Abstract
Two bacterial species with different metabolic features, namely, Pseudomonas aeruginosa and Lactococcus lactis, were used as a comparative experimental model to investigate the antimicrobial target and mechanism of transferrins. In anaerobiosis, P. aeruginosa cells were not susceptible to lactoferrin (hLf) or transferrin (hTf). In aerobiosis, the cells were susceptible but O2 consumption was not modified, indicating that components of the electron transport chain (ETC) were not targeted. However, the respiratory chain inhibitor piericidin A significantly reduced the killing activity of both proteins. Moreover, 2,6-dichlorophenolindophenol (DCIP), a reducing agent that accepts electrons from the ETC coupled to H+ extrusion, made P. aeruginosa susceptible to hLf and hTf in anaerobiosis. These results indicated that active cooperation of the cell was indispensable for the antimicrobial effect. For L. lactis cells lacking an ETC, the absence of a detectable transmembrane electrical potential in hLf-treated cells suggested a loss of H+-ATPase activity. Furthermore, the inhibition of ATPase activity and H+ translocation (inverted membrane vesicles) provided direct evidence of the ability of hLf to inhibit H+-ATPase in L. lactis. Based on these data, we propose that hLf and hTf also inhibit the H+-ATPase of respiring P. aeruginosa cells. Such inhibition thereby interferes with reentry of H+ from the periplasmic space to the cytoplasm, resulting in perturbation of intracellular pH and the transmembrane proton gradient. Consistent with this hypothesis, periplasmic H+ accumulation was prevented by anaerobiosis or by piericidin A or was induced by DCIP in anaerobiosis. Collectively, these results indicate that transferrins target H+-ATPase and interfere with H+ translocation, yielding a lethal effect in vitro.
Transferrins comprise a family of proteins which include iron-binding polypeptides of diverse phylogenetic groups. Two well-studied representative members of this family are transferrin and lactoferrin. These polypeptides have multiple biological functions in blood and mucosal surfaces, respectively. It is thought that both proteins contribute to defense against microbial infection in different host settings, and they are considered components of innate immunity (5, 14).
The antimicrobial activities of transferrin and lactoferrin have been attributed to various causes, including nutritional deprivation of essential iron, catalytic potential in Haber-Weiss-Fenton chemistry, and outer membrane damage in Gram-negative bacteria (1, 13, 27, 33). Alternatively, we have proposed a specific interaction of human lactoferrin with a protein constituent of the microbial cytoplasmic membrane, thus explaining physiological changes associated with lactoferrin-induced cell death (24). This hypothesis derives from the fact that the antimicrobial activity of lactoferrin is tightly linked to cellular bioenergetics and is not due to a permeabilization of cell membranes (6, 24, 25). In addition, the antimicrobial effect may be prevented by extracellular Na+ and K+ (24, 25), suggesting the involvement of the homeostatic processes associated with regulation of cytoplasmic ion concentrations.
In the present study, we assessed the antimicrobial activities of lactoferrin and transferrin on Pseudomonas aeruginosa and Lactococcus lactis cells to identify a putative and common target for transferrins. These species were selected in light of previously observed influences of microbial physiology on the antimicrobial activity of lactoferrin (4, 6, 24, 25) and due to their different metabolic features. P. aeruginosa is a Gram-negative and carbohydrate-nonfermenting species that preferentially uses oxygen (aerobic) or nitrate (anaerobic) as a terminal electron acceptor in respiratory metabolism (32). In the absence of electron acceptors, this opportunistic pathogen may ferment l-arginine, generating sufficient ATP for growth by using substrate-level phosphorylation (11, 23). L. lactis is a Gram-positive, acid-tolerant, and homofermentative organism that utilizes a short, oxygen-dependent respiratory chain solely when hemin is present in the growth medium, thus generating a transmembrane proton gradient through aerobic electron transport (7, 8, 12).
In respiring bacterial species (e.g., P. aeruginosa), the electron transport chain generates a transmembrane proton gradient (ΔpH) necessary for ATP synthesis by the F1Fo-ATPase and for transport of various solutes. However, the F1Fo-ATPase complex is a reversible proton-translocating pump that may extrude protons from the cytoplasm by use of energy provided by ATP hydrolysis. Such proton efflux enhances the proton gradient and assists in regulating the cytoplasmic pH (10, 18). For example, maintenance of optimal intracellular pH (pHi) is an essential function of F1Fo-ATPase for the survival of carbohydrate-fermenting lactic acid bacteria (e.g., L. lactis) (7, 10). These distinct mechanisms of ΔpH maintenance and pHi regulation used by P. aeruginosa and L. lactis were finally used as comparative models to gain insights into the in vitro antibacterial mechanism of human lactoferrin and transferrin. The results suggest that these host defense molecules selectively inhibit the H+-ATPase complex in such bacteria. Based on this finding, we propose a model to explain the antimicrobial mechanism of action of transferrins in vitro under respiratory and fermentative conditions.
MATERIALS AND METHODS
Materials.
Recombinant human apo-lactoferrin (rhLf) and human apo-transferrin (hTf) were obtained from Ventria Bioscience (Sacramento, CA) and Sigma-Aldrich Chemicals (St. Louis, MO), respectively. The peptide Lfpep was obtained from Bio-Synthesis (Lewisville, TX). The reagents 9-amino-3-chloro-7-methoxyacridine (ACMA) and 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)] were purchased from Invitrogen-Molecular Probes (Eugene, OR). Antimycin A, Na2ATP, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), 2,6-dichlorophenolindophenol (DCIP), N,N′-dicyclohexylcarbodiimide (DCCD), 2-(N-morpholino)ethanesulfonic acid (MES), phenylmethylsulfonyl fluoride (PMSF), piericidin A, propidium iodide (PI), Tris, and valinomycin were obtained from Sigma-Aldrich. An ATPase assay kit and PiBind resin were purchased from Innova Biosciences (Cambridge, United Kingdom). M17 broth, tryptic soy broth (TSB), and yeast extract (YE) media were obtained from Difco Laboratories (Detroit, MI).
Bacterial strains and growth conditions.
Cells were grown to mid-log phase in TSB at 37°C (Pseudomonas aeruginosa PAO1) or in M17 broth supplemented with 0.5% glucose (M17G) at 30°C without shaking (Lactococcus lactis subsp. lactis IL1403), as described previously (21). Anaerobic growth and assays under strict anaerobic conditions (80% N2, 10% CO2, and 10% H2) were performed in an anaerobic chamber (model 1024; Forma Scientific, Marietta, OH). The arginine deiminase pathway was induced under anaerobic conditions by growing P. aeruginosa cells in oxygen-sensitive (OS) medium supplemented with 0.4% YE and 40 mM l-arginine (YEA medium), as described previously (23). When required, the denitrification system of P. aeruginosa was stimulated in 0.1× TSB containing 50 mM KNO3 (TSBN) as previously described (16). All media and buffers used for anaerobiosis were held in the anaerobic chamber for a minimum of 48 h prior to use.
Antimicrobial assays.
Bacterial suspensions grown to mid-log phase in appropriate media were washed twice and resuspended in Tris buffer (10 mM Tris-HCl, pH 7.4). The cell suspensions (106 cells/ml) were incubated for 90 min at 37°C (P. aeruginosa) or 30°C (L. lactis) with different concentrations (ranging from 0.03 to 25 μM) of rhLf or hTf, and aliquots were diluted in the same buffer and cultured quantitatively. The influence of extracellular pH on the antibacterial activity was tested using cells resuspended in 10 mM MES (pH 5.5) or 10 mM Tris-HCl (pH 7.4). Ninety percent and 50% inhibitory concentrations (IC90 and IC50) were defined as the protein concentrations that reduced bacterial survival by 90% and 50%, respectively, of that observed in untreated controls. Microbicidal kinetic assays were carried out by incubating cells (106 cells/ml) of P. aeruginosa in the presence of rhLf (0.125, 0.5, and 1 μM) or hTf (1, 2, and 4 μM) and cells of L. lactis with rhLf (0.03, 0.06, and 0.125 μM) or hTf (4 and 25 μM) at 37°C. Aliquots were taken at preselected time intervals, serially diluted in the same buffer, and cultured quantitatively on appropriate media. When required, the assay mixtures contained 5 mM KNO3, 40 mM l-arginine (P. aeruginosa), or 0.5% glucose (L. lactis), and respective quantitative culture plates contained identical concentrations, with the exception of 50 mM KNO3 in nitrate assay plates. The plates were incubated for 24 h at 37°C (P. aeruginosa) or 30°C (L. lactis) for aerobiosis or anaerobiosis, except for YEA and TSBN plates, which were incubated for 2 to 5 days in the anaerobic chamber. After appropriate incubation, colonies were counted to determine the percent survival of treated organisms compared to controls.
Permeabilization.
Cytoplasmic membrane permeabilization was investigated by using the cell-impermeant fluorescent probe propidium iodide in flow cytometric analysis. Cells grown to mid-exponential phase were washed, resuspended (106 cells/ml) in Tris buffer, and incubated with or without the above concentrations of rhLf and hTf or with Lfpep (50 μM) for 90 min at 37°C. The cell suspensions then were reincubated with PI (1 μg/ml) for 5 min. Fluorescence data were acquired as monoparametric histograms by use of a Cytoron Absolute flow cytometer (Ortho Diagnostics Systems Inc., Raritan, NJ). Results are expressed as the percent PI-positive cells with respect to the unstained cells (control).
Transmembrane potential.
Transmembrane electrical potential (Δψ) was monitored using the potential-sensitive fluorescent probe DiSC3(5) as described previously (20). L. lactis cells grown to mid-exponential phase in M17G were washed and resuspended in 50 mM potassium phosphate (pH 5.0). Bacterial suspensions (approximately 107 cells/ml) were incubated (30 min, 30°C) with 2 μM rhLf, 20 μM hTf, or 1 mM DCCD and then reincubated with 3 μM DiSC3(5) to equilibrium of the fluorescent signal. The Δψ was generated upon addition of glucose (15 mM final concentration) as a source of metabolic energy. Fluorescence signals of DiSC3(5) were measured (λex = 651 nm; λem = 675 nm) using a Perkin Elmer LS50B spectrofluorometer. The K+-ionophore valinomycin (2 μM) was added at the end of the reaction to elicit complete collapse of the membrane potential.
Oxygen consumption.
Dissolved oxygen in cell suspensions was measured polarographically by use of a Clark-type electrode (dual digital model 20; Rank Brothers Ltd., Cambridge, United Kingdom) at 25°C. The apparatus consisted of a twin oxygen chamber which enabled a control experiment to be conducted concurrently. Logarithmic-phase P. aeruginosa cells were prepared in Tris buffer as described above. The assays were performed in 1.5 ml of Tris buffer at 25°C. Cell suspensions (107 cells/ml) were preincubated for 15 min at 37°C with rhLf (1, 2, and 4 μM) or hTf (4, 8, and 12 μM). The viability of the cells was determined at 30 min by removing aliquots from the oxygen chambers and plating the subsequent dilutions on TSB agar plates.
DCIP reducing activity.
Cultures of P. aeruginosa were grown in TSBN medium to mid-log phase in anaerobiosis. To maintain strict anaerobic conditions, all of the following steps were performed inside an anaerobic chamber. Briefly, the cells were harvested using a microcentrifuge (Spectrafuge 24D; Labnet International, Edison, NJ), washed in Tris buffer, and adjusted to ∼107 cells/ml in the same buffer. Cells were preincubated (3 min) with or without 3 μM rhLf or 12 μM hTf before the addition of DCIP (0.2 mM final concentration). Next, 1.2-ml aliquots were removed at predetermined times, and the cells were harvested by centrifugation. The supernatant (1 ml) was retained, transferred to a sterile cuvette, and sealed in the anaerobic chamber. The absorbance at 600 nm (A600) of DCIP at 25°C was recorded immediately by use of a Shimadzu UV-1700 PharmaSpec spectrophotometer. The concentration of oxidized DCIP was calculated using a molar extinction coefficient of 11.1 mM−1 cm−1. All colorimetric measurements were repeated in three independent studies for each sample. The DCIP absorbance was also measured in the presence of 3 μM rhLf or 12 μM hTf without cells.
ATPase activity.
The influence of rhLf or hTf on ATPase activity was determined using a colorimetric ATPase assay according to the manufacturer's recommendations. The assays were performed using membrane preparations (40 to 90 μg/100 μl) previously incubated with PiBind resin to remove the free inorganic phosphate (Pi). The membrane samples were preincubated for 10 min at 37°C with rhLf or hTf (10, 20, and 30 μM) or with 0.5 mM DCCD to measure the effects of these compounds on the ATPase activity. The amount of Pi released was calculated by spectrophotometry (A650). For all experiments, calibration was performed using a standard range of Pi concentrations, and data were determined for a minimum of three independent assays.
ATP-dependent proton translocation.
Inverted membrane vesicles were prepared as described previously (7). Briefly, L. lactis cells were grown in M17G medium to late exponential phase, washed, resuspended in MMK buffer (20 mM MOPS-KOH, 10 mM MgCl2, and 300 mM KCl, pH 7.3), and treated with 0.1 mg/ml lysozyme for 18 h. Next, cell suspensions containing 0.2 mM PMSF and 100 μg/ml RNase were passed twice through a French press (15,000 lb/in2) and centrifuged (13,000 × g, 30 min, 4°C) to remove cell debris. The formed vesicles were harvested by ultracentrifugation (125,000 × g, 1 h, 4°C), resuspended in 10 mM HEPES-KOH buffer (pH 7.5) supplemented with 10% glycerol, and stored at −80°C.
Net ATP-dependent proton translocation was assessed using membrane vesicles preincubated for 30 min on ice with rhLf or hTf (10, 20, and 30 μM) or with 0.5 mM DCCD (positive control) to determine the effects of these proteins on H+ translocation. ATP-dependent translocation was monitored by fluorescence quenching of ACMA as described previously (7). Reaction mixtures contained membrane vesicles (∼10 mg/ml total protein) in MMK buffer. After addition of ACMA (0.25 μM), the reaction was initiated by the addition of 1 mM ATP. ACMA fluorescence (λex = 410 nm; λem = 490 nm) was then recorded for 8 min in a spectrofluorometer. The proton ionophore CCCP (2 μg/ml) was added at the end of the reaction to correct for nonspecific quenching.
Other procedures.
Lactoferrin and transferrin were saturated with iron as described by Kalmar and Arnold (17). The saturation status of lactoferrin (holo-rhLf) and transferrin (holo-hTf) was estimated by the ratio of A465 to A280 (15) and was found to be 93% and 87%, respectively. The protein concentration was determined by Bradford assay, with bovine serum albumin as the standard. Nitrate utilization and arginine deaminase activity were determined by use of a nitrate-specific electrode (Mettler-Toledo model DX262-NO3) and by monitoring the production of citrulline from arginine, respectively (3, 16).
Statistical analysis.
Data are expressed as means ± standard deviations (SD), and significance was determined by using Student's t test. P values of <0.05 were considered significant.
RESULTS
Antibacterial activity.
The concentrations of lactoferrin and transferrin that resulted in 90% (IC90) and 50% (IC50) reductions in cell viability were determined and are summarized in Table 1. Data from killing experiments performed at pH 5.5 and pH 7.4 indicated the greatest efficacy of lactoferrin on P. aeruginosa and L. lactis at pH 5.5. A similar result was obtained with transferrin and P. aeruginosa cells (Table 1).
TABLE 1.
Inhibitory concentrations of lactoferrin and transferrina
| Microorganism | pH | Lactoferrin (μM) |
Transferrin (μM) |
||
|---|---|---|---|---|---|
| IC90 | IC50 | IC90 | IC50 | ||
| P. aeruginosa | 5.5 | 0.5 | ND | 2 | ND |
| 7.4 | 1 | 0.5 | 4 | 1 | |
| 7.4 | 2* | 1* | NS* | NS* | |
| L. lactis | 5.5 | 0.062 | ND | NS | NS |
| 7.4 | 0.125 | ND | NS | NS | |
| 7.4 | NS* | NS* | NS* | NS* | |
P. aeruginosa or L. lactis cells were incubated (90 min) with different concentrations (ranging from 0.03 to 25 μM) of lactoferrin or transferrin in 10 mM MES (pH 5.5) or 10 mM Tris-HCl (pH 7.4). Cell viability was calculated with respect to the nontreated cells (control), using a plate count method. ND, not determined; NS, not susceptible at ≥25 μM transferrin. *, values obtained with iron-saturated proteins.
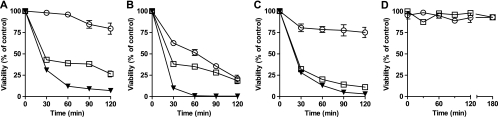
P. aeruginosa cells suspended in Tris buffer were susceptible to rhLf and hTf, in a dose- and time-dependent manner (Fig. 1). Notably, L. lactis cells were susceptible to lactoferrin but not to transferrin (Table 1). Such resistance to the killing activity of hTf remained unchanged even at high hTf concentrations (25 μM) and with an extended (3 h) incubation time (Fig. 1D), as well as at acidic (pH 5.6) and alkaline (pH 7.4) pHs. The antimicrobial activities of the iron-free (apo) proteins were higher than those of the iron-saturated (holo) proteins (Table 1).
FIG. 1.
Kinetics of bactericidal activity of transferrins. P. aeruginosa cells (106 cells/ml) were incubated (37°C) with 0.125 μM (○), 0.5 μM (□), or 1 μM (▾) lactoferrin (A) or with 1 μM (○), 2 μM (□), or 4 μM (▾) transferrin (B). L. lactis cells (106 cells/ml) were incubated (30°C) with 0.031 μM (○), 0.062 μM (□), or 0.125 μM (▾) lactoferrin (C) or with 4 μM (○) or 25 μM (□) transferrin (D). At the given time points, aliquots were plated, and colonies were counted after 24 h. The results are means ± SD for duplicates of at least three independent experiments.
Impact of environmental and metabolic conditions on antimicrobial activity.
Previous studies suggested an energy dependence for lactoferrin killing that might reflect a requirement for active bacterial metabolism during the bactericidal effect (5, 6, 25). We therefore performed bactericidal assays with P. aeruginosa and L. lactis cells under different environmental and metabolic conditions. The influence of anaerobic respiration on the apo-rhLf and apo-hTf antimicrobial effects was determined using P. aeruginosa cells maintained in anaerobiosis. The antimicrobial assays were performed using cells resuspended in Tris buffer with or without 5 mM KNO3 under anaerobic conditions. Cell suspensions containing KNO3 were susceptible to rhLf and hTf (37% ± 9% and 52% ± 8% cell viability, respectively). Interestingly, in the absence of KNO3, the numbers of viable cells treated with rhLf and hTf were not substantially modified versus those of controls (Table 2).
TABLE 2.
Effects of environmental conditions and substrates on bactericidal activity of transferrinsa
| Organism | Incubation conditions | Substrate (concn) | Cell viability (% of control)b |
|
|---|---|---|---|---|
| rhLf | hTf | |||
| P. aeruginosa | Aerobic | 18 ± 7 | 25 ± 6 | |
| Anaerobic | 95 ± 3 | 98 ± 2 | ||
| Anaerobic | KNO3(5 mM) | 37 ± 9 | 52 ± 8 | |
| Anaerobic | l-Arginine (40 mM) | 92 ± 4 | 90 ± 2 | |
| L. lactis | Anaerobic | 10 ± 3 | 96 ± 4c | |
| Anaerobic | Glucose (1%) | 8 ± 4 | 93 ± 8c | |
P. aeruginosa or L. lactis cells were incubated (90 min) in 10 mM Tris-HCl (pH 7.4) with or without the indicated substrates under the specified environmental conditions.
Values are means ± SD for duplicates from at least three independent experiments. The IC90s of lactoferrin (rhLf) and transferrin (hTf) were used.
An hTf concentration of 25 μM was used.
The efficacy of rhLf and hTf on nonrespiring P. aeruginosa cells deriving energy (i.e., ATP) under conditions of strict anaerobiosis was also tested. Arginine-consuming P. aeruginosa cells were not susceptible to rhLf or hTf (Table 2). In contrast, glucose-fermenting L. lactis cells were susceptible to a bactericidal concentration of rhLf (0.125 μM) but not to 25 μM hTf (Table 2).
Control assays performed in parallel in 10 mM Tris-HCl (pH 7.4) further demonstrated that P. aeruginosa cells were able to utilize nitrate and l-arginine under these experimental conditions (data not shown), as previously described (3, 16).
Effect of transferrins on cytoplasmic membrane integrity and function.
Since the antimicrobial activity on P. aeruginosa cells was dependent on the environmental conditions (external pH, aerobiosis versus anaerobiosis) but independent of the cellular energetic state, we performed experiments to determine whether membrane integrity or the function of some associated membrane elements (i.e., electron respiratory chain components) was modified by apo-transferrins.
(i) Permeabilization.
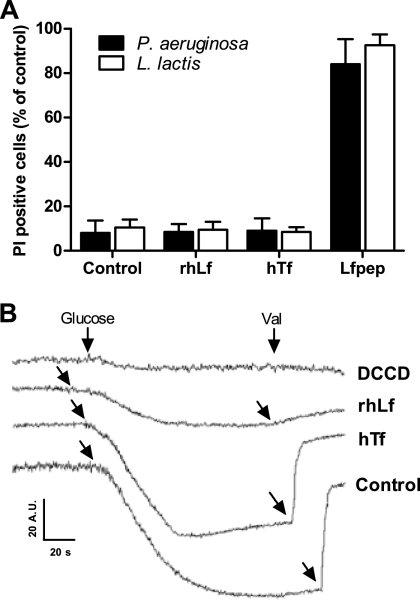
To determine whether cells exhibited elevated membrane permeability following lactoferrin or transferrin treatment, measurements of intracellular propidium iodide accumulation were undertaken in P. aeruginosa and L. lactis cells. The frequencies of P. aeruginosa cells showing PI accumulation after incubation with bactericidal concentrations of rhLf (1 μM) and hTf (4 μM) were 6% ± 5% and 8% ± 5%, respectively (Fig. 2 A). L. lactis cells also showed a low level of PI accumulation, as observed by increased fluorescence intensity in 9% ± 3% and 8% ± 2% of cells following treatment with 0.125 μM rhLf and 4 μM hTf, respectively. This degree of permeabilization was not significantly different from that of untreated controls (Fig. 2A). However, in positive-control assays, the percentages of permeabilized cells after exposure to a bactericidal concentration of the peptide Lfpep (50 μM) were high (82% ± 6% for P. aeruginosa and 84% ± 8% for L. lactis), indicating a disruption of the cytoplasmic membrane sufficient for PI permeabilization (Fig. 2A). Lfpep is an antimicrobial cationic peptide derived from lactoferrin that permeabilizes bacterial and fungal cytoplasmic membranes (24).
FIG. 2.
Effect of transferrins on the cytoplasmic membrane. (A) P. aeruginosa and L. lactis cells were incubated with or without (control) 1 μM lactoferrin (rhLf), 0.125 μM transferrin (hTf), or 50 μM Lfpep (positive control) and then stained with 1 μg/ml propidium iodide (PI). (B) Effect of transferrins on Δψ of L. lactis cells. The addition of glucose to cell suspensions (107 cells/ml) resulted in the generation of a membrane potential, observed as a decrease in DiSC3(5) fluorescence, in control assays (control). Cells were preincubated (30 min, 30°C) with 2 μM lactoferrin (rhLf), 20 μM transferrin (hTf), or 1 mM DCCD. Additions of glucose and 2 μM valinomycin (Val) are indicated (arrows). Fluorescence is expressed in arbitrary units (A.U.).
(ii) Transmembrane potential.
Since changes in the transmembrane electrical potential (Δψ) may reflect ion movements through cell membranes, we next studied whether the previously reported ability of lactoferrin to modify the transmembrane electrical potential (Δψ) of P. aeruginosa cells and other microorganisms may also occur with L. lactis cells (2, 24, 25).
In L. lactis cells metabolizing glucose, Δψ generation depends on the H+ extrusion mediated by FoF1-ATPase (8). In our assays, the addition of glucose to L. lactis cells increased the transmembrane Δψ, resulting in the accumulation of the dye DiSC3(5) into the cells and in decreased fluorescence (Fig. 2B). As expected, the Δψ was negligible for cells preincubated with 1 mM DCCD, a specific inhibitor of H+-ATPase used as a positive control. In a similar way, cells pretreated with a bactericidal concentration of rhLf (2 μM) were unable to generate a detectable transmembrane electrical potential. However, in agreement with the observed resistance of L. lactis to transferrin, a Δψ was detected for cells preincubated with 20 μM transferrin. The subsequent addition of valinomycin (2 μM) dissipated the transmembrane Δψ, resulting in the release of the dye from the cells and in increased fluorescence (Fig. 2B).
(iii) Effect of transferrins on P. aeruginosa respiration.
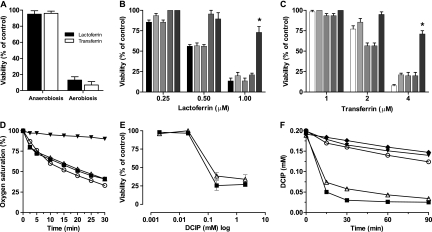
P. aeruginosa cells were not susceptible to rhLf or hTf under anaerobic conditions (Fig. 3 A). The effects of respiratory chain inhibitors, such as antimycin A, piericidin A, and sodium azide, and of the uncoupler CCCP on the bactericidal activity of rhLf and hTf were different (Fig. 3B and C). The antimicrobial activities of rhLf and hTf were not inhibited in cells previously incubated with antimycin A (10 μM) or sodium azide (1 mM) for 15 min at 37°C. However, P. aeruginosa preincubated (15 min, 37°C) with piericidin A (32 μM), an inhibitor of the bacterial type I NADH dehydrogenase (complex I), was significantly (P < 0.05) less susceptible to the killing activity of 1 μM rhLf and 4 μM hTf (73% ± 7% and 71% ± 4% cell survival, respectively). Preincubation (15 min, 37°C) with the proton ionophore CCCP (50 μM) also prevented the antimicrobial activity, but only when the concentrations of the proteins were less than or equal to the IC50 (Fig. 3B and C).
FIG. 3.
Influence of respiration on bactericidal activity of lactoferrin. (A) Viability of P. aeruginosa cells (106 cells/ml) incubated for 90 min at 37°C with 1 μM rhLf or 4 μM transferrin during aerobiosis and anaerobiosis. (B and C) Experimental conditions were the same as those described for panel A, except that the assays were performed aerobically in the presence of the following respiration inhibitors and uncouplers: 1 mM azide (lightest gray bars), 10 μM antimycin (light gray bars), 50 μM CCCP (dark gray bars), and 32 μM piericidin A (darkest gray bars). The activities of different concentrations of rhLf (black bars) and hTf (white bars) alone were also tested. (D) Consumption of oxygen in P. aeruginosa resting cells (○) or cells incubated with 3 μM rhLf (▪), 12 μM hTf (▵), or 32 μM piericidin A (▾), as a positive control, added 15 min before the O2 consumption measurement. (E) Viability of P. aeruginosa cells treated with rhLf (▪) or hTf (▵) in the presence of different concentrations of the electron acceptor DCIP under anaerobic conditions. (F) Concentration of oxidized DCIP in control cell suspensions (○), after addition of 3 μM rhLf (▪) or 12 μM hTf (▵), and in the presence of 3 μM rhLf (▾) or 12 μM hTf (⧫) without cells. *, P < 0.05 versus untreated controls.
P. aeruginosa cells consumed oxygen in the absence of an exogenous energy source (Fig. 3D). Oxygen utilization data obtained for cell suspensions exposed to high bactericidal concentrations of rhLf (3 μM) and hTf (12 μM) indicated rates of consumption of oxygen similar to that of the untreated cells. Similarly, the respiration of cells energized by the addition of succinate (33 mM) was not inhibited by these proteins (data not shown). In control assays, the preincubation of the cells with 32 μM piericidin A resulted in an immediate decrease in respiration (Fig. 3D).
Importantly, cells maintained in strict anaerobiosis were susceptible to rhLf and hTf only when they were preincubated (5 min) with DCIP, an artificial electron acceptor (Fig. 3E). In the presence of 0.2 mM DCIP, the cell viabilities of rhLf- and hTf-treated cells decreased significantly (P < 0.05), to 27% ± 3% and 34% ± 6%, respectively. The absorbance of DCIP in P. aeruginosa cell suspensions treated with 3 μM rhLf or 12 μM hTf was also significantly (∼70% at 90 min) decreased with respect to that in control cells assayed identically in the absence of either protein (Fig. 3F). DCIP was not reduced by rhLf or hTf (Fig. 3F) and was not cytotoxic at the assayed concentrations (data not shown). These data indicate a correlation between the loss of viability of rhLf-treated cells and DCIP reduction.
In conclusion, changes in the transmembrane electrical potential observed in P. aeruginosa (2) and L. lactis cells were not correlated with a permeabilization of cell membranes by transferrins, suggesting an alteration of proton homeostasis. Supporting this notion, respiratory activity of P. aeruginosa cells was an indispensable condition for the antimicrobial effect of transferrins. This requirement was interpreted as an event associated with changes of the pH gradient and intracellular pH due to these proteins being unable to inhibit cellular respiration.
Effect of transferrins on H+-ATPase.
To substantiate the assertion that cytoplasmic proton homeostasis could be modified by apo- and holo-transferrins, the ATPase activity was assayed using isolated membranes. The assessment of the ATPase activity was chosen because under our experimental conditions, the ATPase complex is the only functional and common membrane component involved in proton flux through membranes in both studied species. This function has a critical role in the maintenance of the proton gradient and intracellular pH in many bacterial species.
(i) Effect of transferrins on ATPase-coupled H+ transport.
Inhibition of ATP-dependent proton translocation by rhLf and hTf was determined by fluorescence quenching of ACMA, using inverted membrane vesicles from L. lactis cells (Table 3). The percentages of fluorescence in vesicle assays containing 20 μM apo-lactoferrin or holo-lactoferrin were almost three times higher than those in the absence of the protein, suggesting a decrease of the transmembrane proton gradient generated at the expense of ATP. A decrease in fluorescence quenching (∼32%) was also observed in control assays using the ATPase inhibitor DCCD (0.5 mM). No significant decrease in ACMA quenching was detected in the presence of transferrin (Table 3).
TABLE 3.
Effects of transferrins on ATP-driven proton translocation and ATPase activity of L. lactisa
| Protein | Concn (μM, unless stated otherwise) | Final ACMA fluorescence (% of initial fluorescence) | ATPase activity (%) |
|---|---|---|---|
| Control | 32 ± 0.7 | 8.8 ± 0.6 (100) | |
| DCCD | 0.5 mM | 64 ± 0.5 | 2.3 ± 0.3* (26) |
| Lactoferrin | 10 | 56 ± 0.8 | 2.1 ± 0.9* (24) |
| 20 | 86 ± 0.4 | 2.4 ± 0.5* (27) | |
| 20b | 77 ± 0.7 | 3.1 ± 0.8* (35) | |
| 30 | ND | 2.2 ± 1.5* (25) | |
| Transferrin | 10 | 37 ± 0.9 | 8.3 ± 0.9 (94) |
| 20 | ND | 8.2 ± 0.7 (93) | |
| 20b | ND | 8.1 ± 0.2 (92) | |
| 30 | 39 ± 0.2 | 8.7 ± 1.2 (99) |
ATPase activity is reported as μmol Pi min−1 (mg protein)−1 at 30°C. ND, not determined. Values are means ± SD for at least three experiments. *, P < 0.05.
Iron-saturated protein.
(ii) Effect of transferrins on ATPase activity.
The effect of lactoferrin or transferrin on ATP hydrolysis catalyzed by the membrane fraction of L. lactis cells is shown in Table 3. ATP hydrolysis decreased to approximately 24%, 27%, and 25% of the initial rate after preincubation with 10, 20, and 30 μM rhLf, respectively (Table 3). Similarly, the amount of hydrolyzed ATP decreased to ∼35% after preincubation with 20 μM holo-lactoferrin. In the presence of different concentrations (10, 20, and 30 μM) of hTf, ATPase activity was similar to that of the control (Table 3) and was not modified at different pH values (5.5 and 6.5) or MgCl2 concentrations (1, 2.5, and 3 mM) (data not shown). In control assays, ATPase activity was significantly inhibited (∼74% inhibition) by 0.5 mM DCCD, a specific inhibitor of the bacterial F1Fo-ATPase.
DISCUSSION
The bactericidal activity of lactoferrin, independent of iron withholding, was first reported by Arnold et al. (5), but the antimicrobial mechanism has not been elucidated. In this study, we assumed that in two susceptible but different bacterial species differing in their structural and metabolic features, the possibility of a common target for transferrins is limited to the number of characteristics that these microorganisms share. Consequently, we used P. aeruginosa and L. lactis as a comparative experimental model to identify the antimicrobial target of these innate immune proteins.
Data from distinct experiments indicated that the antimicrobial activities of lactoferrin and transferrin were independent of the energetic state of the cells (i.e., ATP synthesis). This interpretation was substantiated by the observation that P. aeruginosa cells obtaining energy (i.e., ATP) by anaerobic respiration (NO3) were susceptible to these proteins, while cells deriving energy from substrate-level phosphorylation (l-arginine) were resistant. In addition, it seemed unlikely that interference with the energy metabolism was a common mechanism of lactoferrin action due to the fact that glucose-fermenting and resting L. lactis cells were both susceptible to lactoferrin. This result is in agreement with previous observations showing differences in lactoferrin susceptibility for the same bacterial strains growing under different nutritional conditions (4).
Interestingly, the bactericidal activity on P. aeruginosa cells was observed for respiring cells only when terminal electron acceptors (O2 or NO3−) were available. Although these observations pointed to a blocking effect on the respiratory chain, the cellular O2 consumption rates in the presence and absence of rhLf and hTf were similar. Moreover, L. lactis cells lacking a respiratory chain were also susceptible to rhLf, indicating that components of the respiratory chain were not the targets of these proteins. These findings prompted us to investigate if the bactericidal effect was associated with the proton flux mediated by the respiratory chain, which is involved in the generation of a transmembrane pH gradient and in intracellular pH regulation. Since changes in the transmembrane electrical potential (Δψ) may reflect modifications of the pH gradient (ΔpH), the previously reported ability of rhLf to modify the Δψ of P. aeruginosa cells (2) supported the above suggestion.
Despite the consumption of oxygen being similar in rhLf- or hTf-treated and untreated P. aeruginosa cells, the functionality of all or part of the respiratory chain was essential for the antimicrobial activity. This was supported by the fact that piericidin A, a specific inhibitor of the type I NADH dehydrogenase (NADHd type I), inhibited the bactericidal effect on respiring P. aeruginosa cells. The NADHd complex (complex I) couples the transfer of electrons from NADH to ubiquinone (CoQ) or menaquinone (mQ), facilitating the translocation of protons across the cytoplasmic membrane (26). However, antimycin A- or azide-treated cells were susceptible to rhLf and hTf, which may be explained by the possible employment of alternative respiratory pathways (e.g., the cyanide-insensitive respiratory pathway) to circumvent inhibition by such inhibitors (9, 28). Supportive evidence showing that respiratory function was indispensable for the antimicrobial activity was provided by the observation that under anaerobic conditions, the resistance to rhLf and hTf of P. aeruginosa cells was reverted by an artificial electron acceptor (DCIP). This chemical agent is reduced by electrons donated from components of the respiratory chain (i.e., NADH dehydrogenase and cytochromes) coupling H+ translocation from the cytoplasm. Therefore, we concluded that the loss of viability of P. aeruginosa cells treated with rhLf or hTf correlated with H+ extrusion mediated by the respiratory chain, suggesting an alteration of pH gradient and internal pH.
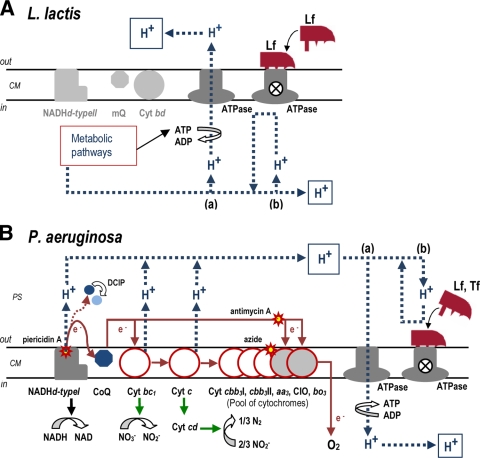
Since L. lactis lacks a functional respiratory chain, both proton gradient and internal pH regulation depends on the H+ extrusion mediated by H+-ATPase (7, 10, 12, 19). Consequently, this complex was investigated as the possible common element in both bacterial species that might be targeted by transferrins. We assumed that the susceptibility of L. lactis cells to lactoferrin could be due to a lethal perturbation of the intracellular pH and the proton gradient due to an inhibition of the ATPase complex (Fig. 4 A). This supposition was inferred from experiments in which the Δψ generation abilities of rhLf-treated and untreated cells metabolizing glucose were compared. Lactoferrin-treated cells were unable to generate a membrane potential, suggesting an inhibition of H+-ATPase. Finally, direct evidence of inhibition of the ATPase complex by lactoferrin was provided by inhibition of ATPase activity and H+ translocation by use of plasma membrane fractions and inverted membrane vesicles of L. lactis cells, respectively.
FIG. 4.
Hypothetical mechanism of antimicrobial action of transferrins. (A) In L. lactis glucose-fermenting cells, H+ pumping through the ATPase (a) is essential for generation of a proton motive force across the cytoplasmic membrane (CM) and for intracellular pH homeostasis, and the latter is critical for cell survival. The blocking effect of lactoferrin (Lf) on H+-ATPase (b) causes an intracellular H+ accumulation, and then the acidification of the bacterial cytoplasm reaches levels incompatible with cell life. L. lactis cells lack a functional respiratory chain (NADHd type II, mQ, and cytochrome bd [Cyt bd]) under these experimental conditions. (B) Respiration of P. aeruginosa coupled to ATP synthase-mediated phosphorylation (a) is uncoupled by the blocking effect of lactoferrin (Lf) or transferrin (Tf) on the ATPase complex (b). Under these experimental conditions, the H+ accumulation in the periplasmic space (PS) leads to cell death. Cellular protection was observed when protons were not pumped to the PS (i.e., anaerobiosis or inhibition of NADHd type I with piericidin A). In anaerobiosis, the presence of the electron acceptor DCIP promotes the H+ pumping mediated by electron transport chain components (e.g., NADHd type I), yielding a lethal effect in Lf- or Tf-treated cells. In P. aeruginosa, electrons are donated from Cyt c to either Cyt cbb3 I, Cyt cbb3 II, or Cyt aa3 (white). Cyt CIO and Cyt ba3 (gray) directly accept electrons from CoQ (9). Relevant steps of anaerobic respiration are indicated (green arrows).
The identification of H+-ATPase as the target of lactoferrin in L. lactis cells suggested a similar mode of action in P. aeruginosa, and the above results obtained with this species were interpreted as follows. The susceptibility of DCIP-treated P. aeruginosa cells to rhLf or hTf in the absence of terminal electron acceptors (O2 or NO3−) was compatible with DCIP reduction, performed by components of the respiratory chain, coupled to the extrusion of protons which were unable to return to the cytoplasm via ATPase (Fig. 4B). The reduction of DCIP was observed only in rhLf- and hTf-treated cells, further implying that this reagent was reduced by a component(s) of the respiratory chain as a response to the uncoupling effect caused by rhLf or hTf.
The suggested selective inhibition of the ATPase complex by lactoferrin and transferrin may also explain the respective susceptibility and resistance observed in respiring and nonrespiring P. aeruginosa cells. We hypothesize that in respiring P. aeruginosa cells treated with rhLf or hTf, reentry of some protons previously extruded by components of the respiratory chain (i.e., NADH dehydrogenase and cytochromes) is blocked by interactions of rhLf or hTf with the H+-ATPase complex. In this model, the subsequent loss of intracellular pH regulation and modification of the proton gradient ultimately lead to cell death (Fig. 4B). A similar requirement of a functional H+-ATPase to recover the protons translocated during NADH oxidation coupled with O2 reduction was recently proposed (22).
Our findings indicate that bacterial cell death induced by transferrins is not caused by cell damage (e.g., membrane permeabilization) but involves an active cooperation of the cell. This observation suggests that the final antimicrobial effectiveness of transferrins depends on the local context where these proteins are secreted. For example, in lung infections of patients with cystic fibrosis, the anaerobic conditions present in the bacterial biofilms could protect P. aeruginosa from the high lactoferrin concentration accumulated in the mucosal fluid, avoiding the eradication of this opportunistic pathogen from the airways of cystic fibrosis patients.
Since the bacterial cell wall is an effective barrier to large proteins, the way that transferrins (>78 kDa) reach a target (i.e., H+-ATPase) on the cytoplasmic membrane has yet to be elucidated. A possible explanation could be the enzymatic generation of lactoferrin-derived peptides with antimicrobial activity. This suggestion is supported by our recent data showing that kaliocin-1, a human lactoferrin-derived antimicrobial peptide which corresponds to the common γ-core motif found in all antimicrobial cysteine peptides (24, 29, 30), may be obtained from lactoferrin by enzymatic digestion. Interestingly, mass spectrometry analysis has shown that this fragment maintains an intact tridimensional structure of the γ-core motif similar to that predicted for the native hLf molecule, suggesting that its potential antimicrobial activity could also be preserved (unpublished results).
It is known that apo-lactoferrin exhibits a higher bactericidal activity than that of holo-lactoferrin (4, 17), and this was also observed in our bacterial killing assays. However, measurements of the ATPase activity in vitro suggest that the mechanism of action proposed here could be independent of the iron saturation state of lactoferrin. Both ATPase activity and proton translocation on membrane vesicles were inhibited less efficiently (but not significantly so) by holo-lactoferrin. The absence of a correlation between the low antimicrobial activity of holo-lactoferrin and our data from ATPase activity experiments suggests the involvement of a concomitant unknown effect associated with the iron saturation state of lactoferrin that requires further investigation.
Beyond the current studies, work is in progress to characterize the potential molecular interactions between the transferrin proteins and the H+-ATPase complex. In this respect, the transferrin resistance of L. lactis but not P. aeruginosa would be an interesting starting point to determine the H+-ATPase domain(s) involved in such a ligand-receptor interaction. These findings also support recent discoveries in evolutionary and phylogenetic relationships among transferrins and other endogenous host defense proteins. For example, for the transferrin protein family, we have reported the presence of evolutionarily conserved three-dimensional structures (i.e., γ-core motifs) previously associated with the antimicrobial activity of all classes of cysteine-stabilized host defense peptides (29, 30). Given the recent discovery that γ-core motifs may mediate targeting of ion channels in microbial pathogens (31), this convergent structural motif could be involved in peptide interactions with one or more specific domains of H+-ATPase and in inhibition of this essential cellular component.
In summary, the data presented herein suggest that the in vitro bactericidal effect of mucosal human lactoferrin and its serum counterpart, transferrin, involves selective inhibition of the H+-ATPase complex. As a result, the H+-ATPase-mediated flux of protons is impaired, yielding effects principally relating to deficiencies in intracellular pH homeostasis inducing cell death. To our knowledge, this is the first description of the interaction of an extracellular human protein with a bacterial H+-ATPase. These findings suggest new opportunities to target energetic systems in bacterial pathogens as a means to discover and develop novel anti-infective agents and therapeutic strategies.
Acknowledgments
We thank M. R. Yeaman (David Geffen School of Medicine at UCLA) for critical reviews and helpful discussions and S. Lory (Harvard Medical School, Boston, MA) for kindly providing P. aeruginosa PAO1.
This work was supported by Laboratorio de Microbiología Oral, University of Oviedo (CN-04-005), Fundación Sira Carrasco para la Ayuda a la Fibrosis Quística (SV-03-FSCARRASCO), and Fondo de Investigación Sanitaria (FIS PI05/2588), Madrid, Spain.
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Ambruso, D. R., and R. B. Johnston. 1981. Lactoferrin enhances hydroxyl radical production by human neutrophils, neutrophil particulate fractions, and an enzymatic generating system. J. Clin. Invest. 67:352-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrés, M. T., M. Viejo-Díaz, F. Pérez, and J. F. Fierro. 2005. Antibiotic tolerance induced by lactoferrin in clinical Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 49:1613-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archibald, R. M. 1944. Determination of citrulline and allantoin and demonstration of citrulline in blood plasma. J. Biol. Chem. 156:121-142. [Google Scholar]
- 4.Arnold, R. R., M. Brewer, and J. J. Gauthier. 1980. Bactericidal activity of human lactoferrin: sensitivity of a variety of microorganisms. Infect. Immun. 28:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold, R. R., M. F. Cole, and J. R. McGhee. 1977. A bactericidal effect for human lactoferrin. Science 197:263-265. [DOI] [PubMed] [Google Scholar]
- 6.Arnold, R. R., J. E. Russell, W. J. Champion, and J. J. Gauthier. 1981. Bactericidal activity of human lactoferrin: influence of physical condition and metabolic state of the target microorganisms. Infect. Immun. 32:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blank, L. M., B. J. Koebmann, O. Michelsen, L. K. Nielsen, and P. R. Jensen. 2001. Hemin reconstitutes proton extrusion in an H+-ATPase-negative mutant of Lactococcus lactis. J. Bacteriol. 183:6707-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooijmans, R. J. W., B. Poolman, G. K. Schuurman-Wolters, W. M. de Vos, and J. Hugenholtz. 2007. Generation of a membrane potential by Lactococcus lactis through aerobic electron transport. J. Bacteriol. 189:5203-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, M., G. R. Tavankar, and H. D. Williams. 2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 149:1275-1284. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duwat, P., S. Sourice, B. Cesselin, G. Lamberet, K. Vido, P. Gaudu, Y. Le Loir, F. Violet, P. Loubière, and A. Gruss. 2001. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison, R. T., T. J. Giehl, and F. M. LaForce. 1988. Damage of the outer membrane of enteric Gram-negative bacteria by lactoferrin and transferrin. Infect. Immun. 56:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnaud, S., and R. W. Evans. 2003. Lactoferrin—a multifunctional protein with antimicrobial properties. Mol. Immunol. 40:395-405. [DOI] [PubMed] [Google Scholar]
- 15.Hashizume, S., K. Kuroda, and H. Murakami. 1987. Cell culture assay of biological activity of lactoferrin and transferrin. Methods Enzymol. 147:302-314. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, D., and J. J. Rowe. 1987. Oxygen regulation of nitrate uptake in denitrifying Pseudomonas aeruginosa. Appl. Environ. Microbiol. 53:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalmar, J. R., and R. R. Arnold. 1988. Killing of Actinobacillus actinomycetemcomitans by human lactoferrin. Infect. Immun. 56:2552-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, H. 1985. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J. Biol. Chem. 260:72-76. [PubMed] [Google Scholar]
- 19.Koebmann, B. J., D. Nilson, O. P. Kuipers, and P. R. Jensen. 2000. The membrane-bound H+-ATPase complex is essential for growth of Lactococcus lactis. J. Bacteriol. 182:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krijgsveld, J., S. A. J. Zaat, J. Meeldijk, P. A. van Veelen, G. Fang, B. Poolman, E. Brandt, J. E. Ehlert, A. J. Kuijpers, G. H. M. Engbers, J. Feijen, and J. Dankert. 2000. Thrombocidins, microbicidal proteins from human blood platelets, are C-terminal deletion products of CXC chemokines. J. Biol. Chem. 275:20374-20381. [DOI] [PubMed] [Google Scholar]
- 21.Poolman, B., D. Molenaar, E. J. Smid, T. Ubbink, T. Abee, P. P. Renault, and W. N. Konings. 1991. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran, S. L., and G. M. Cook. 2005. The F1F0-ATP synthase of Mycobacterium smegmatis is essential for growth. J. Bacteriol. 187:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Wauven, C., A. Piérard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viejo-Díaz, M., M. T. Andrés, J. Pérez-Gil, M. Sánchez, and J. F. Fierro. 2003. Potassium-efflux induced by a new lactoferrin-derived peptide mimicking the effect of native human lactoferrin on the bacterial cytoplasmic membrane. Biochemistry (Moscow) 68:217-227. [DOI] [PubMed] [Google Scholar]
- 25.Viejo-Díaz, M., M. T. Andrés, and J. F. Fierro. 2004. Modulation of in vitro fungicidal activity of human lactoferrin against Candida albicans by extracellular cation concentration and target cell metabolic activity. Antimicrob. Agents Chemother. 48:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, J. E. 1992. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25:253-324. [DOI] [PubMed] [Google Scholar]
- 27.Weimberg, E. D. 1974. Iron and susceptibility to infectious disease. Science 184:952-956. [DOI] [PubMed] [Google Scholar]
- 28.Williams, H. D., J. E. A. Zlosnik, and B. Ryall. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52:3-71. [DOI] [PubMed] [Google Scholar]
- 29.Yount, N. Y., and M. R. Yeaman. 2004. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. U. S. A. 101:7363-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yount, N. Y., M. T. Andrés, J. F. Fierro, and M. R. Yeaman. 2007. The γ-core motif correlates with antimicrobial activity in cysteine-containing kaliocin-1 originating from transferrins. Biochim. Biophys. Acta 1768:2862-2872. [DOI] [PubMed] [Google Scholar]
- 31.Yount, N. Y., D. Kupferwasser, A. Spisni, S. M. Dutz, Z. H. Ramjan, S. Sharma, A. J. Waring, and M. R. Yeaman. 2009. Selective reciprocity in antimicrobial activity versus cytotoxicity of hBD-2 and crotamine. Proc. Natl. Acad. Sci. U. S. A. 106:14972-14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zannoni, D. 1989. The respiratory chains of pathogenic pseudomonads. Biochim. Biophys. Acta 975:299-316. [DOI] [PubMed] [Google Scholar]
- 33.Zhao, X. Y., and T. W. Hutchens. 1994. Proposed mechanisms for the involvement of lactoferrin in hydrolysis of nucleic acid. Adv. Exp. Med. Biol. 357:271-278. [DOI] [PubMed] [Google Scholar]