Abstract
Aspergillus fumigatus sterol 14-α demethylase (CYP51) isoenzymes A (AF51A) and B (AF51B) were expressed in Escherichia coli and purified. The dithionite-reduced CO-P450 complex for AF51A was unstable, rapidly denaturing to inactive P420, in marked contrast to AF51B, where the CO-P450 complex was stable. Type I substrate binding spectra were obtained with purified AF51B using lanosterol (Ks, 8.6 μM) and eburicol (Ks, 22.6 μM). Membrane suspensions of AF51A bound to both lanosterol (Ks, 3.1 μM) and eburicol (Ks, 4.1 μM). The binding of azoles, with the exception of fluconazole, to AF51B was tight, with the Kd (dissociation constant) values for clotrimazole, itraconazole, posaconazole, and voriconazole being 0.21, 0.06, 0.12, and 0.42 μM, respectively, in comparison with a Kd value of 4 μM for fluconazole. Characteristic type II azole binding spectra were obtained with AF51B, whereas an additional trough and a blue-shifted spectral peak were present in AF51A binding spectra for all azoles except clotrimazole. This suggests two distinct azole binding conformations within the heme prosthetic group of AF51A. All five azoles bound relatively weakly to AF51A, with Kd values ranging from 1 μM for itraconazole to 11.9 μM for fluconazole. The azole binding properties of purified AF51A and AF51B suggest an explanation for the intrinsic azole (fluconazole) resistance observed in Aspergillus fumigatus.
The sterol pathways of eukaryotes are highly conserved and are part of a larger biosynthetic pathway that includes the formation of dolichols, coenzyme Q, heme A, and isoprenylated proteins. In Saccharomyces cerevisiae, the first step exclusively involved in sterol synthesis is the formation of squalene, with the first sterol intermediate in the pathway being lanosterol, culminating in ergosterol some 15 enzymatic steps later. In fungi, these reactions are governed by individual enzymes, but closer examination of other fungal genome sequences has revealed that there is often duplication (29) and, in some instances, triplicate versions of the same gene (1). We are interested as to why fungi have kept multiple copies of these genes, the roles of the proteins in vivo, and their contribution to both sterol biosynthesis and fungal resistance.
14-α Demethylase (CYP51) is an ancestral activity of the cytochrome P450 superfamily, which is also the target of azole antifungals (18). The isolation of CYP51 was initially from Saccharomyces cerevisiae (17), and in the fungal pathogen Aspergillus fumigatus, cytochrome P450 was first observed in 1990 (5, 6). Evidence that alteration of CYP51 activity might contribute to azole resistance first emerged in 1997 (11). In this particular study, we have looked in detail at the biochemical properties of the two CYP51 forms in Aspergillus fumigatus (29) encoded by CYP51A (Afu4g06890) and CYP51B (Afu7g03740). A comparison of the deduced amino acid sequences show 63% identity between them; and both orthologues in A. fumigatus have been shown to act in a compensatory manner in the ergosterol pathway; i.e., neither is essential individually, but a double knockdown is lethal (13). It is postulated that CYP51A may encode the major sterol 14-α demethylase activity required for growth on the basis of accumulation of multiple missense mutations linked to azole resistance (31), with CYP51B either being functionally redundant or having an alternative function under particular growth conditions still to be defined. We expressed both proteins in Escherichia coli to investigate their azole binding properties.
MATERIALS AND METHODS
Construction of pSPORT AF51A and pSPORT AF51B expression vectors.
The coding regions of the A. fumigatus (strain Af293 [http://www.aspergillusgenome.org/gbrowse/afum_af293]) CYP51 isoenzyme A (AF51A) and B (AF51B) genes (ExPASy protein database accession numbers Q4WNT5 and Q96W81, respectively) were synthesized commercially by GeneCust Europe (Dudelange, Luxembourg) in pUC57 with codon optimization for expression in E. coli. NdeI and HindIII sites were included at the 5′ and 3′ ends, respectively, of the coding sequence with an additional DNA sequence encoding 6 histidine residues prior to the stop codons. The AF51A and AF51B genes were excised from pUC57 and cloned into the modified pSPORT expression vector (22) using NdeI/HindIII. Positive recombinants were selected by growth on LB agar plates containing 0.1 mg·ml−1 sodium ampicillin, and DNA sequencing confirmed the presence of the correct inserts.
Heterologous expression in E. coli and isolation of recombinant AF51A and AF51B proteins.
Overnight cultures (10 ml) of pSPORT AF51A and pSPORT AF51B were used to inoculate 1-liter volumes of Terrific broth supplemented with 20 g·liter−1 peptone and 0.1 mg·ml−1 sodium ampicillin. Cultures were grown at 37°C and 230 rpm for 7 h, prior to induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and expression at 27°C and 170 rpm for 18 h in the presence of 1 mM 5-aminolevulenic acid. The AF51A and AF51B proteins were isolated according to the method of Arase et al. (3), except that 2% (wt/vol) sodium cholate and no Tween 20 were used in the sonication buffer. The solubilized AF51A and AF51B proteins were purified by affinity chromatography using nickel-nitrilotriacetic acid (Ni2+-NTA) agarose, as described previously (8), with the modification that 0.1% (wt/vol) l-histidine in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol were used to elute nonspecifically bound E. coli proteins after the salt washes. The AF51A and AF51B proteins were eluted with 1% (wt/vol) l-histidine in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol. The Ni2+-NTA agarose-purified AF51A and AF51B proteins were used for all subsequent spectral determinations. The AF51A protein was also isolated as a membrane suspension for sterol binding experiments by omitting the sodium cholate from the sonication buffer and then resuspending the membrane pellet recovered after ultracentrifugation in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol. Protein purity was assessed by SDS-polyacrylamide gel electrophoresis using staining intensity analysis with the UTHSCSA ImageTool (version 3.0) program (http://ddsdx.uthscsa.edu/dig/itdesc.html).
Determination of cytochrome P450 protein concentrations.
Reduced carbon monoxide difference spectra (12) were used to determine cytochrome P450 concentrations using an extinction coefficient of 91 mM−1·cm−1 (34) for the absorbance difference between 448 and 490 nm. In this method, carbon monoxide is passed through the cytochrome P450 solution prior to the addition of sodium dithionite to the sample cuvette. Absolute spectra between 380 and 700 nm were determined using 1 μM native AF51A and AF51B in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol for the oxidized protein, the 10 mM sodium dithionite reduced protein, and the reduced carbon monoxide-P450 complex, as described previously (8). The spin state of P450 samples was estimated from the ratio ΔA393-470/ΔA417-470 as previously described by Lepesheva et al. (25). All spectral determinations were made using an Hitachi U-3310 UV-visible spectrophotometer (San Jose, CA), and quartz semimicrocuvettes with a light path of 4.5 mm were used for all spectra except the CO-difference spectra. A light path of 10 mm was used for the CO difference spectra.
Sterol binding properties.
Lanosterol and eburicol (0.05%, wt/vol) were solubilized in a 1:10 mixture of chloroform-acetone containing 0.5% (vol/vol) Tween 80, prior to evaporation to dryness under nitrogen. The Tween 80-sterol residue was then dissolved in water to produce a stock 0.05% (wt/vol) aqueous sterol solution used for substrate binding experiments. Lanosterol and eburicol were progressively titrated against 10 μM AF51B and 2 μM AF51A in 50 mM potassium phosphate (pH 7.5) and 10% (wt/vol) glycerol in the sample cuvette, and equivalent amounts of 5% (vol/vol) Tween 80 were added to the reference cuvette also containing 10 μM AF51B or 2 μM AF51A. The absorbance difference spectrum between 500 and 350 nm was determined after each incremental addition of sterol. The sterol saturation curves were constructed from the difference in the absorption between 385 and 419 nm (ΔA385-419) including corrections for changes in sample volume. The substrate binding constants (Ks) were determined by nonlinear regression (Levenberg-Marquardt algorithm) using the Hill equation: ΔA = ΔAmax/(1 + Ks/[sterol]n), where n is the apparent Hill number. Sterol binding experiments using E. coli membrane suspensions containing 1 μM AF51A were also performed.
Azole binding properties of AF51A and AF51B proteins.
Binding of azole antifungal agents to the AF51A and AF51B proteins was performed as described previously (23) using split cuvettes with a 4.5-mm path length and with dimethyl sulfoxide (DMSO) also added to the cytochrome P450-containing compartment of the reference cuvette. Stock 0.05-mg·ml−1 solutions of clotrimazole, fluconazole, itraconazole, posaconazole, and voriconazole were prepared in DMSO. The azole was progressively titrated against 1 μM either AF51A or AF51B protein in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol. Difference spectra between 500 and 350 nm were determined after each incremental addition of azole. Binding saturation curves were constructed from the change in the absorbance between the spectral peak and the trough against the azole concentration. A rearrangement of the Morrison equation [ΔA = ΔAmax × {(Et + azole concentration + Kd) − [(Et + azole concentration + Kd)2 − (4 × Et × azole concentration)]0.5}/(2 × Et) (26, 32), where ΔAmax is the maximum change in absorbance difference and Et is the total concentration of CYP51 available to bind azole] was used to determine Kd (dissociation constant) values when ligand binding was tight. Tight binding is observed when the Kd for the ligand is similar to or less than the concentration of CYP51 present (10). The Michaelis-Menten equation [ΔA = (ΔAmax × azole concentration)/(Kd + azole concentration)] was used where the ligand binding was no longer tight, and the Hill equation [ΔA = ΔAmax/(1 + Kd/[azole]n)] was used when binding appeared to be allosteric.
Sequence alignment and phylogenetic analysis of fungal CYP51 proteins.
An alignment of 37 selected fungal CYP51 protein sequences deposited in the ExPASy protein database (http://www.expasy.ch/) was constructed using the ClustalX (version 1.8) program (http://www.clustal.org/) and a phylogenetic tree from the Phylip-dnd file using the Treeview (version 1.6.1) program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The CYP51 sequences (ExPASy protein database accession number) used were those of Antrodia cinnamomea (A8DBU6), Ashbya gossypii (Q759W0), Aspergillus clavatus CYP51A (A1CGZ8), Aspergillus clavatus CYP51B (A1CD61), Aspergillus flavus CYP51A (B8N2C8), Aspergillus flavus CYP51B (B8NFL5), Aspergillus fumigatus CYP51A (Q4WNT5), Aspergillus fumigatus CYP51B (Q96W81), Aspergillus nidulans CYP51A (Q9P462), Aspergillus nidulans CYP51B (Q5ATU7), Blumeria graminis (Q7Z7R3), Blumeriella jaapii (Q1KX12), Botryotinia fuckeliana (Q9P428), Candida albicans (P10613), Candida glabrata (P50859), Candida tropicalis (P14263), Coprinopsis cinerea (A8NUS3), Coprinus cinereus (Q68HC4), Cryptococcus neoformans (AAF35366), Malassezia globosa (A8Q3I7), Monilinia fructicola (B3VMU9), Mycosphaerella graminicola (Q5XWE5), Neosartorya fischeri CYP51A (A1CXT9), Neosartorya fischeri CYP51B (A1DC38), Oculimacula acuformis (Q9UV04), Oculimacula yallundae (Q9HDH9), Penicillium digitatum (Q9P340), Penicillium italicum (Q122664), Penicillium marneffei CYP51B (B6Q8Q4), Phanerochaete chrysosporium (B6DX27), Puccinia graminis (PGTG07202; http://www.broad.mit.edu/annotation/genome/puccinia_graminis), Saccharomyces cerevisiae (P10614), Schizosaccaromyces pombe (Q09736), Talaromyces stipitatus (B8M0V2), Uncinula necator (O14442), Ustilago maydis (P49602), and Venturia inaqualis (Q9P330).
Data analysis.
Curve fitting of the numerical data was performed using the computer program ProFit (version 5.0.1; QuantumSoft, Zurich, Switzerland). Protein-targeting signal peptide prediction was performed using Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/), and TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/) software.
Chemicals.
All chemicals, unless otherwise stated, were obtained from Sigma Chemical Company (Poole, United Kingdom). Voriconazole was supplied by Discovery Fine Chemicals (Bournemouth, United Kingdom). Growth media, sodium ampicillin, IPTG, and 5-aminolevulenic acid were obtained from Foremedium Ltd. (Hunstanton, United Kingdom). Ni2+-NTA agarose affinity chromatography matrix was obtained from Qiagen (Crawley, United Kingdom). Eburicol was synthesized and purified by W. David Nes (Texas Tech University). Posaconazole was a gift from Schering-Plough Ltd. (Welwyn Garden City, United Kingdom).
RESULTS AND DISCUSSION
Sequence alignment and phylogenetic analysis of fungal CYP51 proteins.
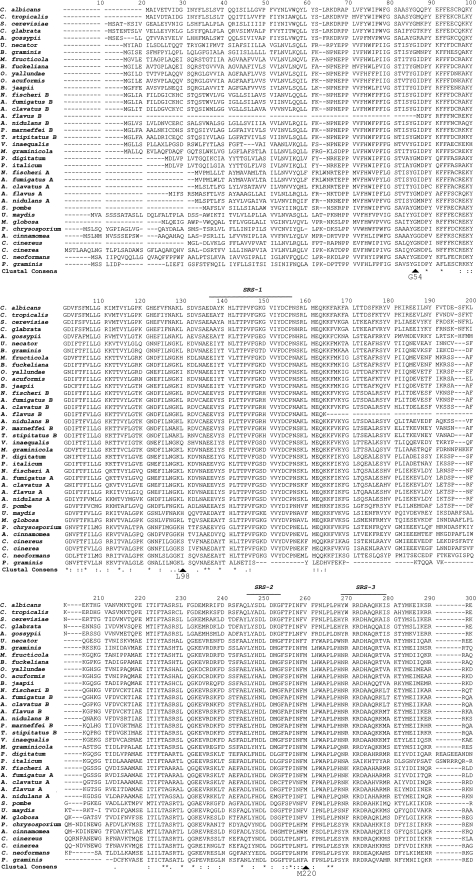
Alignment of 37 fungal CYP51 protein sequences (Fig. 1) indicated that 47 residues were conserved, with the heme-binding domain (S442 to A460 in AF51A and S451 to A469 in AF51B) being the most highly conserved region. The level of sequence conservation among the putative fungal CYP51 substrate recognition sites (SRSs) was relatively low for all sites with the exception of SRS-4, suggesting that sequence alignment may not necessarily be a good means of predicting the residues associated with substrate binding. Mutation of the G54 residue in AF51A is associated with itraconazole and posaconazole cross-resistance (27), whereas mutation of the G448 residue confers voriconazole and ravuconazole cross-resistance (37). Mutation of the L98 and M220 residues in AF51A is associated with cross-resistance to all four azoles (37). G448 is conserved in all 37 fungal CYP51 proteins, with G54 being conserved in all fungal CYP51 proteins except A. flavus CYP51B. L98 is conserved in 26 of the 37 fungal CYP51 proteins, including AF51B and C. albicans CYP51, while M220 is conserved in only 24 of the fungal CYP51 proteins, including AF51B but not C. albicans CYP51. Further investigations are required to resolve the apparent contradiction that nearly all the residues outside the heme-binding domain associated with azole resistance in C. albicans CYP51 (28) are not widely conserved among other fungal CYP51 proteins, in contrast to the case for the residues associated with azole resistance in AF51A, which are more widely conserved. This observation suggests that in C. albicans CYP51, altering the internal three-dimensional structure of the substrate/inhibitor binding pocket and access channel is important in conferring azole resistance, whereas for AF51A, the two most frequent mutation sites (G54 and M220) are thought to interfere with the entry of azole drugs into the hydrophobic access tunnel (9, 27, 37). Such a hydrophobic access tunnel (9) immersed in the endoplasmic reticulum membrane would allow highly lipophilic sterol substrates and azoles to enter the CYP51 active site directly from the lipid bilayer and restrict access to foreign metabolites from the cytosol. The amino acids that interact with either bound azole drug or sterol substrate differ between CYP51 proteins (9, 36, 38), with most involving hydrophobic interactions between substrate/azole and the amino acid side chains lining the internal substrate binding pocket and access channel in conjunction with weaker van der Waals interactions; an exception is the direct coordination of the azole ring nitrogen to the heme ferric iron. The degree of conformational change induced by ligand binding also differs between CYP51 proteins, with azole binding to human CYP51 causing a large structural change in the relative positions of the B′, F, F′, and G helices (38), whereas azole binding to the Trypanosoma brucei and Trypanosoma cruzi CYP51 proteins induces far fewer structural changes (10).
FIG. 1.
Sequence alignment of fungal CYP51 proteins. The ClustalX (version 1.8) program was used to construct the sequence alignment from selected fungal CYP51 proteins deposited in the ExPASy protein database (http://www.expasy.ch/), as detailed in the Materials and Methods section. SRSs were annotated on the basis of an alignment of the CYP51 proteins performed by Strushkevich et al. (38). The four main AF51A mutation sites associated with azole resistance (G54, L98, M220, and G448) in A. fumigatus are labeled along with the heme-thiolate-forming cytseine residue (CH454). Clustal consensus sequence indicates absolutely conserved residues (*), conserved strong (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW) groups (:), and conserved weaker (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY) groups (.) (http://www.clustal.org/).
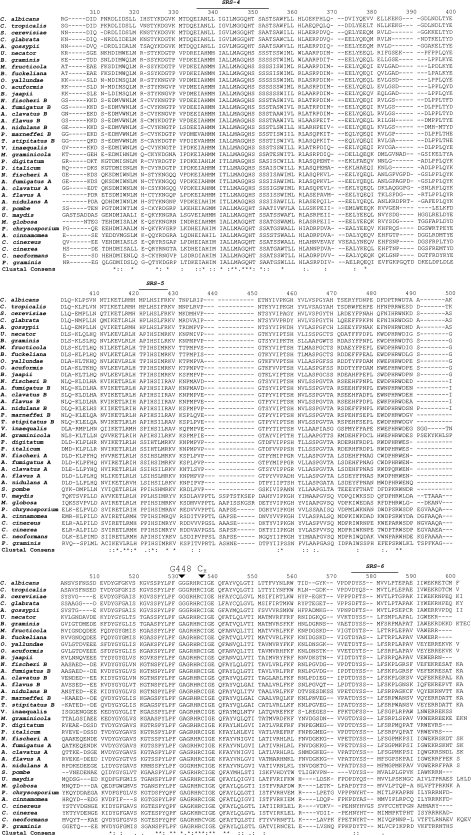
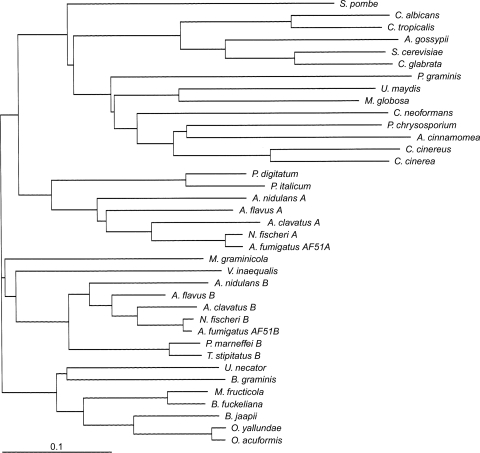
Phylogenetic analysis of the 37 fungal CYP51 proteins included (Fig. 2) indicated that AF51A had the greatest homology with the CYP51A proteins of other Aspergillus species, with which it had 74 to 81% sequence identities, and to the closely related species N. fischeri CYP51A, with which it shared 96% sequence identity. Similarly, AF51B had the greatest homology with the CYP51B proteins of other Aspergillus species (79 to 89% sequence identity) and shared the greatest sequence identity (98%) with N. fischeri CYP51B. AF51B had 78% and 79% identities with the CYP51B proteins of the closely related eurotiomycetes P. marneffei and T. stipitatus. AF51A had 66 and 68% identities with the CYP51 proteins of the closely related eurotiomycetes P. italicum and P. digitatum. However, the sequence identity between AF51A and AF51B was only 63%. Sequence identities with the S. cerevisiae and C. albicans CYP51 proteins were 49 and 47%, respectively, for AF51A and 50 and 47%, respectively, for AF51B, whereas the sequence identities with human CYP51 were only 38 and 39% for AF51A and AF51B, respectively. Before the discovery of CYP51 isoenzymes in A. fumigatus (29) and among other ascomycete filamentous fungi (http://www.expasy.ch/), the presence of more than one CYP51 gene was a characteristic of higher plant species, such as arabidopsis and poplar (33, 35; http://www.expasy.ch/). Recently, the genomes of three Fusarium species (F. oxysporum, F. verticilloides, and F. graminearum) have been published (http://www.broadinstitute.org/annotation/genome/fusarium_graminearum/GenomesIndex.html). Preliminary annotation has indicated the presence of three CYP51 genes in each Fusarium species, with one gene being closely related to AF51A (65 to 68% identity), a second gene being closely related to AF51B (65% identity), and a third gene (CYP51C) having 49 to 51% identity to AF51A and 52 to 57% identity to AF51B.
FIG. 2.
Phylogenetic tree of fungal CYP51 proteins. The ClustalX (version 1.8) and Treeview (version 1.6.1) programs were used to construct the phylogenetic tree from selected fungal CYP51 protein sequences deposited in the ExPASy proteomics database (http://www.expasy.ch/), as detailed in the Materials and Methods section.
Heterologous expression and purification of recombinant proteins.
AF51A and AF51B were expressed in E. coli with the protein localized in the membrane fraction. This was in agreement with predictions made using the Predotar, SignalP3.0, and TargetP1.1 programs that AF51A and AF51B were membrane-bound proteins located in the endoplasmic reticulum with N-terminal membrane anchors of 1 to 26 and 1 to 33 amino acid residues, respectively. Protein isolation by cholate extraction using sonication (3) gave yields of 17 (±6) and 134 (±31) nmol per liter culture for AF51A and AF51B, respectively. The levels of expression of AF51B were comparable to those achieved for C. albicans CYP51 in the pCWori+ vector (7, 24); however, the 8-fold lower yield observed with AF51A was in part due to the instability of the solubilized AF51A protein during the measurement of the reduced CO difference spectrum. Purification by Ni2+-NTA agarose chromatography resulted in 36% and 54% recoveries of native AF51A and AF51B, respectively, with most of the lost CYP51 protein being recovered by inclusion of 2% (wt/vol) sodium cholate in the elution buffer, indicating a hydrophobic interaction with the column matrix. SDS-polyacrylamide gel electrophoresis confirmed that the purities of Ni2+-NTA agarose-purified AF51A and AF51B were greater than 90% when the purity was assessed by the staining intensity. Apparent molecular masses of 58.9 kDa were obtained for both proteins and were close to the predicted values of 58,888 and 59,751 Da for AF51A and AF51B, respectively, including the 6-histidine C-terminal extensions.
Spectral properties of recombinant CYP51 protein.
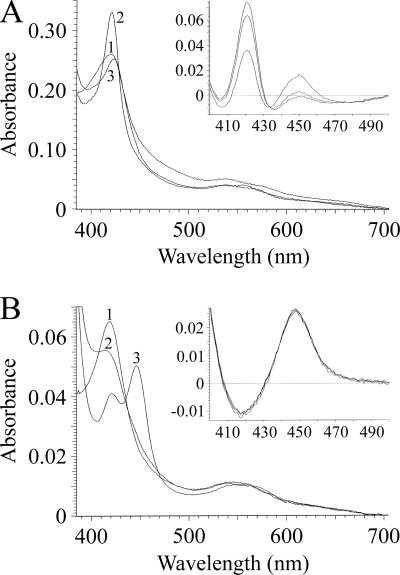
The absolute spectra (Fig. 3) and reduced CO-P450 difference spectra (Fig. 3, insets) of AF51A and AF51B were characteristic of those for a native cytochrome P450 enzyme (6, 14). AF51A was predominantly in the ferric low-spin state with a heme iron Soret (γ) band at 419 nm, in addition to α, β, and δ bands at 575, 539, and 350 nm, respectively. AF51B was also predominantly in the ferric low-spin state with a heme Soret peak at 419 nm, in addition to α, β, and δ bands at 568, 539, and 349 nm, respectively. A dithionite one-electron reduction caused a red shift of the Soret peak to 421 nm for AF51A, whereas it caused a blue shift to 415 nm for AF51B. Binding carbon monoxide to the dithionite-reduced ferrous form of AF51A did not produce the characteristic red shift of the Soret band from 419 to 448 nm but produced a small red shift to 423 nm instead, indicating the formation of the inactive P420 complex with CO. Carbon monoxide binding to ferrous AF51B, in contrast, did result in the characteristic red shift of the Soret peak to 448 nm. The carbon monoxide difference spectra (12), where dithionite was added to the sample cuvette just prior to measurement, did give characteristic Soret spectral peaks at 450 and 448 nm for the AF51A and AF51B proteins, respectively (Fig. 3, insets), although the AF51A spectral peak at 450 nm quickly degraded to inactive P420 over 3 min, whereas the reduced AF51B-CO complex remained stable.
FIG. 3.
Absolute and reduced carbon monoxide spectra of AF51A and AF51B proteins. The absolute spectra of purified AF51A (A) and AF51B (B) were determined in the oxidized resting state (line 1), the one-electron dithionite reduced state (line 2), and the reduced state bound to carbon monoxide (line 3). Carbon monoxide difference spectra (12) are shown as insets to the main panels. Protein dilutions equivalent to 1 μM CO-P450red were chosen for comparison. Spectral determinations were made using quartz semimicrocuvettes with path lengths of 4.5 mm for absolute spectra and 10 mm for CO difference spectra.
Substrate binding properties.
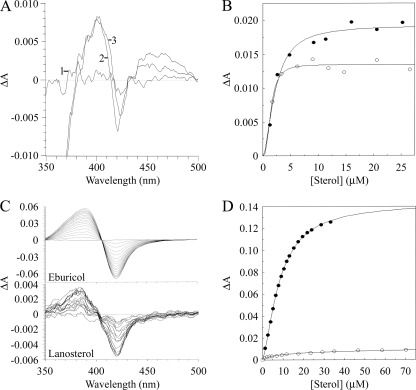
Progressive titration of 10 μM AF51B with eburicol gave a strong type I difference spectrum (Fig. 4C) with a peak at 385 nm and a trough at 419 nm, indicating a change in spin state from low to high spin. The absolute spectra of AF51B indicated that the presence of 35 μM eburicol (data not shown) caused a 10% increase in the high-spin fraction of AF51B from 35 to 45%. The spin state change occurs due to substrate binding to the apoprotein displacing the water molecule coordinated as the sixth ligand to the low-spin hexacoordinated heme, resulting in the adoption of the high-spin pentacoordinated heme conformation (14). The substrate molecule does not coordinate with the heme prosthetic group. The Hill equation best fitted the eburicol saturation curve (Fig. 4D), yielding a Ks value of 22.6 ± 0.9 μM. The relatively small change in spin state caused by the binding of sterol was previously observed for other CYP51 enzymes (2, 7, 16, 25), with spin state changes usually not exceeding 10%. Lanosterol binding to AF51B gave a much weaker type I binding spectrum, although with a higher affinity (Ks, 8.6 ± 1.02 μM). The absolute spectra of AF51B in the presence and absence of 70 μM lanosterol indicated that a less than 1% change in spin state had occurred. The Ks value obtained for lanosterol was comparable to the Km value of 7.4 μM for lanosterol reported for C. albicans CYP51 in metabolism studies (39) but 2-fold higher than the Km value for S. cerevisiae CYP51 (20). Further investigations are required to fully characterize the substrate binding specificity of AF51B. No reproducible type I binding spectra could be obtained with either lanosterol or eburicol using 2 μM purified AF51A, in part probably due to the apparent instability, as judged by the CO difference spectrum, of AF51A in free solution. However, weak type I binding spectra were obtained with both eburicol (Ks, 4.1 ± 1.2 μM) and lanosterol (Ks, 3.1 ± 1.3 μM) using an E. coli membrane suspension containing 1 μM recombinant AF51A protein (Fig. 4A and B), suggesting that incorporation into a lipid membrane stabilizes the AF51A tertiary structure. This result for AF51A and AF51B is in agreement with the sterol pathway producing eburicol before sterol 14-α demethylation by CYP51.
FIG. 4.
Eburicol and lanosterol binding properties of AF51A and AF51B. Eburicol and lanosterol bound weakly to 1 μM AF51A in an E. coli membrane suspension (A). Line 1, the baseline; line 2, difference spectrum obtained with 4.8 μM eburicol; line 3, difference spectrum obtained with 6.2 μM lanosterol. (B) The eburicol (filled circles) and lanosterol (hollow circles) saturation curves for AF51A were constructed from the difference in absorbance at 395 and 419 nm. (C) Eburicol and lanosterol were progressively bound to 10 μM purified AF51B to produce type I difference spectra. (D) The eburicol (filled circles) and lanosterol (hollow circles) saturation curves for AF51B were constructed from the difference in absorbance at 385 and 419 nm. The Hill equation was used to fit the binding data. No reproducible type I difference spectra were obtained using 2 μM purified AF51A with either eburicol or lanosterol.
Azole binding properties.
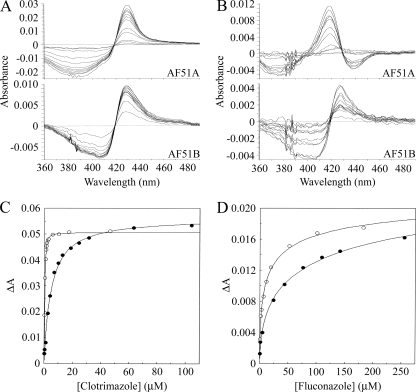
Clotrimazole bound to AF51A and AF51B, producing type II difference spectra (Fig. 5A) caused by the imidazole ring N-3 nitrogen coordinating as the sixth ligand with the heme iron (15). However, spectral differences between AF51A and AF51B were evident, with the spectral trough of AF51A being at 386 nm and that for AF51B being at 407 nm (Table 1). The trough at 386 nm indicated that AF51A was predominantly in the high-spin conformation prior to binding to the azole ligand to form the low-spin CYP51-azole complex (14). This suggests that clotrimazole preferentially binds to the high-spin conformation of the AF51A protein. We have previously shown that voriconazole preferentially binds to the high-spin form of Mycobacterium smegmatis CYP164A2 (40). The trough at 407 nm indicated that AF51B was predominantly in the low-spin conformation prior to binding to the clotrimazole ligand (14). Fluconazole bound to AF51A and AF51B, producing spectra that, while being type II in nature, were significantly different from each other (Fig. 5B). AF51B gave the characteristic type II spectrum of a predominantly low-spin P450 binding to an azole ligand with a peak at 428 nm and a trough at 403 nm. Similar spectra were observed with AF51B binding to itraconazole and posaconazole (data not shown). AF51A, however, had a spectral peak that became progressively blue shifted from 427 to 418 nm as the fluconazole concentration increased from 1 to 250 μM and two troughs at 376 and 438 nm, respectively. The first trough at 376 nm indicated that prior to binding to fluconazole, AF51A was predominantly in the high-spin conformation, and the second trough at 438 nm in conjunction with the progressively blue-shifted spectral peak indicated the existence of two distinct perturbations of the heme environment within AF51A during fluconazole binding, suggesting the possibility of two binding orientations for fluconazole molecules with the AF51A heme prosthetic group. Chen et al. (9) have recently demonstrated two binding conformations of fluconazole and posaconazole in T. brucei and T. cruzi CYP51 proteins. Itraconazole and posaconazole gave spectra (data not shown) with AF51A similar to the spectrum of fluconazole. The azole binding saturation curves (Fig. 5C and D) indicated profound differences between AF51A and AF51B. AF51A had relatively low affinities for clotrimazole, itraconazole, and posaconazole, obeying the Michaelis-Menten binding model with Kd values of 4.8, 1.0, and 2.7 μM, respectively. In contrast, AF51B tightly bound to clotrimazole, itraconazole, and posaconazole, obeying the Morrison quadratic tight-binding model (26, 32), with Kd values of 0.10, 0.03, and 0.07 μM, respectively (Table 2). Although itraconazole and posaconazole are structurally similar, both AF51A and AF51B had a 2.5-fold higher affinity for itraconazole than posaconazole. The difference in azole binding mechanisms between AF51A and AF51B is exemplified for clotrimazole in Fig. 5C. AF51A bound to fluconazole weakly (Kd, 11.9 μM) and voriconazole relatively weakly (Kd, 3.6 μM), with the Hill equation best describing the resulting binding saturation curves. The apparent Hill numbers of 0.53 and 0.70 obtained for fluconazole and voriconazole, respectively, suggest that negative cooperativity was occurring between AF51A monomers during the binding of these two azole drugs. AF51B also bound to fluconazole weakly (Kd, 4 μM) with an apparent Hill number of 0.51, indicating negative cooperativity between AF51B monomers. However, AF51B bound to voriconazole tightly (Kd, 0.42 μM) with an apparent Hill number close to 1, suggesting no cooperativity between AF51B monomers during voriconazole binding. Mechanistically, AF51B is more analogous to C. albicans CYP51 than to AF51A, as AF51B displayed tight binding toward clotrimazole, itraconazole, and voriconazole like C. albicans CYP51 (41), whereas AF51A did not exhibit tight binding with any of the azoles examined. Weak fluconazole binding (Kd, 19 μM) was also observed with M. smegmatis CYP51 (40). The fluconazole saturation curves for AF51A and AF51B (Fig. 5D) account for the intrinsic fluconazole resistance of wild-type A. fumigatus (11, 30) in comparison with the resistance of wild-type C. albicans CYP51, which has a 250-fold higher affinity (Kd 0.045 μM) for fluconazole than AF51A (41).
FIG. 5.
Azole binding properties of AF51A and AF51B proteins. Azole antifungals were progressively titrated against AF51A (filled circles) and AF51B (hollow circles). The resultant type II difference spectra obtained with clotrimazole (A) and fluconazole (B) are shown. Saturation curves for clotrimazole (C) and fluconazole (D) were constructed. The Hill equation was used to fit the fluconazole data, the Michaelis-Menten equation was used to fit the AF51A clotrimazole data, and the Morrison equation was used to fit the tight binding observed for clotrimazole with AF51B. Native P450 concentrations of 1 μM, as determined by CO-P450 difference spectroscopy (12), were used.
TABLE 1.
Type II difference spectra characteristics of AF51A and AF51B bound to azole antifungal agents
| Azole | Type II difference spectrum (nm) |
||||
|---|---|---|---|---|---|
| AF51A |
AF51B |
||||
| λpeak | λtrough1 | λtrough2a | λpeak | λtrough | |
| Clotrimazole | 429 | 386 | None | 428 | 407 |
| Fluconazole | 427 → 418 | 376 | 438 | 428 | 403 |
| Itraconazole | 421 → 418 | 375 | 437 | 426 | 397 |
| Posaconazole | 424 → 419 | 376 | 438 | 427 | 404 |
| Voriconazole | 419 → 417 | 373 | 436 | 427 | 406 |
λtrough2 is a second spectral trough obtained only when AF51A was bound to triazole antifungal compounds.
TABLE 2.
Azole binding properties of AF51A and AF51B proteins
| Azole | AF51A |
AF51B |
|||||
|---|---|---|---|---|---|---|---|
| ΔAmaxa | Kd (μM) | Apparent Hill no.a,b | ΔAmax | Kd (μM) | Apparent Hill no. | Tight-binding Kd (μM)c | |
| Clotrimazole | 0.05628 | 4.79 (±0.27) | 1.00 | 0.01679 | NAd | NA | 0.103 (±0.007) |
| Fluconazole | 0.02697 | 11.93 (±0.99) | 0.53 | 0.01143 | 4.03 (±0.25) | 0.51 | NA |
| Itraconazole | 0.01880 | 1.01 (±0.08) | 1.00 | 0.01222 | NA | NA | 0.031 (±0.005) |
| Posaconazole | 0.02494 | 2.69 (±0.11) | 1.00 | 0.01263 | NA | NA | 0.073 (±0.006) |
| Voriconazole | 0.02379 | 3.63 (±0.12) | 0.70 | 0.00990 | 0.423 (±0.031) | 0.99 | 0.429 (±0.057) |
The standard deviations of the ΔAmax and apparent Hill number values were less than 5%.
An apparent Hill number of 1.00 indicates that the Michaelis-Menten equation gave the best fit to the binding data.
Tight-binding Kd values for AF51B were calculated using the Morrison equation.
NA, not applicable.
MIC values for azole antifungal drugs with wild-type and CYP51 gene-knockout mutants of A. fumigatus (Table 3) indicate the importance of AF51A in conferring enhanced resistance to azole antifungal agents compared with that which would otherwise be the case if AF51B was the sole CYP51 present. MIC results are influenced by a variety of factors, including inoculum density and the medium used, besides cell permeability, cellular efflux rates, drug lipophilicity, and the availability and affinity of a drug for the target. This makes direct comparisons between observed MIC and Kd values for CYP51 difficult. However, the observed Kd values for itraconazole, posaconazole, and voriconazole with AF51A and AF51B were broadly comparable to the MIC values reported previously (Table 3), whereas fluconazole gave a MIC value for the wild-type organism far higher than the CYP51 Kd value. The direct ligand binding studies described here had correctly identified fluconazole to be the weakest antifungal agent and itraconazole, posaconazole, and voriconazole to be effective inhibitors of A. fumigatus CYP51 proteins A and B. Previous MIC determinations using AF51A- and AF51B-knockout A. fumigatus mutants (13) have shown that AF51A was responsible for conferring resistance to fluconazole and itraconazole (Table 3), with the AF51A knockout causing a 16-fold increase in azole susceptibility. The finding that the wild-type A. fumigatus strains with AF51A-knockout mutations were 17 and 40 times more sensitive than the parental strain to fluconazole and ketoconazole, respectively (31), agrees with our finding that AF51B is more sensitive to azole antifungal agents than AF51A and supports a rationale for the appearance of mutations in CYP51A as the main cause of resistance.
TABLE 3.
MICs for azole antifungals with A. fumigatusa
| Azole | MIC (μg·ml−1) |
|||
|---|---|---|---|---|
| Mutant |
Wild type |
|||
| ΔAF51A | ΔAF51B | CEA10 and CM-237 | AF293 | |
| Fluconazole | 6.2-12.5b (20-41)c | >100b (>327) | >100b (>327) | 32-64d (105-210) |
| Itraconazole | 0.037-0.075b (0.053-0.107) | 0.6b (0.85) | 0.6b (0.85) | 0.5e (0.71) |
| Posaconazole | NDf | ND | ND | 0.5g (0.72) |
| Voriconazole | 0.12-0.50h (0.34-1.44) | ND | 0.52h (1.49) | 0.25e-0.50g (0.72-1.44) |
Previously published MICs for wild-type A. fumigatus strains and CYP51-knockout mutants were correlated. The molecular weights of fluconazole, itraconazole, posaconazole, and voriconazole were 306, 706, 701, and 349, respectively.
Data for Δerg11A and Δerg11B mutants constructed from the wild-type CEA10 A. fumigatus strain (13).
Values in parentheses are the equivalent MICs in μM.
MIC reported by Kim et al. (19).
MIC reported by Balajee et al. (4).
ND, not determined.
MIC reported by Lamaris et al. (21).
ΔAF51A mutants constructed from the wild-type CM-237 A. fumigatus strain (31).
We are presently investigating the azole binding properties of AF51A mutations at the three mutation hot spots Gly54, Leu98, and Met220 to gain further insight into the azole resistance mechanism of this CYP51 isoenzyme. Preliminary results have indicated that the point mutations so far examined yielded AF51A proteins that, while being relatively stable in the E. coli membrane fractions, were unstable when they were solubilized. We are currently optimizing a methodology to produce these mutant proteins.
Acknowledgments
We are grateful to the European Union for support by the FP6 EURESFUN STREP and the Biotechnology and Biological Science Research Council of the United Kingdom, the National Institutes of Health of the United States (grant NSF-MCB 0920212 to W.D.N.), and the Welch Foundation (grant D-1276 to W.D.N.) for supporting this work.
We thank Schering-Plough Ltd. (Welwyn Garden City, United Kingdom) for the kind gift of the posaconazole used in this study.
Footnotes
Published ahead of print on 26 July 2010.
REFERENCES
- 1.Alcazar-Fuoli, L., E. Mellado, G. Garcia-Effron, M. J. Buitrago, J. F. Lopez, J. O. Grimalt, J. M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 50:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoyama, Y., Y. Yoshida, Y. Sonoda, and Y. Sato. 1987. Metabolism of 32-hydroxy-24,25-dihydrolanosterol by purified cytochrome P-45014DM from yeast: evidence for contribution of the cytochrome to whole process of lanosterol 14 alpha-demethylation. J. Biol. Chem. 262:1239-1243. [PubMed] [Google Scholar]
- 3.Arase, M., M. R. Waterman, and N. Kagawa. 2006. Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 344:400-405. [DOI] [PubMed] [Google Scholar]
- 4.Balajee, S. A., J. Gribskov, M. Brandt, J. Ito, A. Fothergill, and K. A. Marr. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43:5996-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard, S. A., S. W. Ellis, S. L. Kelly, and P. F. Troke. 1990. A novel method for studying ergosterol biosynthesis by a cell-free preparation of Aspergillus fumigatus and its inhibition by azole antifungal agents. J. Med. Vet. Mycol. 28:335-344. [PubMed] [Google Scholar]
- 6.Ballard, S. A., S. L. Kelly, S. W. Ellis, and P. F. Troke. 1990. Interaction of microsomal cytochrome P-450 isolated from Aspergillus fumigatus with fluconazole and itraconazole. J. Med. Vet. Mycol. 28:327-334. [PubMed] [Google Scholar]
- 7.Bellamine, A., G. I. Lepesheva, and M. R. Waterman. 2004. Fluconazole binding and sterol demethylation in three CYP51 isoforms indicate differences in active site topology. J. Lipid Res. 45:2000-2007. [DOI] [PubMed] [Google Scholar]
- 8.Bellamine, A., A. T. Mangla, W. D. Nes, and M. R. Waterman. 1999. Characterisation and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:8937-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C.-K., S. S. F. Leung, C. Guilbert, M. P. Jacobson, J. H. McKerrow, and L. M. Podust. 2010. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Neglect. Trop. Dis. 4:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland, R. A. 2005. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists, p. 178-213. Wiley-Interscience, New York, NY. [PubMed]
- 11.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estabrook, R. W., J. A. Peterson, J. Baron, and A. G. Hildebrandt. 1972. The spectrophotometric measurement of turbid suspensions of cytochromes associated with drug metabolism, p. 303-350. In C. F. Chignell (ed.), Methods in pharmacology, vol. 2. Appleton-Century-Crofts, New York, NY. [Google Scholar]
- 13.Hu, W., S. Sillaots, S. Lemieux, J. Davison, S. Kauffman, A. Breton, A. Linteau, C. Xin, J. Bowman, J. Becker, B. Jiang, and T. Roemer. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefcoate, C. R. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 52:258-279. [DOI] [PubMed] [Google Scholar]
- 15.Jefcoate, C. R., J. L. Gaylor, and R. L. Calabrese. 1969. Ligand interactions with cytochrome P450. I. Binding of primary amines. Biochemistry 8:3455-3463. [DOI] [PubMed] [Google Scholar]
- 16.Kahn, R. A., S. Bak, C. E. Olsen, I. Svendsen, and B. L. Møller. 1996. Isolation and reconstitution of the heme-thiolate protein obtusifoliol 14α-demethylase from Sorghum bicolor (L.) Moench. J. Biol. Chem. 271:32944-32950. [DOI] [PubMed] [Google Scholar]
- 17.Kalb, V. F., C. W. Woods, T. G. Turi, C. R. Dey, T. R. Sutter, and J. C. Loper. 1987. Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae DNA-A. J. Mol. Cell. Biol. 6:529-537. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. E. Kelly, C. J. Jackson, A. G. Warrilow, and D. C. Lamb. 2005. The diversity and importance of microbial cytochromes P450, p. 585-617. In P. R. Ortiz de Montellano (ed.), Cytochrome P450—structure, mechanism, and biochemistry. Kluwer Academic/Plenum Publishers, New York, NY.
- 19.Kim, J., B. Campbell, N. Mahoney, K. Chan, R. Molyneux, and G. May. 2008. Chemosensitization prevents tolerance of Aspergillus fumigatus to antimycotic drugs. Biochem. Biophys. Res. Commun. 372:266-271. [DOI] [PubMed] [Google Scholar]
- 20.Kitahama, Y., M. Nakamura, Y. Yoshida, and Y. Aoyama. 2009. The construction and characterization of self-sufficient lanosterol 14-demethylase fusion proteins consisting of yeast CYP51 and its reductase. Biol. Pharm. Bull. 32:558-563. [DOI] [PubMed] [Google Scholar]
- 21.Lamaris, G. A., R. Ben-Ami, R. E. Lewis, and D. P. Kontoyiannis. 2008. Does pre-exposure of Aspergillus fumigatus to voriconazole or posaconazole in vitro affect its virulence and the in vivo activity of subsequent posaconazole or voriconazole, respectively? A study in a fly model of aspergillosis. J. Antimicrob. Chemother. 62:539-542. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, D. C., M. Cannieux, A. G. S. Warrilow, S. Bak, R. A. Kahn, N. J. Manning, D. E. Kelly, and S. L. Kelly. 2001. Plant sterol 14 alpha-demethylase affinity for azole fungicides. Biochem. Biophys. Res. Commun. 284:845-849. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, D. C., D. E. Kelly, K. Venkateswarlu, N. J. Manning, H. F. Bligh, W. H. Schunck, and S. L. Kelly. 1999. Generation of a complete, soluble, and catalytically active sterol 14 alpha-demethylase-reductase complex. Biochemistry 38:8733-8738. [DOI] [PubMed] [Google Scholar]
- 24.Lepesheva, G. I., L. M. Podust, A. Bellamine, and M. R. Waterman. 2001. Folding requirements are different between sterol 14α-demethylase (CYP51) from Mycobacterium tuberculosis and human or fungal orthologs. J. Biol. Chem. 276:28413-28420. [DOI] [PubMed] [Google Scholar]
- 25.Lepesheva, G. I., C. Virus, and M. R. Waterman. 2003. Conservation in the CYP51 family: role of the B′ helix/BC loop and helices F and G in enzyme function. Biochemistry 42:9091-9101. [DOI] [PubMed] [Google Scholar]
- 26.Lutz, J. D., V. Dixit, C. K. Yeung, L. J. Dickmann, A. Zelter, J. A. Thatcher, W. L. Nelson, and N. Isoherranen. 2009. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem. Pharmacol. 77:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann, P. A., R. M. Parmegiani, S.-Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (erg11p, cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 29.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, W. J. G. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellado, E., G. Garcia-Effron, M. J. Buitrago, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2005. Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49:2536-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, J. F. 1969. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185:269-286. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, D. R. 1999. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 369:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Omura, T., and R. Sato. 1964. The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 239:2379-2385. [PubMed] [Google Scholar]
- 35.Paquette, S. M., S. Bak, and R. Feyereisen. 2000. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19:307-317. [DOI] [PubMed] [Google Scholar]
- 36.Podust, L. M., J. Stojan, T. L. Poulos, and M. R. Waterman. 2001. Substrate recognition sites of 14α-sterol demethylase from comparative analysis of amino acid sequences and X-ray structure of Mycobacterium tuberculosis CYP51. J. Inorg. Biochem. 87:227-235. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Tudela, J. L., L. Alcazar-Fuoli, E. Mellado, A. Alastruey-Izquierdo, A. Monzon, and M. Cuenca-Estrella. 2008. Epidemiological cut-offs and cross-resistance to azole drugs in Aspergillus fumigatus. Antimicrob. Agents Chemother. 52:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strushkevich, N., S. A. Usanov, and H. W. Park. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397:1067-1078. [DOI] [PubMed] [Google Scholar]
- 39.Trösken, E. R., M. Adamska, M. Arand, J. A. Zarn, C. Patten, W. Völkel, and W. K. Lutz. 2006. Comparison of lanosterol-14α-demethylase (CYP51) of human and Candida albicans for inhibition by different antifungal azoles. Toxicology 228:24-32. [DOI] [PubMed] [Google Scholar]
- 40.Warrilow, A. G. S., C. J. Jackson, J. E. Parker, T. H. Marczylo, D. E. Kelly, D. C. Lamb, and S. L. Kelly. 2009. Identification, characterization, and azole-binding properties of Mycobacterium smegmatis CYP164A2, a homolog of ML2088, the sole cytochrome P450 gene of Mycobacterium leprae. Antimicrob. Agents Chemother. 53:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warrilow, A. G. S., C. M. Martel, J. E. Parker, N. Melo, D. C. Lamb, W. D. Nes, D. E. Kelly, and S. L. Kelly. 2010. Azole binding properties of sterol 14-α demethylase (CaCYP51). Antimicrob. Agents Chemother. 54:XXX-XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]