Abstract
Genetic mutations are one of the major mechanisms by which bacteria acquire drug resistance. One of the known mechanisms for inducing mutations is the SOS response system. We investigated the effect of disrupting recA, an inducer of the SOS response, on resistance development using an in vitro hollow-fiber infection model. A clinical Staphylococcus aureus isolate and a laboratory wild-type strain of Escherichia coli were compared to their respective recA-deleted isogenic daughter isolates. Approximately 2 × 105 CFU/ml of bacteria were subjected to escalating levofloxacin exposures for up to 120 h. Serial samples were obtained to ascertain simulated drug exposures and total and resistant bacterial burdens. Quinolone resistance determining regions of gyrA and grlA (parC for E. coli) in levofloxacin-resistant isolates were sequenced to confirm the mechanism of resistance. The preexposure MICs of the recA-deleted isolates were 4-fold lower than those of their respective parents. In S. aureus, a lower area under the concentration-time curve over 24 h at steady state divided by the MIC (AUC/MIC) was required to suppress resistance development in the recA-deleted mutant (an AUC/MIC of >23 versus an AUC/MIC of >32 was necessary in the mutant versus the parent isolate, respectively), and a prominent difference in the total bacterial burden was observed at 72 h. Using an AUC/MIC of approximately 30, E. coli resistance emergence was delayed by 24 h in the recA-deleted mutant. Diverse mutations in gyrA were found in levofloxacin-resistant isolates recovered. Disruption of recA provided additional benefits apart from MIC reduction, attesting to its potential role for pharmacologic intervention. The clinical relevance of our findings warrants further investigations.
Resistance to antibacterial agents is a major health concern that is spreading widely. Increasingly, common drugs like β-lactams and fluoroquinolones have been rendered ineffective in the clinic. Consequently, it is necessary that new therapeutic interventions be developed to combat resistance and meet the medical demands of treating infections due to pathogens resistant to available antibacterial agents.
Resistance to antibacterial agents occurs frequently via genetic mutations or the acquisition of genetic material carrying resistance determinants. Evidence suggests that genetic mutations are one of the main mechanisms of defense employed by bacteria against many antibacterial agents (1, 2). Bacteria can escape the lethal effect of antimicrobial agents by acquiring mutations in target binding site, genes regulating efflux pumps, and genes encoding porin synthesis (3, 12, 13, 20). Needless to say, mutations are important for acquiring resistance, and finding ways to reduce mutation rates in bacteria might help us to combat resistance.
Bacteria commonly develop spontaneous mutations as a consequence of error-prone DNA replication. Some of these random mutations may confer resistance to antibiotics. Lately, studies have suggested that bacteria also play an active role in inducing mutations in the event of exposure to certain classes of antibiotics or other environmental stresses (4, 16). One of the mechanisms by which these mutations are induced is the SOS response system (8). The SOS response system is a global response to DNA damage in which DNA repair and mutagenesis are induced. A central part of the SOS response is the derepression of various SOS genes, which are under the direct and indirect transcription control of a repressor, LexA. Another important protein involved in regulating the SOS response is RecA (the sensor of the system). RecA is a recombinase protein that has coprotease activity, functions in DNA strand exchange, and facilitates replicative bypass of DNA lesions. In a nontriggered state, LexA binds as a dimer in the promoter region of SOS genes and downregulates its own expression and that of other SOS genes. Formation of single-stranded DNA or a stalled replication fork resulting from DNA damage acts as a trigger for the SOS system and activates RecA. Upon activation, coprotease activity of RecA causes autocatalytic cleavage of the LexA dimer and leads to derepression of the SOS genes. The LexA regulon in most cases includes the recombination and repair genes recA, recN, and ruvAB, the nucleotide excision repair genes uvrAB and uvrD, the error-prone DNA polymerase (Pol) genes dinB (encoding Pol IV) and umuDC (encoding Pol V), and DNA polymerase II, in addition to other functions. Thus, induction of the SOS response trades long-term fidelity for short-term viability by mediating such processes as cell division inhibition, excision repair, upregulation of the tricarboxylic acid cycle, and error-prone replication. Overall, this process leads to a higher rate of mutant formation.
Since spontaneous mutations are an integral part of the bacterial adaptation and do not provide an obvious means to target, the pathways that lead to mutations linked with antibiotic resistance (such as the SOS response) seems to be a logical and potentially lucrative target for pharmacological intervention. Hence, we attempted to investigate the effect of RecA (one of the key regulatory proteins involved in the SOS response and a potential pharmacological target) on resistance development. Two clinically important pathogens, Staphylococcus aureus and Escherichia coli, were selected as our test microorganisms. S. aureus is a Gram-positive bacterium that is known to cause pneumonia and skin infections. E. coli is a Gram-negative bacterium that is a common cause of intra-abdominal and urinary tract infections.
MATERIALS AND METHODS
Antimicrobial agent.
Levofloxacin hydrochloride was purchased from Waterstone Technologies (Carmel, IN). A stock solution was prepared by dissolving the powder in water and was stored in aliquots at −70°C. Prior to each investigation, an aliquot of the stock solution was thawed and diluted accordingly with cation-adjusted Mueller-Hinton broth (Ca-MHB) (BBL, Sparks, MD) or sterile water.
Microorganisms.
The community-associated methicillin-resistant S. aureus isolate (ASAU021) that was used in the study was obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (strain NRS384). Previous molecular investigations revealed that this isolate was positive for mecA and belonging to the USA300 genotype (10). PCR analysis (detailed below) revealed a point mutation in grlA (S80F). Using a temperature-sensitive construct (pMAD), a recA-deleted isogenic daughter isolate (ASAU022) was derived (5). A recA-deleted derivative (MG1655ΔrecA) was also obtained from a wild-type E. coli (MG1655) strain. The bacteria were stored at −70°C in Protect storage vials (Key Scientific Products, Round Rock, TX). Fresh isolates were subcultured twice on 5% blood agar plates (Hardy Diagnostics, Santa Maria, CA) and incubated for 24 h at 35°C prior to each experiment.
Susceptibility studies.
The levofloxacin MIC was determined by a modified broth macrodilution method as described by the CLSI (6). The final bacterial concentration in each macrodilution tube was approximately 5 × 105 CFU/ml of Ca-MHB. Serial 2-fold dilutions of levofloxacin were used. The MIC was defined as the lowest concentration of drug that resulted in no visible growth after 24 h of incubation at 35°C in ambient air. The studies were conducted in triplicate and were repeated at least once on separate days.
Time-kill studies.
To elucidate the impact of recA deletion on the bacterial response to static levofloxacin exposures, time-kill studies were conducted with escalating concentrations of levofloxacin (placebo control, 0.5× MIC and 2× MIC). On the day of the experiment, an overnight culture of each isolate was inoculated into prewarmed Ca-MHB and incubated further at 35°C until log-phase growth was attained. The bacterial suspension was diluted with Ca-MHB based on absorbance at 630 nm so that each flask had 16 ml of bacterial suspension at approximately 2 × 105 CFU/ml. The experiments were conducted in a shaking water bath set at 35°C. Serial samples (500 μl) were obtained over 24 h (baseline, 1, 2, 4, 8, 12, and 24 h) from each flask in duplicate, and the viable bacterial population was determined by quantitative culture. Before plating, the bacterial samples were centrifuged at 10,000 × g for 15 min at 4°C and reconstituted with sterile normal saline to their original volumes in order to minimize the drug carryover effect. Total bacterial populations were quantified by spirally plating (Spiral Biotech, Bethesda, MD) serial 10× dilutions of the samples on Mueller-Hinton agar (MHA) plates (BD Diagnostics, Sparks, MD). The MHA plates were incubated in a humidified incubator (35°C) for up to 24 h, and the bacterial density was quantified by visual enumeration of the colonies. The theoretical lower limit of detection was 400 CFU/ml.
Hollow-fiber infection model.
To further elucidate the impact of recA deletion on resistance development under more clinically relevant drug exposures, an in vitro hollow-fiber infection model was used. This model simulates a fluctuating drug concentration as opposed to the static concentration in time-kill studies. The schematics of the infection model have been described in detail previously (19). The inocula (20 ml) were prepared as described above, and a final inoculum of approximately 2 × 105 CFU/ml was used for each isolate. Based on the mutation frequency of the bacteria (approximately 1 in 107 to 108), the starting inocula were deemed to be homogenous and preexisting mutants were not anticipated at baseline. The isogenic pair of S. aureus isolates (ASAU021 and ASAU022) was subjected to escalating levofloxacin exposures (an area under the concentration-time curve over 24 h at steady state divided by the MIC [AUC/MIC ratio] ranging from 0 to 70, adjusted for their respective MICs) for up to 5 days; repeated doses were given once every 24 h with a targeted elimination half-life of 5 to 7 h (14). To provide some generalizability of the results, an isogenic pair of E. coli isolates (MG1655 and MG1655ΔrecA) was also exposed to one target (the most informative) AUC/MIC, based on the results from experiments conducted with S. aureus. To ascertain the pharmacokinetic profiles simulated in the infection models, serial samples were obtained on alternate days from the circulating loop of the system. The levofloxacin concentration in these samples was assayed by a validated method outlined below, and a one-compartment linear model was fit to the observed concentration-time profiles using the ADAPT II software program (7). In addition, serial samples were also obtained daily in duplicate (baseline, 4, 8, 24, 28, 48, 52, 72, 96, and 120 h, predose when applicable) from the infection models to determine the viable bacterial burden over time. The samples were washed once in sterile saline and diluted 10× serially before plating (50 μl) on drug-free MHA and MHA supplemented with levofloxacin (at 3× and 12× MIC). Levofloxacin-supplemented plates were made in two concentrations to detect isolates with different magnitudes of reduced susceptibility (drug resistance). The generally accepted interday deviation for MIC testing is a 2-fold difference in the MIC value; hence, a 3× MIC-supplemented plate would allow reliable detection of a population with reduced susceptibility. In addition, 12× MIC levofloxacin-supplemented plates were used to further delineate the mechanism of resistance. The MHA plates were incubated at 35°C in humidified ambient air for up to 72 h before the CFU were enumerated visually.
Analysis of drug concentration.
Samples were analyzed using a high-performance liquid chromatography (HPLC) system consisting of a Shimadzu SIL-HTC autosampler, degasser, and binary pumps. Separation was performed using a Zorbax SB-C3 5-μm, 50- by 2.1-mm column. The mobile phase was delivered at 0.7 ml/min as a 1.5-min linear gradient from 95A:5B to 2A:98B, where A was water with 0.5% formic acid and B was acetonitrile with 0.5% formic acid. The mass spectrometer (API3000) was operated in electrospray (ES) positive ionization mode. Data were acquired using a precursor Q1 mass at 362.08 and a fragment Q3 mass at 318.1. The samples (50 μl) were mixed with 10 μl of internal standard (propranolol, 15 μg/ml in water) and cleaned up by addition of 300 μl of methanol, followed by vortexing and centrifugation. The supernatant (300 μl) was diluted with 200 μl water. Levofloxacin standards (5 μl of stock solution added to 45 μl of Ca-MHB) were prepared similarly. The assay was linear in the concentration range from 0.001 to 5 mg/liter.
Confirmation of resistance.
Two random isolates from levofloxacin-supplemented plates (at each concentration, 3× MIC and 12× MIC) were recovered at the end of each experiment in which regrowth was observed. Susceptibility testing was repeated to ascertain levofloxacin resistance. The mechanism of levofloxacin resistance was elucidated by PCR of the quinolone resistance determining regions (QRDRs) of the gyrA and grlA (or parC for E. coli) genes. For S. aureus, the thermocycling conditions consisted of an initial denaturing step of 94°C for 15 min, followed by 35 cycles of 94°C for 1 min, 51°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. Primer sequences used are as detailed in Table 1. For E. coli, the thermocycling conditions and the primers used have been detailed previously (17).
TABLE 1.
Forward and reverse primers used for the S. aureus genes gyrA and grlA
RESULTS
Susceptibilities.
The MICs of ASAU021 and ASAU022 to levofloxacin were found to be 1 mg/liter and 0.25 mg/liter, respectively. In addition, the levofloxacin MICs for MG1655 and MG1655ΔrecA were 0.016 mg/liter and 0.004 mg/liter, respectively. Generally speaking, the deletion of recA in a variety of bacteria conferred a reduction in the levofloxacin MIC by 2- to 8-fold (data not shown).
Time-kill studies.
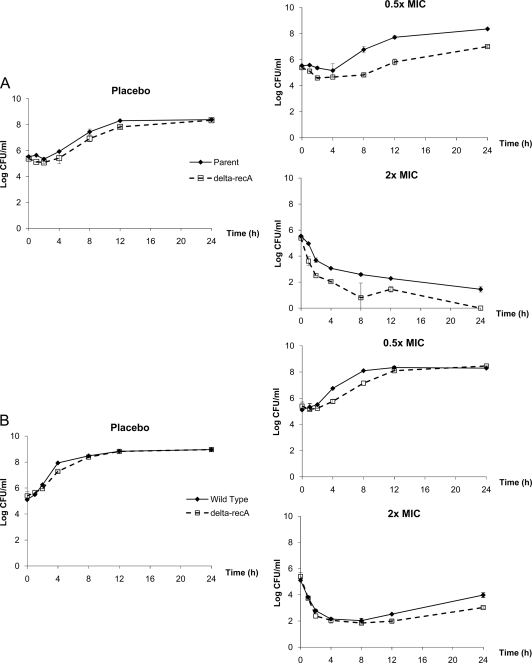
The responses observed for the two isogenic strains of S. aureus and E. coli using static levofloxacin concentrations are shown in Fig. 1. For each pair of isolates, a comparison was made after normalizing for their respective MICs to determine if there was an additional benefit from the recA deletion beyond a decrease in the MIC. Deletion of recA did not appear to change the growth rates considerably for either pair in the placebo control experiments. The bacterial burdens observed for the parent isolates were either identical to or higher than those for the recA-deleted mutants at all times. The recA-deleted S. aureus isolate showed a slower regrowth rate (at 0.5× MIC) and a steeper kill rate (at 2× MIC) than the parent isolate. In contrast, the recA-deleted E. coli isolate demonstrated an only marginally lower regrowth rate; the difference (if any) between the isogenic isolates was much less pronounced.
FIG. 1.
Observed bacterial response in time-kill studies: S. aureus (A) or E. coli (B). Data are shown as means ± standard deviations.
Hollow-fiber infection model.
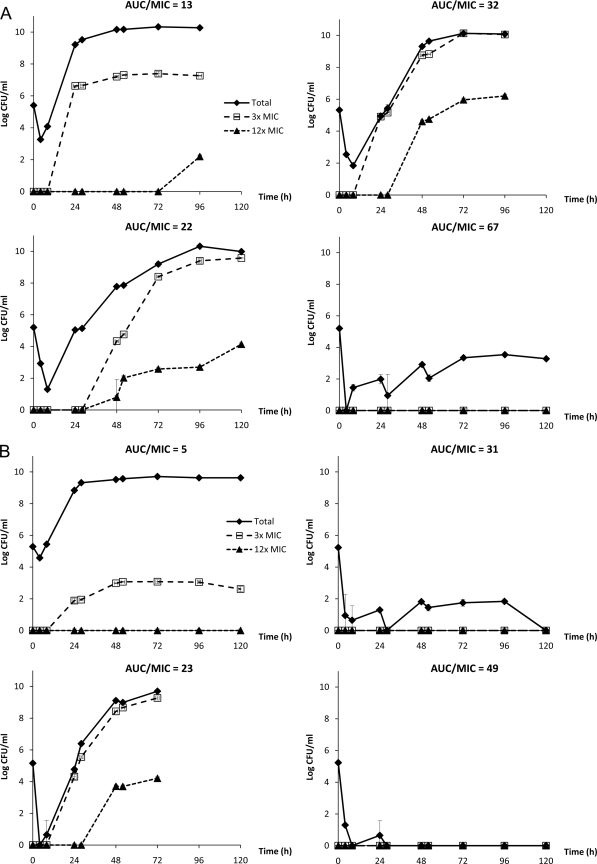
Simulated drug exposures in the infection models were satisfactory. All the simulated drug exposures had an r2 value of >0.87, and half-lives were within the target of 5 to 7 h (data not shown). Bacterial (S. aureus) responses to selective levofloxacin AUC/MIC exposures are shown in Fig. 2. Overall, the placebo did not exert a selective pressure on the bacterial population; the total population was not suppressed, but no resistance emergence was observed (data not shown). With low drug exposures, there was initially a significant reduction in the bacterial burden, followed by regrowth and the emergence of resistance. In tracking the proportion of bacterial subpopulations with reduced susceptibilities to levofloxacin (with different levofloxacin-supplemented plates), it was demonstrated that the bacterial population was gradually replaced by mutants with reduced levofloxacin susceptibility over time. In contrast, sustained suppression of the bacterial population was observed with elevated drug exposures.
FIG. 2.
Observed bacterial responses to various levofloxacin exposures in hollow-fiber infection models: parent S. aureus ASAU021 (A) or recA-deleted mutant ASAU022 (B). Data are presented as means ± standard deviations. The experiments were performed up to 120 h or when the hollow-fiber cartridge could no longer confine the bacteria, whichever occurred earlier.
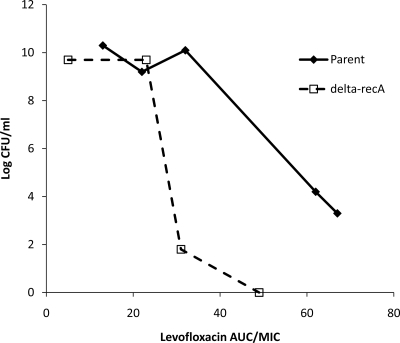
To provide a direct assessment of the impact of RecA on resistance development, the experimental data were analyzed in 2 ways. To suppress resistance development over time, a levofloxacin AUC/MIC ratio greater than 32 was necessary for the S. aureus parent isolate. In contrast, an AUC/MIC ratio greater than 23 was required for the recA-deleted mutant; no resistance emergence was observed with an AUC/MIC ratio of 31. At this drug exposure, bacterial population eradication was observed for the recA-deleted mutant but not for the parent isolate. In another analysis, the total bacterial burdens observed for the 2 isogenic S. aureus isolates after 72 h of drug exposure were compared, as shown in Fig. 3. After adjusting for the MIC, there was a “left shift” observed in the relationship between the bacterial burden of the recA-deleted mutant and the levofloxacin AUC/MIC. In both analyses, the most drastic difference between the isogenic pair appeared to be around an AUC/MIC of 30. Consequently, an additional set of experiments (with a similar design) was performed with an isogenic pair of E. coli strains using an AUC/MIC of 30 as the target.
FIG. 3.
Comparison of observed total bacterial burdens for the parent and recA-deleted strains of S. aureus after 72 h of levofloxacin exposure.
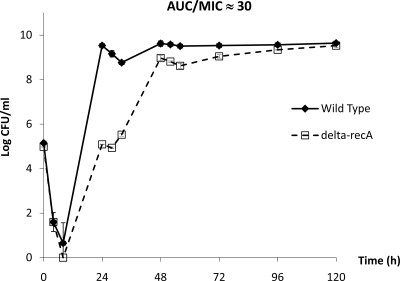
Bacterial (E. coli) responses to a targeted levofloxacin AUC/MIC exposure were also evaluated. Surprisingly, regrowth and resistance emergence were observed with both isolates under almost identical pharmacodynamic exposures. Of note, there was a delay of approximately 24 h in the bacterial burden time course of the recA-deleted mutant, as shown in Fig. 4. The less-pronounced difference in the bacterial response between the E. coli wild type and the recA-deleted mutant was consistent with the results in time-kill studies (Fig. 1B).
FIG. 4.
Comparison of observed total bacterial burdens of wild-type and recA-deleted E. coli with similar levofloxacin exposures in hollow-fiber infection models. Data are presented as means ± standard deviations.
Mechanisms of resistance.
The parent S. aureus isolate (ASAU021) was found to have a point mutation (S80F) encoded in grlA, despite exhibiting a susceptible phenotype. All S. aureus isolates recovered from levofloxacin-supplemented plates were found to have elevated levofloxacin MICs (4- to >48-fold increases) compared to that of their parent strain. Additional mutations encoded in gyrA (e.g., S84A, S84L, S84P, E88G, and E88K) were found in all the levofloxacin-resistant isolates, but no one predominant point mutation was noted. Only one additional mutation was found encoded in grlA (V82L); this resistant isolate was derived from the parent isolate exposed to a levofloxacin AUC/MIC ratio of 32, the most resistance-selecting drug exposure examined.
Similarly, E. coli isolates recovered from levofloxacin-supplemented plates were found to have elevated levofloxacin MICs (8- to 96-fold increases) compared to their parent strain. In contrast to S. aureus, only mutations encoded in gyrA (e.g., S83L, D87G, and D87Y) were found. No mutation in the QRDR of parC was detected.
DISCUSSION
Mutations are one of the primary mechanisms of resistance to antibacterial agents. The SOS system is a stress response system which generates mutants by means of activating error-prone DNA polymerases. The two key regulatory proteins of this pathway are LexA and RecA. Our study attempted to provide insights into the SOS system as a potential target for therapeutic intervention to suppress the emergence of resistance. To our knowledge, this is the first study to examine RecA as a pharmacological target using simulated human drug exposures.
Results from a previous study targeting the SOS response system to suppress resistance were promising. Cirz et al. studied the induction of mutation by fluoroquinolones and the impact of LexA cleavage on the emergence of resistance (4). It was demonstrated that ciprofloxacin induced a 106-fold increase in mutational frequency in E. coli. It was further shown an E. coli strain having a noncleavable LexA protein (and thus unable to activate the SOS response) was less amenable to resistance emergence than its parent wild-type strain in a neutropenic murine thigh infection model. While 3% of the wild-type population developed resistance, there were no resistant mutants observed in the lexA-deleted population, when a dose of 0.5 mg/kg of body weight (approximately equal to an AUC/MIC of 35) of ciprofloxacin was administered. Interestingly, this was the range of AUC/MIC (∼30) where we observed the greatest effect of recA deletion in our study.
In this study, we further extended the concept of inhibiting the SOS response system. We targeted the second regulatory protein (RecA) to investigate the effect of SOS inhibition on resistance emergence. In contrast to the study by Cirz et al., where only one dose was tested in the animal infection model, multiple simulated human drug exposures in an in vitro hollow-fiber infection model were used. This experimental setup also allowed investigations for a longer duration of exposure and the direct effect of inhibition of the SOS response without the potential interference of the (residual) immune system.
Our study revealed RecA as a potential pharmacologic target in several aspects. First, recA deletion itself resulted in a 4-fold reduction in the levofloxacin MIC. Using the same dose of an antibacterial agent would result in a higher pharmacodynamic exposure (AUC/MIC ratio) if RecA were fully inhibited (pharmacologically or otherwise). For example, a clinical dose of levofloxacin (500 mg daily) would provide an AUC/MIC of approximately 40 for the clinical S. aureus isolate (for which resistance emergence could occur over time), but the same dose of levofloxacin would provide an AUC/MIC of approximately 160 when RecA is inhibited (with which resistance development would be unlikely). Second, we also attempted to explore if there was any additional benefit of recA deletion after normalizing drug exposure to MIC. Our results suggested that recA deletion delayed the onset of resistance emergence in E. coli apart from conferring a reduction in the MIC. Third, we observed a marked difference between the S. aureus parent and its recA-deleted isolate (Fig. 3). As the AUC/MIC ratio exceeded 30 (a selective pressure of adequate intensity), there was a pronounced difference in the total bacterial burden after 72 h. Keeping in mind this decrease in the bacterial burden and delay in the onset of resistance, a “hit hard and early” approach could be applied to maximize the benefit of RecA inhibition. Finally, from the molecular aspect, since no preexisting mutants were expected to be present at baseline, it was believed that isolates with diverse mutation patterns in QRDR arose as a result of the SOS response pathways. Resistance development was most favored and likely to be observed with a low to intermediate drug exposure. The resistance selection pattern in relation to pharmacodynamic exposure was consistent with the inverted-U phenomenon reported previously (18).
It was evident that mutations (via the SOS response or otherwise) could not be completely obliterated with recA deletion. RecA is an important and early step in the SOS response (11); bacteria cannot rely on this primary rescue pathway when this gene is disrupted. However, another gene(s) may take over after some time delay. Possible reasons for this postulation could involve redundancy in the pathways that regulate the SOS response. It has been suggested that there are LexA- and RecA-independent pathways to trigger the SOS response (15). For example, several β-lactams can induce translesion synthesis and mutagenesis by activating dinB, which is independent of the LexA/RecA regulatory system. In addition, fluoroquinolones have also been suggested to simulate intra- and interchromosomal recombination in E. coli through a mechanism that does not require LexA cleavage (9). Results of our study indicated that in comparison to S. aureus, results in E. coli were less pronounced for the target AUC/MIC tested. However, a conclusive judgment cannot be made since only one dose exposure was tested. A wider range of experimental drug exposures would be necessary to confirm the true difference (if any).
It should be noted that our results could have been biased by imprecision of MIC determination. We used a geometric dilution series of drug concentrations in the study, which means that a 4-fold MIC difference between wild-type and recA-deleted isolates could actually be anywhere between 2- and 8-fold. The observed difference in bacterial responses after normalizing to an “identical AUC/MIC” was relatively small (within a 2-fold difference) and could have been due entirely to MIC variations. However, similar trends observed in two microorganisms were compelling enough to conclude that RecA inhibition was beneficial, since the probability of a systemic bias toward true lower MIC measurements is not anticipated to be high.
In conclusion, our study provided useful insights into a potential target to combat the looming danger of antibiotic resistance, and further investigations are warranted. As suggested earlier, based on our results, one possible therapeutic strategy could be to hit hard and early with a fluoroquinolone when RecA is inhibited, before any secondary pathways to induce mutation could be activated. An interesting future study could be an evaluation of the impact of other SOS response mutants (including double mutants) in resistance emergence. Further in vivo experiments to investigate the impact of RecA inhibition in the presence of an intact immune system would also be valuable.
Acknowledgments
This work was supported by contract HDTRA1-07-C-0005 from the Defense Threat Reduction Agency (DTRA) and Transformational Medical Technologies (TMT).
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the U.S. Department of Defense or the U.S. Government.
We thank Christine D. Hardy, Ryan T. Cirz, and Heinz E. Moser (Achaogen, South San Francisco, CA) for their technical assistance in this study and a critical review of the manuscript.
Footnotes
Published ahead of print on 26 July 2010.
REFERENCES
- 1.Almahmoud, I., E. Kay, D. Schneider, and M. Maurin. 2009. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 64:284-293. [DOI] [PubMed] [Google Scholar]
- 2.Chen, J. Y., L. K. Siu, Y. H. Chen, P. L. Lu, M. Ho, and C. F. Peng. 2001. Molecular epidemiology and mutations at gyrA and parC genes of ciprofloxacin-resistant Escherichia coli isolates from a Taiwan medical center. Microb. Drug Resist. 7:47-53. [DOI] [PubMed] [Google Scholar]
- 3.Chuanchuen, R., W. Wannaprasat, K. Ajariyakhajorn, and H. P. Schweizer. 2008. Role of the MexXY multidrug efflux pump in moderate aminoglycoside resistance in Pseudomonas aeruginosa isolates from Pseudomonas mastitis. Microbiol. Immunol. 52:392-398. [DOI] [PubMed] [Google Scholar]
- 4.Cirz, R. T., J. K. Chin, D. R. Andes, V. de Crecy-Lagard, W. A. Craig, and F. E. Romesberg. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirz, R. T., M. B. Jones, N. A. Gingles, T. D. Minogue, B. Jarrahi, S. N. Peterson, and F. E. Romesberg. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing: 17th informational supplement. CLSI document M100-S17. CLSI, Wayne, PA.
- 7.D'Argenio, D. Z., and A. Schumitzky. 1997. ADAPT II user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical simulations resource. University of Southern California, Los Angeles, LA.
- 8.Erill, I., S. Campoy, and J. Barbe. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637-656. [DOI] [PubMed] [Google Scholar]
- 9.Lopez, E., M. Elez, I. Matic, and J. Blazquez. 2007. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol. Microbiol. 64:83-93. [DOI] [PubMed] [Google Scholar]
- 10.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesak, L. R., V. Miao, and J. Davies. 2008. Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3394-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan-Linnell, S. K., L. Becnel Boyd, D. Steffen, and L. Zechiedrich. 2009. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 53:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oteo, J., A. Delgado-Iribarren, D. Vega, V. Bautista, M. C. Rodriguez, M. Velasco, J. M. Saavedra, M. Perez-Vazquez, S. Garcia-Cobos, L. Martinez-Martinez, and J. Campos. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32:534-537. [DOI] [PubMed] [Google Scholar]
- 14.Pea, F., E. Di Qual, A. Cusenza, L. Brollo, M. Baldassarre, and M. Furlanut. 2003. Pharmacokinetics and pharmacodynamics of intravenous levofloxacin in patients with early-onset ventilator-associated pneumonia. Clin. Pharmacokinet. 42:589-598. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Capilla, T., M. R. Baquero, J. M. Gomez-Gomez, A. Ionel, S. Martin, and J. Blazquez. 2005. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J. Bacteriol. 187:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riesenfeld, C., M. Everett, L. J. Piddock, and B. G. Hall. 1997. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41:2059-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh, R., K. R. Ledesma, K. T. Chang, J. G. Hou, R. A. Prince, and V. H. Tam. 2009. Pharmacodynamics of moxifloxacin against a high inoculum of Escherichia coli in an in vitro infection model. J. Antimicrob. Chemother. 64:556-562. [DOI] [PubMed] [Google Scholar]
- 18.Tam, V. H., A. Louie, M. R. Deziel, W. Liu, and G. L. Drusano. 2007. The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob. Agents Chemother. 51:744-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam, V. H., A. Louie, T. R. Fritsche, M. Deziel, W. Liu, D. L. Brown, L. Deshpande, R. Leary, R. N. Jones, and G. L. Drusano. 2007. Impact of drug-exposure intensity and duration of therapy on the emergence of Staphylococcus aureus resistance to a quinolone antimicrobial. J. Infect. Dis. 195:1818-1827. [DOI] [PubMed] [Google Scholar]
- 20.Von Groll, A., A. Martin, P. Jureen, S. Hoffner, P. Vandamme, F. Portaels, J. C. Palomino, and P. A. da Silva. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob. Agents Chemother. 53:4498-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]