 | ACGATTCAAAGCTAACTG | |
| ddl F2 | TAGAGACATTGAATATGCC | 550 |
| ddl R2 | TCGAATGTGCTACAATC | |
| ORF18F | GGCAAATATAGTCAATTTTACTGAC | 660 |
| ORF19R | GACAAAACAGCTTAGCTACAGC | |
| ORF27F | CAGATGTAATTACAAATACTGTTGG | 549 |
| ORF28R | AAGTCCTGGAAGTTTAGGTGTTTT |
In phase 1, PCR for single genes was performed in a total volume of 50 μl consisting of 1.6 mM each deoxynucleoside triphosphate (dNTP; Applied Biosystems, Foster City, CA), 400 nM each primer, 1× buffer, 1 mM MgCl2, 0.5 units of AmpliTaq gold enzyme (Applied Biosystems), and 2 μl of DNA extract (equaling 100 to 500 ng). PCRs were carried out in a GeneAmp PCR system 9700 (Applied Biosystems) with reaction cycles as follows: an initial denaturation step of 2 min at 94°C, 30 cycles of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C; and a final elongation at 72°C for 7 min. In phase 2, a multiplex PCR assay for traA, repR, and the ddl genes from E. faecalis and E. faecium was performed using a Qiagen multiplex PCR kit (Valencia, CA) according to the manufacturer's instructions. PCRs were performed in a GeneAmp PCR system 9700 (Applied Biosystems) with the following reaction cycles: an initial denaturation step of 2 min at 94°C, 30 cycles of 15 s at 95°C, 90 s at 55°C, and 90 s at 72°C; and a final elongation for 7 min at 72°C. The PCR products were visualized on a 1% agarose gel. In phase 3, a multiplex PCR assay for traA and repR was performed using the conditions described for the phase-2 multiplex assay. For traA and repR PCR assays, lysates of an E. faecalis JH2-2 transconjugant containing the Inc18-like plasmid pIP501 and E. faecalis JH2-2 without plasmid were used as positive and negative controls, respectively.
The Tn1546 vanA element arrangement was analyzed by restriction fragment length polymorphism (RFLP) as described by Clark et al. (4).
Susceptibility testing.
Susceptibility to vancomycin and other antimicrobial agents was determined by reference broth microdilution (BMD) using in-house-prepared panels with cation-adjusted Mueller-Hinton broth (BD Biosciences, Sparks, MD). Susceptibility methods and interpretation were performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines (6).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed by digesting genomic DNA with SmaI restriction enzyme using standard procedures for enterococci (28). TIFF images of PFGE gels were analyzed with BioNumerics, version 5.1 (Applied Maths, Austin, TX) using Dice coefficients plus unweighted-pair group method using average linkages (UPGMA) clustering.
Conjugation.
Five milliliters of brain heart infusion (BHI) broth containing 25 μg/ml vancomycin or 25 μg/ml fusidic acid was inoculated with traA-positive donor isolates or the recipient E. faecalis JH2-2, respectively, and incubated overnight at 37°C. One hundred microliters of each culture was added to a new 5-ml BHI broth and incubated for 5 h before being filtered through a 0.45-μM Nalge filter under vacuum. The filter was then placed on a BHI agar plate and incubated for 18 h. The filter with overlying colonies was then removed from the agar and placed in BHI broth. Serial dilutions were prepared and plated on BHI selective plates containing 25 μg/ml each of vancomycin and fusidic acid. Controls were performed to detect breakthrough growth of the donor strain on fusidic acid-containing agar and of the recipient strain on vancomycin-containing agar. For three conjugation experiments, breakthrough growth of the donor was identified on fusidic acid-containing agar. These conjugations were repeated using 50 μg/m1 fusidic acid and 25 μg/ml rifampin to increase inhibition of the donor strains, and no breakthrough growth of the donor strains was identified.
DNA sequencing and analysis.
To understand the genetic organization of the Inc18-like vanA plasmids from Michigan VRE isolates, three plasmids from three VRSA-associated VRE isolates were sequenced. The plasmids were sequenced on Applied Biosystem 3730XL sequencers with BigDye Terminator version 3.1 cycle sequencing kits (Applied Biosystems, Inc.). Plasmid sequencing was performed by primer walking with primers either purchased from IDT (Integrated DNA Technologies, Inc., San Diego, CA) or made at the CDC's core facility. Reaction mixtures were cleaned with Agencourt Cleanseq beads (Agencourt Bioscience Corporation, Beverly, MA). PhredPhrap and Consed software (University of Washington, WA) were used for base-calling of the sequence data and assembly of the data into continuous DNA sequences. DNA sequences were analyzed with Clone Manager, version 9 (Sci-Ed Software, Cary, NC).
Nucleotide sequence accession numbers.
The nucleotide sequence data for plasmids pWZ7140 (47,277 bp), pWZ909 (42,602 bp), and pWZ1668 (48,365 bp) from VRSA cases 1, 4, and 5 associated with E. fiecalis were submitted to the National Center for Biotechnology Information Data Libraries (GenBank), and the sequences have been assigned accession numbers GQ484955, GQ484954, and GQ484956, respectively.
RESULTS
Completed sequence of plasmid pWZ909.
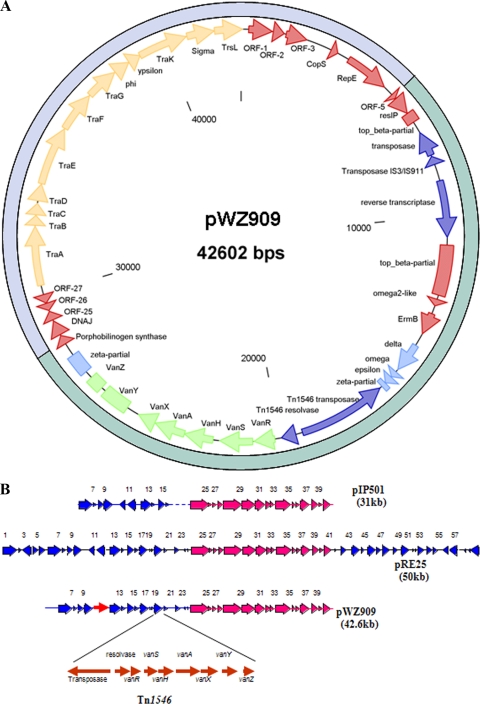
In side-by-side comparisons, we found that plasmids PWZ7l40, pWZ909, and pWZ1668 have the same genetic organization and sequences, with few exceptions. The three plasmids share 100% homology over 42 kb of sequence; however, when compared to pWZ909, pWZ7140 has a 4.7-kb insertion (putative transposase gene) and pWZ1668 has two insertions of 5.4 kb and 0.6 kb (putative transposase gene and IS456, respectively). These plasmids also have a genetic organization similar to that of Inc18 plasmids pIP501 and pRE25 (Fig. 1). The map of a representative plasmid, pWZ909 (from VRSA case 4), is shown in Fig. 1. This plasmid has 44 putative open reading frames, and 16 are 100% homologous to the conjugative transfer region of pIP501. The major difference between pIP501 and pWZ909 is the insertion of a Tn1546 vanA transposon in pWZ909 (Fig. 1B). The DNA sequence of the transfer (tra) region, specifically traA, which encodes the nickase, is conserved in the Inc18 plasmid family (16, 24). The traA gene and another conserved gene, repR (coding for an essential replication protein) were chosen as plasmid-specific genetic targets for a PCR assay to detect Inc18-like plasmids.
FIG. 1.
(A) Circular map of the Inc18-like vanA plasmid in the E. faecalis isolate from VRSA case 4. Genes are shown as arrows; certain partial or nonfunctional genes are shown as boxes. The putative conjugative transfer region is in gold, the vancomycin resistance genes are noted in light green, transposon genes are noted in dark blue, and the putative replication genes are in red. ORF, open reading frame. (B) Comparison of genetic organization of pWZ909 with that of Inc18 plasmids pRE25 and pIP501. The conjugative transfer region of pWZ909 is very similar to that of plasmids pRE25 from E. faecalis and pIP501 from Streptococcus agalactiae (24). The putative transfer region is shown in red with black outline, antibiotic resistance genes and replication regions are in blue, and transposon Tn1546 containing vancomycin genes is in red without outline.
The three sequenced plasmids demonstrate Tn1546 characteristics that were common among all VRSA-associated VRE isolates, and assays for these characteristics were used for surveillance of the Inc18-like vanA plasmid in other VRE isolates. One characteristic was the Tn1546 insertion location; a PCR assay was developed to detect the presence of this same insertion location in other plasmids, and 4 of 5 Michigan VRSA-associated VRE isolates were positive by the assay (Table 1). The other common characteristic was a wild-type Tn1546 arrangement that was demonstrated in 4 of 5 Michigan VRSA-associated VRE isolates by RFLP analysis.
Inc18 vanA plasmids in VRE isolates from three geographical locations (phase 1).
The phase-1 isolates consisted of 172 VRE isolates: 88 E. faecalis and 23 E. faecium isolates from three southeastern Michigan laboratories; 22 E. faecalis and 9 E. faecium isolates from one Chicago, IL, laboratory; and 21 E. faecalis and 9 E. faecium isolates from one Durham, NC, laboratory (Table 4). All isolates were tested for vanA, traA, repR, and Enterococcus-specific genes (ddl) by PCR amplification. If the isolates were positive for traA and repR, then the Tn1546 arrangement and insertion sites were analyzed by RFLP and PCR, respectively. Thirteen VRE isolates from metropolitan Detroit (11.7%) were positive for three genes: vanA, traA, and repR (Table 4). Another 77 VRE isolates were vanA positive but negative for both traA and repR. Of the VRE isolates from outside Michigan, none were positive for all three genes, vanA, traA, and repR, although 56 of 61 were positive for vanA. The 13 traA- and repR-positive isolates from Michigan had wild-type Tn1546-like elements, and the transposon was inserted at the same site as in pWZ909. All of the isolates that were positive for an Inc18-like vanA plasmid from Michigan were E. faecalis; none were E. faecium. Each isolate had a vancomycin MIC of at least 512 μg/ml. The results of conjugative experiments showed that all of these isolates were able to transfer the vanA-mediated vancomycin resistance to E. faecalis JH2-2, with traA and repR being cotransferred with vanA (data not shown). This suggests that all three genes were located on the same conjugative plasmid. The frequency of transfer for each conjugation was approximately 10−5 to 10−7 per donor cell; there was no significant difference in conjugation frequency observed among Inc18-like plasmid-carrying isolates.
TABLE 4.
VRE isolates with Inc18-like plasmids from three different locations (phase 1)
| Location | Total no. of isolates | Species (no. of isolates) | No. of isolates positive for: |
||
|---|---|---|---|---|---|
| vanA | traA | repR | |||
| Detroit, MI | 111 | E. faecalis (88) | 68 | 13 | 13 |
| E. faecium (23) | 22 | 0 | 0 | ||
| Durham, NC | 30 | E. faecalis (21) | 19 | 0 | 0 |
| E. faecium (9) | 9 | 0 | 0 | ||
| Chicago, IL | 31 | E. faecalis (22) | 19 | 0 | 0 |
| E. faecium (9) | 9 | 0 | 0 | ||
| Total | 172 | 146 | 13 | 13 | |
Expanded survey of Inc18-like plasmids in VRE isolates (phase 2).
A total of 1,269 VRE isolates from the STAR*ICU trial were tested for the presence of traA, repR, and the Enterococcus ligase genes, using a multiplex PCR assay. Isolates positive for traA and repR were then tested for vanA and the Tn1546 arrangement and insertion site. All VRE isolates from two Michigan health care facilities were tested (n = 386), including 291 E. faecium isolates (75%), 72 E. faecalis isolates (19%), and 23 VRE isolates of other species (6%) (Table 5). Fifteen isolates were positive for the Inc18-like vanA plasmid: 9 were E. faecalis, 3 E. faecium, 2 E. avium, and 1 E. raffinosus. Of these, 14 isolates had plasmid Tn1546 characteristics that were like those of pWZ909 (i.e., the same insertion site and arrangement). A total of 883 VRE isolates from 17 health care facilities in 14 other states were tested. These consisted of 704 E. faecium isolates (80%), 112 E. faecalis isolates (12%), and 67 VRE isolates of other species (8%). Three E. faecalis isolates and 3 E. faecium isolates were positive for an Inc18-like plasmid. One of these E. faecium isolates had Tn1546 characteristics like those of pWZ909 (Table 5). Of the six VRE isolates from outside Michigan that were positive for the Inc18-like plasmid, two were from Ohio, two were from Georgia, one was from Arizona, and one was from Iowa.
TABLE 5.
Characteristics of Inc18-like plasmids in VRE isolates from the STAR*ICU trial (phase 2)
| Location | Species | No. of isolates (%) | No. (%) of isolates with |
||
|---|---|---|---|---|---|
| traA, repR, and vanA | Tn1546 wild-type arrangementa | Tn1546 insertion sitesb | |||
| MI | E. faecalis | 72 (19) | 9 | 8 | 8 |
| E. faecium | 291 (75) | 3 | 3 | 3 | |
| Other spp. | 23 (6) | 3 | 3 | 3 | |
| Total | 386 | 15 (3.9) | 14 | 14 (3.6) | |
| Outside MI | E. faecalis | 112 (12.5) | 3 | 0 | 0 |
| E. faecium | 704 (80) | 3 | 1 | 1 | |
| Other spp. | 67 (7.5) | 0 | 0 | 0 | |
| Total | 883 | 6 (0.7) | 1 | 1 (0.1) | |
The wild-type arrangement of Tn1546 was considered to be the prototype (2).
Tn1546 junction sites are the same as those of plasmids pWZ909, pWZ7140, and pWZ1668.
All the plasmid-positive isolates had vancomycin MICs of at least 512 μg/ml. The results of conjugative assays demonstrated that all of these plasmid-positive isolates were able to transfer the vanA-mediated vancomycin resistance to E. faecalis JH2-2 via Inc18-like plasmids. No significant difference in conjugation frequency was observed between the six isolates and other Inc18-like plasmid-positive isolates.
Inc18-like vanA plasmid distribution within Michigan (phase 3).
To examine the distribution of Inc18-like plasmids within Michigan, 200 isolates of E. faecalis from the Michigan Department of Community Health Laboratory collection were tested with a multiplex PCR assay for traA and repR. Isolates that were positive for these genes were tested for vanA, Tn1546 arrangements, and the Tn1546 insertion site. Of these 200 E. faecalis isolates, 11 (5.5%) were positive for Inc18-like vanA plasmids: 6 from blood samples, 3 from wound samples, and 2 from urine samples. All of the plasmids had pWZ909-like Tn1546 characteristics. All 11 of the isolates were from three health care institutes located within a 30-mile radius of Detroit (Table 6).
TABLE 6.
Distribution of E. faecalis isolates with Inc18-like plasmids from Michigan State Health Department surveillance from 1995 to 2002 (phase 3)
| Distance of source from Detroit (miles) | No. of E. faecalis isolates | No. (%) of plasmid-positive isolates | Geographic distribution (no. of plasmid-positive isolates/total no. of isolates) |
|---|---|---|---|
| <30 | 160 | 11 (6.8) | Detroit (6/78) |
| 2 sites in Oakland county (5/78) | |||
| 2 sites in Wayne county (0/4) | |||
| >30 | 40 | 0 | 14 sites (0/40) |
| Total | 200 | 11 (5.5) | 19 sites (11/200) |
PFGE patterns of VRE isolates with Inc18-like vanA plasmids.
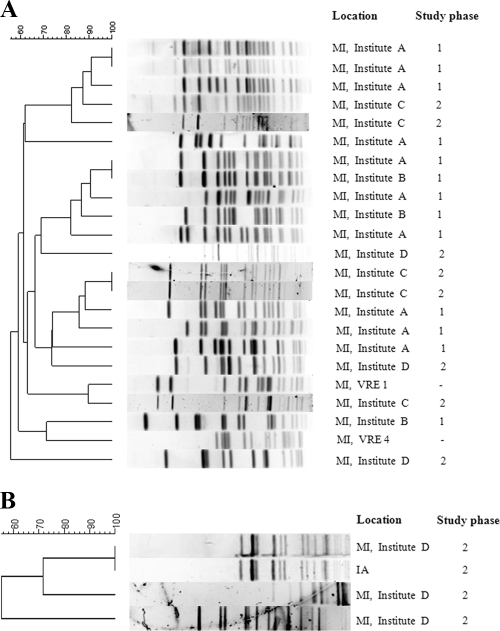
Typing by PFGE indicated that the majority of these VRE isolates were unrelated (Fig. 2). However, three pairs of E. faecalis isolates from the metropolitan Detroit area shared PFGE patterns that were indistinguishable. Also, one E. faecium isolate containing the Inc18-like vanA plasmid was identified outside Michigan and had a PFGE pattern that was indistinguishable from that of a Michigan E. faecium isolate carrying the same plasmid.
FIG. 2.
PFGE analysis of SmaI-digested whole-chromosomal DNA of E. faecalis (A) and E. faecium (B) isolates with the Inc18-like vanA plasmid. The location and study phase of strain isolation are listed on the right. Isolates VRE1 and VRE4 were associated with VRSA cases 1 and 4, respectively.
DISCUSSION
VRE isolates with VRSA-associated Inc18-like vanA plasmids were more common in Michigan than in other states. The best estimate of the occurence of Inc18-like vanA plasmids in VRE isolates, from phase 2, was 3.9% of isolates in Michigan and 0.7% of isolates from outside Michigan. However, there are two limitations of this estimate: first, more VRE isolates from the two Michigan institutions than from the institutions in other states were sampled, and, second, isolates from only 14 states were tested. It is thus possible that there are other geographic foci where VRE isolates with this plasmid are more prevalent. It is important to note that, although the plasmid was more common in Michigan, it was still rare in Michigan VRE isolates.
The exact characteristics of an Inc18-like vanA plasmid that are important for its transfer from Enterococcus spp. to S. aureus are unknown. It could be that not all Inc18-like vanA plasmids detected in this study are potential vanA donors to S. aureus. Because we did not know which characteristics were important, we looked for similarities between Inc18-like vanA plasmids and VRSA-associated plasmids (e.g., pWZ909) by assaying for the Tn1546 insertion site and arrangement that are characteristic among Inc18-like vanA plasmids from VRSA cases. Most of the plasmids detected in Michigan VRE isolates shared these characteristics, while most of the plasmids detected in VRE isolates from outside Michigan did not. Only one isolate from outside Michigan, an E. faecium isolate from Iowa that also had the same PFGE pattern as an E. faecium isolate from Michigan, had pWZ909-like Tn1546 characteristics. Data were not available to determine any possible epidemiologic link between the Iowa patient and Michigan patients.
The conjugation experiments in this study indicate that the Inc18-like vanA plasmids are able to transfer from one enterococcus isolate to another. The diversity of PFGE patterns among plasmid-positive isolates is consistent with these findings and suggests that Inc18-like vanA plasmids are being transferred between isolates in the natural environment as well. Among 21 analyzed Michigan E. faecalis isolates with Inc18-like plasmids, we found only three pairs of isolates with PFGE patterns that were indistinguishable from one another. These results suggest that the Inc18-like vanA plasmid has disseminated among Enterococcus spp. by horizontal dissemination of the plasmid rather than clonal spread of a single strain. This observation is consistent with the reports by Garcia-Migura et al. (12, 13), who reported Tn1546 vanA elements on Inc18-like plasmids in 45 of 150 different E. faecium isolates from farms (12). Their findings suggested that dissemination of vancomycin resistance by horizontal transfer could play an important role within livestock systems. The ability of these plasmids to be transmitted between strains is also supported by our conjugation experiment results, which showed that each of the plasmid-positive isolates tested was able to transfer vancomycin resistance to E. faecalis JH2-2.
Considering that most VRE isolates are E. faecium (3, 31), it was notable that, among VRE isolates collected from Michigan institutions in the STAR*ICU study (phase 2), the Inc18-like vanA plasmid was found more commonly in E. faecalis isolates (12.5%) than in E. faecium isolates (0.6%). The issue does not seem to be that the plasmid is species limited; besides being found in E. faecalis and E. faecium, the plasmid was also identified in two other species of enterococcus. We do not know why this plasmid was more common in E. faecalis isolates in Michigan. It could be that this is a relatively new vanA plasmid and that there are more vanA-negative E. faecalis than vanA-negative E. faecium isolates to serve as recipients. Although several reports have shown that pheromone response plasmids (including vanA and non-vanA plasmids) are more frequently found in E. faecalis isolates than in E. faecium isolates and those of other Enterococcus species (5, 7, 11, 19, 32), Inc18-like plasmids are pheromone independent. We do not know if the species carrying the plasmid is important for vanA transfer from Enterococcus to S. aureus. One factor that may be important for vanA transfer is biofilm formation, and E. faecalis cells are better than cells of other Enterococcus spp. at biofilm formation (18). Regardless, the increased occurrence of the plasmid in E. faecalis isolates is consistent with the identification of vancomycin-resistant E. faecalis colonization or infection in 5 of the Michigan VRSA patients.
The findings in this study may be useful in predicting future VRSA occurrence. It is possible that, if VRE isolates with these plasmids increase in occurrence, the occurrence of VRSA will also increase. Periodic surveillance for these vanA plasmids among VRE isolates would be informative. Considering that VRSA isolates are so rare, there are likely other factors that are important for VRSA to occur. For example, there may be S. aureus and environmental factors that facilitate plasmid transfer. Additional studies are needed to identify those factors. However, regardless of exactly what factors are needed, our findings provide an additional public health rationale for controlling the spread of VRE. In additional to preserving treatment options for enterococcus infections, reducing the spread of VRE may be an important means of preserving the activity of vancomycin for the treatment of S. aureus infections, especially in areas where Inc18-like plasmids have begun to emerge.
Acknowledgments
We thank Ainsley Nicholson for plasmid annotation and David Lonsway for technical help.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the National Institutes of Health.
Footnotes
Published ahead of print on 26 July 2010.
REFERENCES
- 1.Aligholi, M., M. Emaneini, F. Jabalameli, S. Shahsavan, H. Dabiri, and H. Sedaght. 2008. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med. Princ. Pract. 17:432-434. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camins, B. C., M. M. Farley, J. J. Jernigan, S. M. Ray, J. P. Steinberg, and H. M. Blumberg. 2007. A population-based investigation of invasive vancomycin-resistant Enterococcus infection in metropolitan Atlanta, Georgia, and predictors of mortality. Infect. Control Hosp. Epidemiol. 28:983-991. [DOI] [PubMed] [Google Scholar]
- 4.Clark, N. C., L. M. Weigel, J. B. Patel, and F. C. Tenover. 2005. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 49:470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finks, J., E. Wells, T. L. Dyke, N. Husain, L. Plizga, R. Heddurshetti, M. Wilkins, J. Rudrik, J. Hageman, J. Patel, and C. Miller. 2009. Vancomycin-resistant Staphylococcus aureus, Michigan, U. S. A., 2007. Emerg. Infect. Dis. 15:943-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flannagan, S. E., J. W. Chow, S. M. Donabedian, W. J. Brown, M. B. Perri, M. J. Zervos, Y. Ozawa, and D. B. Clewell. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob. Agents Chemother. 47:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803-817. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Migura, L., H. Hasman, C. Svendsen, and L. B. Jensen. 2008. Relevance of hot spots in the evolution and transmission of Tn1546 in glycopeptide-resistant Enterococcus faecium (GREF) from broiler origin. J. Antimicrob. Chemother. 62:681-687. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Migura, L., E. Liebana, and L. B. Jensen. 2007. Transposon characterization of vancomycin-resistant Enterococcus faecium (VREF) and dissemination of resistance associated with transferable plasmids. J. Antimicrob. Chemother. 60:263-268. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, C. F., and B. S. Kunka. 1983. Plasmid transfer in Pediococcus spp.: intergeneric and intrageneric transfer of pIP501. Appl. Environ. Microbiol. 46:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurenbach, B., C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann. 2003. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50:86-93. [DOI] [PubMed] [Google Scholar]
- 17.Langella, P., and A. Chopin. 1989. Conjugal transfer of plasmid pIP501 from Lactococcus lactis to Lactobacillus delbruckii subsp. bulgaricus and Lactobacillus helveticus. FEMS Microbiol. Lett. 51:149-152. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed, J. A., and D. B. Huang. 2007. Biofilm formation by enterococci. J. Med. Microbiol. 56:1581-1588. [DOI] [PubMed] [Google Scholar]
- 19.Paoletti, C., G. Foglia, M. S. Princivalli, G. Magi, E. Guaglianone, G. Donelli, C. Pruzzo, F. Biavasco, and B. Facinelli. 2007. Co-transfer of vanA and aggregation substance genes from Enterococcus faecalis isolates in intra- and interspecies matings. J. Antimicrob. Chemother. 59:1005-1009. [DOI] [PubMed] [Google Scholar]
- 20.Patel, J. B., W. C. Huskins, W. Zhu, J. A. Jernigan, N. C. Clark, K. F. Anderson, L. K. McDougal, C. Chenoweth, G. J. Alangaden, and A. P. R. Murray. 2008. Dissemination of Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus, abstr. K-568. Abstr. 48th Intersci. Conf. Antimicrob. Agents. Chemother., Washington, DC. [DOI] [PMC free article] [PubMed]
- 21.Patel, R., K. E. Piper, M. S. Rouse, J. M. Steckelberg, J. R. Uhl, P. Kohner, M. K. Hopkins, F. R. Cockerill, III, and B. C. Kline. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J. Clin. Microbiol. 36:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha, B., A. K. Singh, A. Ghosh, and M. Bal. 2008. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 57:72-79. [DOI] [PubMed] [Google Scholar]
- 23.Schaberg, D. R., D. B. Clewell, and L. Glatzer. 1982. Conjugative transfer of R-plasmids from Streptococcus faecalis to Staphylococcus aureus. Antimicrob. Agents Chemother. 22:204-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 25.Teixeira, L. M., M. G. Carvalho, and A. R. R. Facklam. 2007. Enterococcus, p. 430-442. In P. R. Murray, E. J. Baron, J. H. Horgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 26.Thompson, J. K., and M. A. Collins. 1988. Evidence for the conjugal transfer of the broad host range plasmid pIP501 into strains of Lactobacillus helveticus. J. Appl. Bacteriol. 65:309-319. [DOI] [PubMed] [Google Scholar]
- 27.Tiwari, H. K., and M. R. Sen. 2006. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect. Dis. 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 30.Weigel, L. M., R. M. Donlan, D. H. Shin, B. Jensen, N. C. Clark, L. K. McDougal, W. Zhu, K. A. Musser, J. Thompson, D. Kohlerschmidt, N. Dumas, R. J. Limberger, and J. B. Patel. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner, G., T. M. Coque, A. M. Hammerum, R. Hope, W. Hryniewicz, A. Johnson, I. Klare, K. G. Kristinsson, R. Leclercq, C. H. Lester, M. Lillie, C. Novais, B. Olsson-Liljequist, L. V. Peixe, E. Sadowy, G. S. Simonsen, J. Top, J. Vuopio-Varkila, R. J. Willems, W. Witte, and N. Woodford. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13:pii19046. [PubMed] [Google Scholar]
- 32.Wirth, R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur. J. Biochem. 222:235-246. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, W., N. C. Clark, L. K. McDougal, J. Hageman, L. C. McDonald, and J. B. Patel. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]