Abstract
Nonreplicating or dormant cells of Mycobacterium tuberculosis constitute a challenge to tuberculosis (TB) therapy because of their tolerance or phenotypic resistance to most drugs. Here, we propose a simple model for testing drugs against nongrowing cells that exploits the 18b strain of M. tuberculosis, a streptomycin (STR)-dependent mutant. Optimal conditions were established that allowed 18b cells to replicate in the presence of STR and to survive, but not multiply, following withdrawal of STR. In the presence of the antibiotic, M. tuberculosis 18b was susceptible to the currently approved TB drugs, isoniazid (INH) and rifampin (RIF), and to the experimental drugs TMC207, PA-824, meropenem (MER), benzothiazinone (BTZ), and moxifloxacin (MOXI). After STR depletion, the strain displayed greatly reduced susceptibility to the cell wall inhibitors INH and BTZ but showed increased susceptibility to RIF and PA-824, while MOXI and MER appeared equipotent under both conditions. The same potency ranking was found against nonreplicating M. tuberculosis 18b after in vivo treatment of chronically infected mice with five of these drugs. Despite the growth arrest, strain 18b retains significant metabolic activity in vitro, remaining positive in the resazurin reduction assay. Upon adaption to a 96-well format, this assay was shown to be suitable for high-throughput screening with strain 18b to find new inhibitors of dormant M. tuberculosis.
Tuberculosis (TB) is one of the most important infectious diseases caused by a single pathogen and afflicts both healthy and immunocompromised individuals. One-third of the human population is reported to be latently infected with Mycobacterium tuberculosis, and millions of lives are lost every year (8). The emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains (10), together with a deadly synergy with HIV, has worsened the global problem and forced the research community to look for new strategies to fight M. tuberculosis. As a consequence, several drug discovery programs have been established worldwide with the goal of finding new therapeutic agents that may complement or even replace the existing directly observed therapy short course (DOTS) for TB. These programs have to cope with the ability of the pathogen to enter the so-called dormant or latent phase (5, 6), characterized by phenotypic drug resistance (11). Complete control of TB will therefore require finding drugs that are effective against the reservoir of nongrowing tubercle bacilli and, hence, the necessity of modeling this state in vitro and in vivo in order to screen for active compounds.
Latent TB is the result of complex host-pathogen interactions (26, 35), and mimicking this state may thus be partial and challenging. Different models have been proposed so far, each one based on a particular stress that the pathogen may encounter in the host. In particular, the oxygen depletion model mimics the hypoxic environment of the granuloma (30, 31) and the nutrient starvation model recapitulates the lack of nutrients (4), whereas treatment with nitric oxide (15) and growth in acidic medium (9) represent two additional stresses M. tuberculosis has to deal with during infection. Most recently, these treatments were combined (7). While these models contributed to unraveling M. tuberculosis phenotypes in dormancy, they are too cumbersome for application to high-throughput screening (HTS) assays. A reproducible and easy-to-use model of nongrowing cells is therefore required.
In this context, we felt that the streptomycin (STR)-dependent M. tuberculosis strain 18b might represent a good candidate for modeling dormant bacteria in a simple yet conditional way. M. tuberculosis 18b was isolated in Japan in 1955 from a patient with streptomycin-resistant tuberculosis (12). The bacterial phenotype was then recognized as streptomycin dependent, with the microorganism being unable to grow unless streptomycin was present. Additionally, the strain did not lose viability when maintained in streptomycin-free cultures for several weeks and grew again upon adding streptomycin back (12). The molecular reason for this unique phenotype was investigated, and insertion of a cytosine in the 530 loop of the 16S rRNA, known to be involved in STR resistance, was found to be associated with the dependency (13). Recent vaccine work exploited M. tuberculosis 18b for establishing dormant infection in mouse and guinea pig models (16, 17). Notably, those authors demonstrated that the strain replicates in the lungs and spleens of animals injected with STR, whereas it no longer grows and it enters the nonreplicating state upon STR withdrawal, thus leading to the conclusion that it mimics dormancy.
In this work, we performed an in-depth characterization of M. tuberculosis 18b growth in vitro and in vivo, with an emphasis on drug susceptibility testing, in order to develop a new model for HTS approaches.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis 18b was grown at 37°C with shaking in 7H9 broth (Difco) supplemented with Middlebrook albumin-dextrose-catalase (ADC) enrichment, 0.2% glycerol, 0.05% Tween 80 and, when necessary, 50 μg/ml STR, or on solid Middlebrook 7H10 medium (Difco) supplemented with 0.5% glycerol, Middlebrook oleic acid-albumin-dextrose-catalase (OADC), and 50 μg/ml STR.
Drugs and chemicals.
Compounds tested in the drug susceptibility assays were pharmaceutical standard (moxifloxacin [MOXI; Avelox] tablets and meropenem [MER] powder) or came from Sigma-Aldrich (isoniazid [INH], rifampin [RIF], STR, clavulanate [CLAV], ethambutol, and pyrazinamide). Experimental drugs were provided by K. Andries (TMC207) and V. Makarov (BTZ043), while PA-824 was synthesized as previously described (27).
Growth curves in the presence and absence of STR.
In order to characterize the growth of M. tuberculosis 18b after STR removal, the strain was grown to mid-log phase (optical density at 600 nm [OD600] of 0.4) in STR-containing medium. Cells were then collected by centrifugation, washed three times with phosphate-buffered saline (PBS) supplemented with 0.05% Tween 80 to remove STR, and resuspended in STR-free medium. Cultures were finally diluted to an OD600 of 0.05 or lower and incubated at 37°C with shaking. The OD was recorded at different time points to obtain growth curves. Different concentrations of STR were tested in order to evaluate the STR-dependent phenotype. To this end, a STR-free culture was prepared as described above and incubated for 10 days at 37°C to stop growth completely. STR was then added at different concentrations, and the OD600 was recorded at different time points to construct growth curves.
Time-lapse microscopy.
A 2-μl aliquot of the STR-starved M. tuberculosis 18b culture was spread on a coverslip and covered by a semipermeable membrane in a microfluidic device as described earlier (3, 21). The inlet and outlet tubes were connected to a syringe pump and a medium waste tank, respectively. Medium flow was at a constant rate of 25 μl/min. For medium exchanges, syringes were filled with either 7H9 medium or 7H9 medium containing 50 μg/ml STR. We used a DeltaVision microscope system (Applied Precision, WA) for time-lapse microscopy and observed multiple fields (50 to 60) in a single device at 60-min intervals. Phase images were acquired at each point by using a 100× phase objective. In the experiment shown below in Fig. 2 and in Movie S1 in the supplemental material, STR was introduced into the medium at 50 h.
Drug susceptibility testing.
Drug treatment was performed both in the presence of STR (actively growing cells) and in the absence of STR (nongrowing cells) at 37°C with shaking. When used for testing drugs against actively growing cells, M. tuberculosis 18b was grown to mid-log phase in STR-containing medium. The culture was then diluted to an OD of 0.05 and split into 10-ml samples, and drugs were added at the concentrations indicated below in Fig. 3, with an untreated sample as control. Serial dilutions of the cultures were plated on 7H10 supplemented with glycerol, OADC, and STR at day 7 after addition of drugs. The STR-dependent phenotype was checked by plating the same dilutions on 7H10 without STR. CFU were counted after 4 to 5 weeks of incubation at 37°C.
To test drug activity against nongrowing cells, bacteria were washed three times with PBS supplemented with 0.05% Tween 80 and resuspended in STR-free medium. The culture was then incubated at 37°C for 10 to 12 days until it stopped growing completely. Drug treatment and plating were performed as described for actively growing cells.
REMA.
A 2-week-old STR-starved culture was prepared as described for the drug susceptibility testing, diluted to an OD600 of 0.2, and used in the resazurin reduction microplate assay (REMA) as previously described (24). Briefly, 3-fold serial dilutions (starting from 10 μg/ml) of each drug were prepared in 96-well plates containing tubercle bacilli in a total volume of 100 μl and then incubated for 7 days at 37°C before addition of 10 μl of 0.025% resazurin. After overnight incubation, fluorescence of the resazurin metabolite resorufin was determined (excitation at 560 and emission at 590 nm, measured by using a TECAN Infinite M200 microplate reader).
In vivo efficacy of drugs.
BALB/c mice (four per group) were infected intravenously, in the lateral tail vein, with 1.2 × 108 bacteria, and STR was given subcutaneously at a dose of 100 mg/kg of body weight for 3 weeks. Drug treatment was started 4 weeks postinfection with the following compounds administered by gavage once daily, six times/week, for 8 weeks: INH at 25 mg/kg, RIF at 10 mg/kg, PA-824 at 100 mg/kg, BTZ043 at 50 mg/kg, MOXI at 100 mg/kg. INH and MOXI were dissolved in water and RIF was suspended in water after grinding in a mortar, whereas PA-824 and BTZ043 were formulated as previously described (21, 29). Control and treated mice were sacrificed, lungs were homogenized, and dilutions were plated on 7H10 plates with STR for enumeration of viable bacilli.
High-throughput screening assays.
To determine if the REMA was suitable for screening large compound libraries, the Z-factor (33) was initially determined for STR-starved 18b cells (OD600, 0.1), 18b cells growing in the presence of STR (OD600, 0.003), and H37Rv cells (OD600, 0.001). The Z-factor is a measure of statistical effect size and is used in HTS experiments, prior to setting up large screens, to judge the quality of the assay by comparing the responses to positive and negative controls. A Z-factor of 1 represents the ideal assay, whereas a Z-factor of 0 means that positive and negative controls give equal signals and the assay is not reliable (33). For this purpose, the strains mentioned above were incubated in 96-well plates, with or without 10 μg/ml RIF (positive and negative controls, respectively). After 1 week of incubation, resazurin was added and fluorescence was determined as described above. Mean values and standard deviations were calculated for both conditions and used to determine the Z-factor according to the previously published equation (33).
A commercially available compound library (NINDS Custom Collection II, containing 1,040 drugs; MicroSource, Gaylordsville, CT) was then used to further evaluate the use of the REMA in compound screening. Briefly, the NINDS library was diluted to 500 μM in dimethyl sulfoxide (DMSO) and 2 μl of each compound was added to 96-well plates in duplicate using the Zephyr dispenser (Caliper). One hundred microliters of either 2-week-old STR-starved 18b cells (OD600, 0.1), 18b cells growing in the presence of STR (OD600, 0.003), or H37Rv cells (OD600, 0.001) was added (MicroFlo Select dispenser; Witec AG, Luzern, Switzerland). As controls, each plate contained one column (eight wells) of DMSO only and one column of 10 μg/ml RIF. Bacterial viability was determined as described above, and results were analyzed as follows. The eight DMSO negative controls and eight RIF positive controls served as references. Mean fluorescence values and standard deviations were calculated for the references, and compounds were considered to have a significant activity if their fluorescence fell outside the mean plus 3 standard deviations of the negative controls. If both replicates for a compound were statistically significant, a score was calculated using the following equation: (compound value − mean negative control)/(mean positive control − mean negative control).
Statistical analysis.
Statistical analysis was performed with unpaired t tests in Prism version 5.0 (GraphPad, San Diego, CA).
RESULTS
Growth of M. tuberculosis 18b in vitro.
Initial characterization of M. tuberculosis 18b involved studying growth and survival in axenic cultures. For this purpose, a STR-starved culture was prepared as described in Materials and Methods, and different concentrations of STR were added. As reported in Fig. 1A, no growth was detected without STR, whereas addition of as little as 5 μg/ml STR allowed bacterial replication, with a calculated generation time of 38 h. In addition, growth rate directly correlated with the amount of STR in the culture, with 50 μg/ml being the most appropriate concentration for achieving optimal growth (generation time of 28 h). The generation time for M. tuberculosis 18b is therefore appreciably longer than that of H37Rv, which was 18.7 h (data not shown). As higher STR concentrations did not affect bacterial growth (data not shown), we used 50 μg/ml as the working concentration in the subsequent experiments.
FIG. 1.
Growth curves of M. tuberculosis 18b upon addition of different concentrations of STR and after STR removal. (A) Different concentrations of STR were added to a STR-starved culture, as detailed in the legend to the graph, and the OD was recorded at different times to obtain growth curves. (B) Two independent M. tuberculosis 18b cultures (indicated by squares and diamonds in the graph) were washed three times to remove STR, diluted to an OD600 of 0.03 or 0.06, and incubated at 37°C with shaking.
Additional insight into the STR-dependent phenotype came from the analysis of the growth curve after removal of STR. As shown in Fig. 1B, strain 18b requires at least 10 days before entering the nonreplicating phase, thus suggesting that residual STR inside cells has to be diluted and/or metabolized in the course of several generations before growth stops. When growth arrest has occurred, the optical density and cell viability (number of CFU) remain stable for over 1 month after STR withdrawal (Table 1). Notably, readdition of STR to starved cells led to an increase in optical density and CFU, proving that strain 18b can grow again when the antibiotic is added to the culture (Table 1). These results confirm the peculiar STR-dependent phenotype of M. tuberculosis 18b and represent the basis for the following work.
TABLE 1.
Correlation between OD600 and CFU after growth arrest and upon addition of streptomycina
| Time after STR removal | OD600 | CFU/ml |
|---|---|---|
| Day 0 (STR removal) | 0.50 | 1.60E+8 |
| 1 wk | 0.45 | 8.50E+7 |
| 2 wks | 0.40 | 5.00E+7 |
| 3 wks | 0.50 | 6.00E+7 |
| 4 wks | 0.44 | 2.30E+7 |
| 5 days after addition of STR | 0.72 | 5.00E+8 |
M. tuberculosis 18b was grown in 7H9 with STR to an OD600 of 0.5 and washed to remove STR. Streptomycin-starved cells were then incubated in 7H9 with 0.2% glycerol and 0.05% Tween 80. The OD600 and CFU were determined at the time points indicated. Four weeks after removal, STR was added back to the culture and the increases in the OD600 and CFU were recorded.
Growth of M. tuberculosis 18b in a microfluidics device.
The data described above, although suggestive that the strain enters a nongrowing state on withdrawal of STR, do not reveal the dynamics of growth during this state. It is conceivable that during the STR starvation period there is continuous elongation and division going on, which is balanced by cell death resulting in no net growth. To further delineate the behavior of strain 18b during this phase, we directly observed STR-starved bacteria installed in a microfluidics device by using automated time-lapse microscopy. In these experiments, where we monitored thousands of cells, no bacteria were observed to undergo elongation or division during the period of STR starvation (Fig. 2; see also Movie S1 in the supplemental material). Two days later, we reexposed the cells to STR to study the recovery of the bacteria. There was a significant lag observed (60 ± 28 h) before the bacteria started elongating and dividing. The mass doubling time of the bacteria in the device was determined to be 21.3 ± 6.1 h. We also observed a significant proportion of cells (90%) that did not resume growth even after exposure to STR for 9 days, suggesting that these cells were probably damaged or dead.
FIG. 2.
M. tuberculosis 18b cells growing in a microfluidics device. Growth was monitored at 60-min intervals by video microscopy (see Movie S1 in the supplemental material). Representative frames shown were obtained at 25-h intervals. STR (50 μg/ml) was added at 50 h and maintained in the culture until the end of the experiment. Bars, 5 μm.
In vitro susceptibility of M. tuberculosis 18b to antituberculosis compounds.
The activities of several front-line and experimental TB drugs were tested against M. tuberculosis 18b. In a first, control experiment, the evaluation was performed on actively growing cells, as described in Materials and Methods. Figure 3A shows the results as well as the drug concentrations, which were either those commonly used or 10 times the MIC against the reference strain H37Rv (1, 2, 4, 14, 21, 28). In the absence of drugs, cells grew with a >1-log increase in the number of CFU over 7 days of incubation, consistent with the 28-h generation time observed. INH treatment caused a 2.5-log decrease in CFU (P = 0.006), thus showing the same killing effect as the newly developed compound BTZ043 (P = 0.009) (21). RIF and MOXI were also active, causing >3-log decreases in CFU (P = 0.0035 for both of them), whereas TMC207 (1) and PA-824 (28) performed less well, with a 2-log decrease in viable counts and P values of 0.0064 and 0.0066, respectively. Similar results were obtained with the combination of a beta-lactam antibiotic (meropenem) and a beta-lactamase inhibitor (clavulanate) (P = 0.0067). Most importantly, no colonies were detected on STR-free plates, thus indicating that revertants did not arise and that the STR-dependent phenotype is extremely stable (the frequency of reversion is <10−8). These data indicate that all drugs are active against exponentially growing M. tuberculosis 18b.
FIG. 3.
Test of drug activity against actively growing and nongrowing M. tuberculosis 18b cells. (A) A STR-containing M. tuberculosis 18b culture was split into several samples and incubated with the different drugs at the concentrations reported. Serial dilutions were made and plated 7 days after addition of drug. The no-drug data represent the untreated control. Results are displayed as means and standard deviations of two independent experiments. (B) A STR-starved M. tuberculosis 18b culture was split into several samples and processed as described for the experiment shown in panel A. MOXI and MER/CLAV were tested in independent experiments with appropriate controls (no-drug, INH, and RIF controls, with results similar to those for the controls included in this graph).
In the following step, the same drugs were tested for their activities on STR-starved cells, and the results are reported in Fig. 3B. Contrary to what was observed when STR was present, the untreated control displayed no growth, thereby confirming that cells were arrested. Addition of INH did not cause a significant reduction in the number of CFU (P = 0.1387). In contrast, BTZ043 had a modest effect on nonreplicating bacteria (<1 log after 7 days of treatment; P = 0.0048). TMC207 performed slightly better than BTZ043, in agreement with previous reports (19, 25), and MOXI also displayed some killing activity, with a 3-log decrease (P = 0.0079 and 0.004, respectively). Interestingly, the combination of meropenem and clavulanate reduced the number of CFU (P = 0.0117). The most striking results were obtained with RIF and PA-824: both of them were highly active, causing >4-log reductions in viable counts (P = 0.0006 and 0.0003, respectively), with PA-824 appearing to be more effective under these conditions than against actively growing bacteria.
In vivo application of M. tuberculosis 18b.
In order to check whether M. tuberculosis 18b mimicked nonreplicating cells in vivo, we established a chronic infection in BALB/c mice receiving STR subcutaneously for 3 weeks to allow bacterial replication. During this period, we observed a 1-log increase in the number of CFU in the lungs (Fig. 4). Ten days after STR withdrawal, treatment was started using INH, RIF, PA-824, BTZ043, or MOXI alone and continued for 8 weeks. Treatment outcome was assessed by counting CFU in the lungs. As shown in Fig. 4, the number of bacteria in the untreated animals remained stable after an initial drop of ∼1 log when STR was no longer administered. INH treatment had a negligible bactericidal effect (P = 0.44), as expected against nongrowing cells, while BTZ043 caused a 0.5-log reduction in CFU (P = 0.036), consistent with its marginal benefit against nongrowing cells in vitro (Fig. 3B). In contrast, RIF (P < 0.0001) and MOXI (P < 0.0001) both displayed good killing activity, with 3.6- and 3.2-log reductions in bacterial load, respectively, at the end of the experiment. Importantly, RIF was effective from the beginning of the treatment, thereby confirming its potency against nongrowing cells, whereas MOXI appeared more efficacious as the treatment progressed. PA-824 exhibited reasonable killing efficiency, resulting in a 1.5-log decrease in the bacterial load after 8 weeks of treatment (P = 0.0002).
FIG. 4.
Evaluation of M. tuberculosis 18b in the mouse model of infection. M. tuberculosis 18b-infected mice were treated for 8 weeks with the indicated drugs, starting from week 4 postinfection. Control and treated animals were sacrificed at 2, 4, and 8 weeks after beginning treatment, and CFU were determined by plating lung homogenates on 7H10 plates with STR.
Can M. tuberculosis 18b be used for high-throughput screening assays?
The data reported above underline the potential of M. tuberculosis 18b for HTS assays. For this purpose, the REMA was tested in the 96-well format.
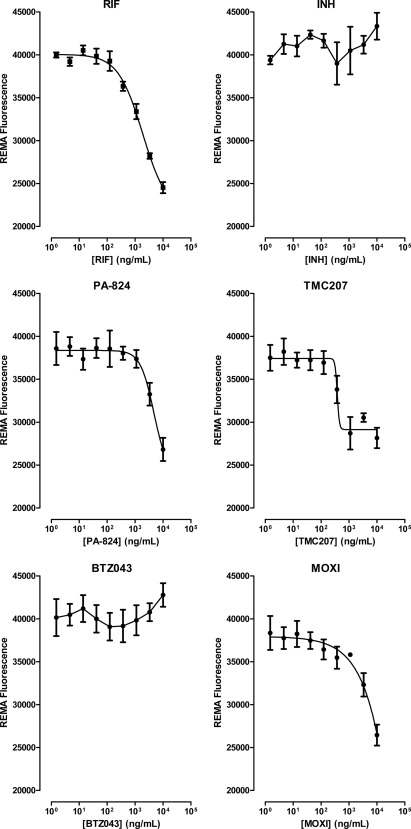
In the REMA, metabolic activity of the cell correlates with the reduction of resazurin to resorufin, and the resulting change in color from blue to pink can be easily detected and quantified by measuring fluorescence. STR-starved 18b cells were therefore incubated in 96-well plates for 7 days with the drugs mentioned above, and resazurin was added thereafter. Neither INH nor BTZ043 affected resazurin reduction, whereas a marked concentration-dependent decrease in fluorescence was seen with RIF (activity starting at >370 ng/ml), MOXI (>1 μg/ml), PA-824 (>1 μg/ml), and TMC207 (>370 ng/ml) (Fig. 5). Importantly, these data agree with the CFU results reported above. Two additional drugs were included in the REMA, ethambutol (EMB) and pyrazinamide (PZA). While EMB did not cause a decrease in fluorescence, PZA had some effect at the highest concentration tested (100 μg/ml) (see Fig. S1 in the supplemental material).
FIG. 5.
Resazurin microplate assay with nongrowing M. tuberculosis 18b. Serial 3-fold dilutions of each drug were prepared in a 96-well plate, and their activities against a STR-starved M. tuberculosis 18b culture were evaluated in a REMA. Results are expressed as means and standard deviations of two independent experiments and show the correlations of drug concentration to fluorescence.
These results, which are consistent with those from viability assays, indicate that M. tuberculosis 18b can be used in HTS assays to measure metabolic activity, such as resazurin reduction.
Application of M. tuberculosis 18b to HTS.
A pilot HTS study was performed using REMA to determine the Z-factor (33), which is reflective of the signal dynamic range and of the statistical data variation, by incubating M. tuberculosis H37Rv and 18b, with and without STR, in the presence or absence of RIF (see Materials and Methods). Satisfactory Z-factors were obtained (33): 0.75 for STR-starved 18b, 0.85 for 18b growing in the presence of STR, and 0.90 for H37Rv, thus indicating that the assay was robust enough for HTS application.
Screening of the 1,040 compounds in the NINDS library revealed a total of 41 active compounds (Fig. 6). Overall there was good concordance between the compounds that inhibited H37Rv and growing 18b cells, while far fewer compounds were found to be active against nongrowing 18b cells. As expected, RIF, rifaximin, another rifamycin, and MOXI were found to be effective under all three conditions. Other fluoroquinolones (gatifloxacin, levofloxacin, cloxiquin, ofloxacin, norfloxacin, lomefloxacin, and pefloxacin) inhibited growing H37Rv and 18b cells but displayed much weaker or no activity against nongrowing 18b cells. In addition to general antibacterial and antiseptic compounds, gramicidin was also shown to be active under all of the conditions tested, suggesting that maintenance of ion gradients across the membrane is essential for nonreplicating cells as well. This observation is indeed supported by the fact that two other ionophores, salinomycin and nigericin, were only active against nongrowing 18b. Two drugs that target thymidylate synthase (sodium p-aminosalicylate and fluorouracil) were effective against replicating bacteria only, and the same result was noticed for some compounds targeting the ribosome (fusidic acid and tetracycline derivatives). Finally, INH and EMB were found to effectively inhibit H37Rv growth, whereas they displayed poor activity against 18b plus STR, probably because of the reduced growth rate, and had no effect when tested on nongrowing cells. These results substantiate the data presented above and demonstrate the feasibility of HTS applications using STR-starved strain 18b cells.
FIG. 6.
Drug activity scores from the pilot HTS study based on the REMA and the NINDS Custom Collection II library. The reported scores were calculated as described in Materials and Methods and represent the mean fluorescence values obtained for each compound after subtraction of the background fluorescence and normalization to the fluorescence observed with the positive control, RIF. A score of 1 indicates that the compound is as effective as RIF, while a score of 0 indicates a compound with no effect. The color coding is as follows: white, score below 0.5; yellow, score between 0.5 and 0.69; orange, score between 0.7 and 0.89; red, score of ≥0.9.
DISCUSSION
Inspired by the pioneering studies performed with M. tuberculosis 18b by Kondo and Kanai (18), we decided to undertake a more extensive analysis of the strain in vitro and in vivo to evaluate its potential in TB drug discovery programs. We confirmed the well-described STR-dependent phenotype (12, 17) and demonstrated that removal of STR from the in vitro culture did not cause immediate growth arrest: cells replicated for at least 1 week before growth stopped. We propose that, during this period, residual antibiotic is closely associated with the ribosome where, according to the model of Honoré et al. (13), it contributes to stabilization of the translation apparatus. The turnover of the ribosome then allows dilution of STR and growth arrest. This was taken into account when we planned experiments of drug sensitivity on nongrowing cells, as these cells were STR starved for 2 weeks. Under these conditions, we observed an ∼1-log decrease in CFU 1 month after STR removal. Single-cell experiments using microfluidics corroborated these findings, demonstrating that the STR starvation phase is in fact characterized by the absence of elongation and growth. However, in this assay too, we observed a significant proportion of cells that did not resume growth after readdition of STR (90%, which corresponds to the 1-log decrease observed in CFU counts). This population may represent bacteria that take longer to respond to STR, remain viable but not culturable, or are in some way damaged. However, propidium iodide staining at the end of the experiment ruled out the possibility that these bacteria had undergone lysis. The use of untreated controls in drug efficacy studies overcomes this problem.
The usefulness of M. tuberculosis 18b as a dormancy model for drug susceptibility testing was assessed by using either currently approved or experimental TB drugs whose efficacies against growing cells were first demonstrated. The in vitro potencies of these compounds were found to be different when tested against STR-starved, nongrowing cells. In the latter case, INH and BTZ043 displayed little or no activity, whereas RIF and PA-824 almost sterilized the samples. These data are in agreement with findings obtained with other dormancy models (4, 27, 30, 32). Additional insight into 18b sensitivity to antituberculosis compounds came from studies with TMC207, MOXI, and meropenem/clavulanate. All of the drugs showed some killing activity and, most importantly, our results confirm and extend those obtained previously with nongrowing cells (14, 15, 19, 22, 25).
Studies performed in vivo revealed that the STR-dependent phenotype is maintained in chronically infected mice (Fig. 4), with cells replicating when the antibiotic is administered and persisting in a nonmultiplying state upon its withdrawal (17). The in vivo efficacy of compounds reflects the cidality seen in vitro with nonmultiplying tubercle bacilli (Fig. 3B), thus supporting the notion of M. tuberculosis 18b mimicking latent TB infection and validating 18b as an appealing model for in vivo studies as well. Different animal models for this condition exist. For instance, the Cornell model relies on INH and PZA treatment to induce a state of nonreplicating persistence (23), but the nonreplicating condition obtained may differ from that occurring in latent human TB. A second approach is to use a low-dose aerosol infection route in Mycobacterium bovis BCG-immunized mice (20), but this requires up to 12 weeks to establish (34). Compared to these animal models, an approach using persisting M. tuberculosis 18b cells has the advantages of being both faster and simpler, as it requires no drug treatment.
The availability of a simple in vitro dormancy model would be an asset for TB drug discovery programs and would complement those that exist already, such as the Wayne or starvation models (4, 30). In proof-of-principle experiments with known drugs, we showed that strain 18b was compatible with the REMA and could be used in a 96-well format successfully. In our screening of the NINDS Custom Collection II library, the hit rate with STR-starved 18b was lower than that with actively growing H37Rv or 18b, and the nongrowing cells were not susceptible to drugs active against replicating bacteria (e.g., INH, fluorouracil, and sodium p-aminosalicylate [Fig. 6]). In contrast to actively growing cells, they were vulnerable to ionophores, underlining the importance of maintaining ion homeostasis across the membrane (25).
There are several advantages to using M. tuberculosis 18b rather than other models for dormancy. First, manipulation is simple and needs no special equipment, making it suitable for most laboratories. In addition, there is no requirement for hypoxia, thus simplifying the logistics for HTS. Finally, excellent agreement was seen between the in vitro and in vivo drug efficacy data sets.
In conclusion, the study presented here provides a comprehensive description of M. tuberculosis 18b in vitro, validates the hypothesis of using this strain as a model for nongrowing cells, underpins its exploitation in HTS assays, and suggests potential in vivo applications to test drugs that target latent tuberculosis.
Supplementary Material
Acknowledgments
We thank K. Andries for providing TMC207, V. Makarov for providing BTZ043, J. McKinney, H. Boshoff, and A. Campos-Neto for advice, and the Biomolecular Screening Facility at EPFL for performance of the HTS assays.
R. C. Hartkoorn was the recipient of a postdoctoral fellowship from the Heiser Program for Research in Leprosy and Tuberculosis of the New York Community Trust. This work was funded in part by the Swiss National Science Foundation (31003A-125061) and the European Commission (LHSP-CT-2005-018923, HEALTH-F3-2007-201762).
Footnotes
Published ahead of print on 2 August 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2008. Handbook of anti-tuberculosis agents. Introduction. Tuberculosis (Edinb.) 88:85-86. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff, H. I., and C. E. Barry III. 2005. Tuberculosis: metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70-80. [DOI] [PubMed] [Google Scholar]
- 6.Chan, J., and J. Flynn. 2004. The immunological aspects of latency in tuberculosis. Clin. Immunol. 110:2-12. [DOI] [PubMed] [Google Scholar]
- 7.Deb, C., C. M. Lee, V. S. Dubey, J. Daniel, B. Abomoelak, T. D. Sirakova, S. Pawar, L. Rogers, and P. E. Kolattukudy. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dye, C. 2006. Global epidemiology of tuberculosis. Lancet 367:938-940. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi, N. R., A. Moll, A. W. Sturm, R. Pawinski, T. Govender, U. Lalloo, K. Zeller, J. Andrews, and G. Friedland. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575-1580. [DOI] [PubMed] [Google Scholar]
- 11.Gomez, J. E., and J. D. McKinney. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 84:29-44. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, T. 1955. Experimental studies on the mechanism of infection and immunity in tuberculosis from the analytical standpoint of streptomycin-dependent tubercle bacilli. 1. Isolation and biological characteristics of a streptomycin-dependent mutant, and effect of streptomycin administration on its pathogenicity in guinea pigs. Kekkaku 30:4-8; 45-46. (In Japanese with English summary.) [PubMed] [Google Scholar]
- 13.Honoré, N., G. Marchal, and S. T. Cole. 1995. Novel mutation in 16S rRNA associated with streptomycin dependence in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:769-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugonnet, J. E., L. W. Tremblay, H. I. Boshoff, C. E. Barry III, and J. S. Blanchard. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain, S., M. Malik, L. Shi, M. L. Gennaro, and K. Drlica. 2009. In vitro model of mycobacterial growth arrest using nitric oxide with limited air. Antimicrob. Agents Chemother. 53:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashino, S. S., D. R. Napolitano, Z. Skobe, and A. Campos-Neto. 2008. Guinea pig model of Mycobacterium tuberculosis latent/dormant infection. Microbes Infect. 10:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashino, S. S., P. Ovendale, A. Izzo, and A. Campos-Neto. 2006. Unique model of dormant infection for tuberculosis vaccine development. Clin. Vaccine Immunol. 13:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo, E., and K. Kanai. 1988. An experimental model of chemotherapy on dormant tuberculous infection, with particular reference to rifampicin. Jpn. J. Med. Sci. Biol. 41:37-47. [DOI] [PubMed] [Google Scholar]
- 19.Koul, A., L. Vranckx, N. Dendouga, W. Balemans, I. Van den Wyngaert, K. Vergauwen, H. W. Gohlmann, R. Willebrords, A. Poncelet, J. Guillemont, D. Bald, and K. Andries. 2008. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 283:25273-25280. [DOI] [PubMed] [Google Scholar]
- 20.Lazarevic, V., D. Nolt, and J. L. Flynn. 2005. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J. Immunol. 175:1107-1117. [DOI] [PubMed] [Google Scholar]
- 21.Makarov, V., G. Manina, K. Mikusova, U. Mollmann, O. Ryabova, B. Saint-Joanis, N. Dhar, M. R. Pasca, S. Buroni, A. P. Lucarelli, A. Milano, E. De Rossi, M. Belanova, A. Bobovska, P. Dianiskova, J. Kordulakova, C. Sala, E. Fullam, P. Schneider, J. D. McKinney, P. Brodin, T. Christophe, S. Waddell, P. Butcher, J. Albrethsen, I. Rosenkrands, R. Brosch, V. Nandi, S. Bharath, S. Gaonkar, R. K. Shandil, V. Balasubramanian, T. Balganesh, S. Tyagi, J. Grosset, G. Riccardi, and S. T. Cole. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324:801-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik, M., and K. Drlica. 2006. Moxifloxacin lethality against Mycobacterium tuberculosis in the presence and absence of chloramphenicol. Antimicrob. Agents Chemother. 50:2842-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCune, R. M., Jr., W. McDermott, and R. Tompsett. 1956. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J. Exp. Med. 104:763-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palomino, J. C., A. Martin, M. Camacho, H. Guerra, J. Swings, and F. Portaels. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2720-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, S. P., S. Alonso, L. Rand, T. Dick, and K. Pethe. 2008. The proton motive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105:11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders, B. M., and W. J. Britton. 2007. Life and death in the granuloma: immunopathology of tuberculosis. Immunol. Cell Biol. 85:103-111. [DOI] [PubMed] [Google Scholar]
- 27.Singh, R., U. Manjunatha, H. I. Boshoff, Y. H. Ha, P. Niyomrattanakit, R. Ledwidge, C. S. Dowd, I. Y. Lee, P. Kim, L. Zhang, S. Kang, T. H. Keller, J. Jiricek, and C. E. Barry III. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover, C. K., P. Warrener, D. R. VanDevanter, D. R. Sherman, T. M. Arain, M. H. Langhorne, S. W. Anderson, J. A. Towell, Y. Yuan, D. N. McMurray, B. N. Kreiswirth, C. E. Barry, and W. R. Baker. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962-966. [DOI] [PubMed] [Google Scholar]
- 29.Tyagi, S., E. Nuermberger, T. Yoshimatsu, K. Williams, I. Rosenthal, N. Lounis, W. Bishai, and J. Grosset. 2005. Bactericidal activity of the nitroimidazopyran PA-824 in a murine model of tuberculosis. Antimicrob. Agents Chemother. 49:2289-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 32.Xie, Z., N. Siddiqi, and E. J. Rubin. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 49:4778-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, T., M. Zhang, I. M. Rosenthal, J. H. Grosset, and E. L. Nuermberger. 2009. Short-course therapy with daily rifapentine in a murine model of latent tuberculosis infection. Am. J. Respir. Crit. Care Med. 180:1151-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y. 2004. Persistent and dormant tubercle bacilli and latent tuberculosis. Front. Biosci. 9:1136-1156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.