Abstract
We report the dissemination of a conjugative IncI1 plasmid carrying blaCTX-M-1, conferring resistance to extended-spectrum cephalosporins, in Salmonella enterica isolates from poultry and humans in France from 2003 to 2008. By IncI1 plasmid subtyping, this plasmid was shown to be genetically related to that found in Escherichia coli isolates from healthy poultry in France.
Food-producing animals are the primary reservoir of zoonotic pathogens, and the prevalence of resistance to extended-spectrum cephalosporins (ESCs) in Escherichia coli and Salmonella enterica has increased in recent years. In Belgium and France, the emergence of resistance to ESCs, due to extended-spectrum β-lactamases (ESBLs), in E. coli and S. enterica from animal (mainly cattle and poultry) and human origins (1, 3, 6, 7, 10, 11, 14, 15, 17) has been reported. Resistance in these bacteria was reported to be conferred mainly by the ESBL genes blaCTX-M-2, blaCTX-M-9, blaCTX-M-15, and blaTEM-52 (1, 3, 6, 11, 14, 15, 17). In addition, the occurrence of CTX-M-1 ESBL resistance in E. coli isolates recovered from food animals (cattle, poultry, and swine) in France was recently reported (7, 10, 11). The ESBL resistance genes were shown to reside on large conjugative plasmids of the IncI1 or IncHI2 incompatibility group (3, 5, 6).
Since 2003, a number of S. enterica strains showing resistance to ESCs by production of an ESBL not previously detected in Salmonella in France and with various additional resistances to other antibiotic families have been isolated from poultry (n = 1) and from humans (n = 9) in France (Table 1). The human isolates consisted of seven serovar Typhimurium (including a monophasic variant) strains, one serovar London strain, and one serovar Newport strain, and the avian isolate was of serovar Llandoff. The purpose of the present study was to identify the ESBL gene and to characterize the plasmid(s) carrying this gene. For this, in addition to other methods, we applied the recently described plasmid multilocus sequence typing method for IncI1 plasmids carrying ESBL genes (5).
TABLE 1.
Characteristics of the Salmonella strains and their transconjugants producing CTX-M-1 used in this studya
| Strainb | Serovar | PFGE type | Origin | Yr | MIC (μg/ml) of: |
Coresistance markersc | IncI1 pMLSTd | SGI1 | |
|---|---|---|---|---|---|---|---|---|---|
| Cro | Caz | ||||||||
| 03-8748 | Newport | NDc | Human | 2003 | >256 | 8 | Su, Tm | − | |
| 03-8748TC1 | 256 | 2 | Su, Tm | 3 | |||||
| 05-9280 | Typhimurium | XTYM-1 | Human | 2005 | >256 | 32 | Cm, Spt, Str, Su, Tc, Tm | + | |
| 05-9280TC1 | >256 | 32 | Su, Tm | 3 | |||||
| 06CEB6542SAL | Llandoff | ND | Poultry | 2006 | 64 | 4 | Su, Tc | − | |
| 06CEB6542SALTC1 | 8 | 0.5 | Su, Tc | 3 | |||||
| 06-6550 | Typhimurium | XTYM-115 | Human | 2006 | 256 | 16 | Su, Tc, Tm | − | |
| 06-6550TC1 | 64 | 4 | Su, Tm | 3 | |||||
| 07-819 | London | ND | Human | 2007 | >256 | 8 | Su, Tm | − | |
| 07-819TC1 | 256 | 4 | Su, Tm | 3 | |||||
| 08-843 | Typhimurium | XTYM-1 | Human | 2008 | 256 | 16 | Cm, Spt, Str, Su, Tc, Tm | + | |
| 08-843TC1 | 256 | 8 | Su, Tm | 3 | |||||
| 08-1537 | Typhimurium | XTYM-117 | Human | 2008 | >256 | 64 | Su, Tm | − | |
| 08-1537TC1 | 128 | 8 | Su, Tm | 3 | |||||
| 08-1745 | Typhimurium | XTYM-117 | Human | 2008 | >256 | 16 | Su, Tm | − | |
| 08-2211 | Typhimurium | XTYM-117 | Human | 2008 | 256 | 8 | Su, Tm | − | |
| 08-2712 | 4,5,12:i:− | XTYM-30 | Human | 2008 | 256 | 16 | Su, Tc, Tm | − | |
| 08-2712TC1 | 64 | 8 | Su, Tm | 3 | |||||
Abbreviations: Caz, ceftazidime; Cm, chloramphenicol; Cro, ceftriaxone; Spt, spectinomycine; Str, streptomycin; Su, sulfonamide; Tc, tetracycline; Tm, trimethoprim; ND, not determined.
Strains ending in “TC1” are E. coli transconjugant strains.
Antibiotics other than β-lactams.
IncI1 plasmid multilocus sequence type (pMLST), according to Garcia-Fernandez et al. (5), of the CTX-M-1 plasmids extracted from transconjugants.
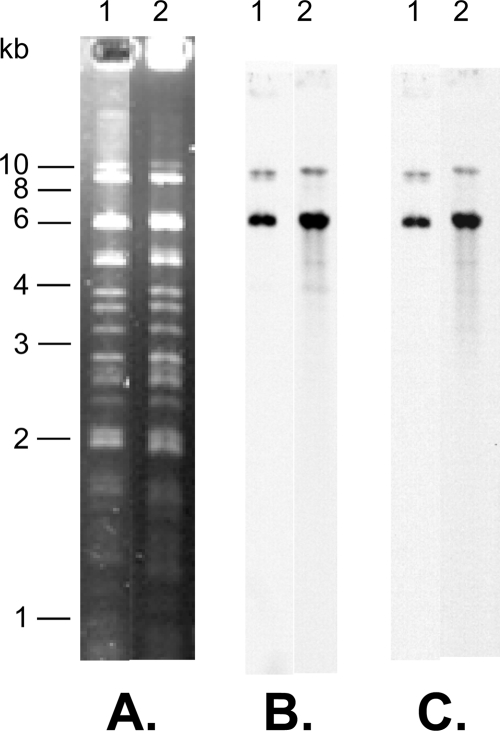
The strains studied are shown in Table 1. Antibiotic susceptibility testing was done by the disc diffusion method, and MICs of ceftriaxone and ceftazidime were determined by Etest as described previously (1, 15, 17). Resistance to ESCs was transferred from all Salmonella strains to E. coli recipient strain J5 (resistant to rifampin) by conjugation as previously described (1, 15, 17). Depending on the strain, other resistances were transferred, mostly sulfonamide together with trimethoprim resistance (Table 1). The other resistances from multidrug-resistant strains were not transferred by conjugation. The levels of resistance to ESCs, as determined by the MIC, were lower in the transconjugant strains than in the parental strains. However, this was also observed in previous studies (3, 15). PCR assays to detect ESBL genes (TEM, SHV, and CTX-M) were performed on parental and transconjugant strains using previously described primers (1, 15, 17), and nucleotide sequencing of the amplicons identified the blaCTX-M-1 resistance gene in all strains (Table 1). Plasmids extracted from the transconjugants were further characterized by PstI restriction analysis, which showed that they were similar, with a size of 100 kb as determined by S1 nuclease experiments (Fig. 1 and data not shown). Interestingly, the plasmid restriction profile was also similar to that from blaCTX-M-1-carrying plasmids from E. coli isolated from healthy poultry in France (Fig. 1 and data not shown) (7) but distinct from blaCTX-M-1-carrying plasmids from E. coli isolates from other animal sources in France (data not shown) (11). This suggested a possible common avian origin for this blaCTX-M-1-carrying plasmid. Southern blot hybridization experiments using a blaCTX-M-1 gene probe revealed one PstI fragment of 6 kb in all plasmids, which is the same size as that found in the E. coli plasmid of avian origin (Fig. 1). The blaCTX-M-1 gene has been previously reported to be linked to the ISEcp1 insertion sequence (7, 11). PCR performed as described previously (11), using a forward primer in the ISEcp1 gene and a reverse primer in the blaCTX-M-1 gene, was positive for all transconjugant plasmids (data not shown). The link with ISEcp1 was further confirmed by Southern blotting with an ISEcp1 probe, which revealed a fragment identical in size to that revealed by the blaCTX-M-1 probe on the PstI-restricted plasmids (Fig. 1).
FIG. 1.
Restriction analysis (PstI) (A) and Southern blot hybridization with a blaCTX-M-1 probe (B) or with an ISEcp1 probe (C) of plasmid DNAs isolated from E. coli transconjugants with E. coli isolates from poultry origin in France (lanes 1) and the Salmonella isolates in this study (lanes 2) as parental strains.
To better clarify the genetic relatedness of the plasmids, the blaCTX-M-1-positive plasmids were typed by the PCR-based replicon typing as previously described (2), demonstrating that they all belong to the IncI1 incompatibility group. IncI1 plasmids were recently observed in E. coli and different serovars of Salmonella isolated in Belgium, France, Germany, Spain, and the United Kingdom and were found to be associated with relevant β-lactamases such as CMY-2, CMY-7, CTX-M-1, CTX-M-15, and TEM-52, suggesting a high prevalence of this kind of plasmid in Europe (3, 5, 8, 9, 13). In addition, further characterization using the recently described pMLST method for the characterization of IncI1 plasmids showed that all Salmonella blaCTX-M-1-carrying plasmids in this study were of sequence type 3, as reported for E. coli isolates from poultry in France (data not shown) (5). This was also confirmed for the E. coli blaCTX-M-1-carrying control plasmids of poultry origin in this study. Thus, like restriction typing, replicon typing and pMLST indicated a possible common avian origin for the blaCTX-M-1-carrying plasmid emerging in E. coli and Salmonella in France.
Among the ESC-resistant Salmonella strains studied, two serovar Typhimurium isolates showed an additional multidrug resistance profile with resistances to chloramphenicol, streptomycin, spectinomycin, sulfonamide, tetracycline, and trimethoprim (Table 1). This multidrug resistance profile is characteristic of the Salmonella genomic island 1 (SGI1) antibiotic resistance gene cluster commonly found in the serovar Typhimurium DT104 epidemic clone (12). Identification of SGI1 and mapping of its antibiotic resistance gene cluster performed as described previously (4) confirmed that the two isolates possessed SGI1, with its classical antibiotic resistance gene cluster consisting of a complex class 1 integron (12). This combination of SGI1-mediated multidrug resistance in Salmonella strains associated with ESC resistance has rarely been reported, and further surveillance of such strains is thus warranted (3).
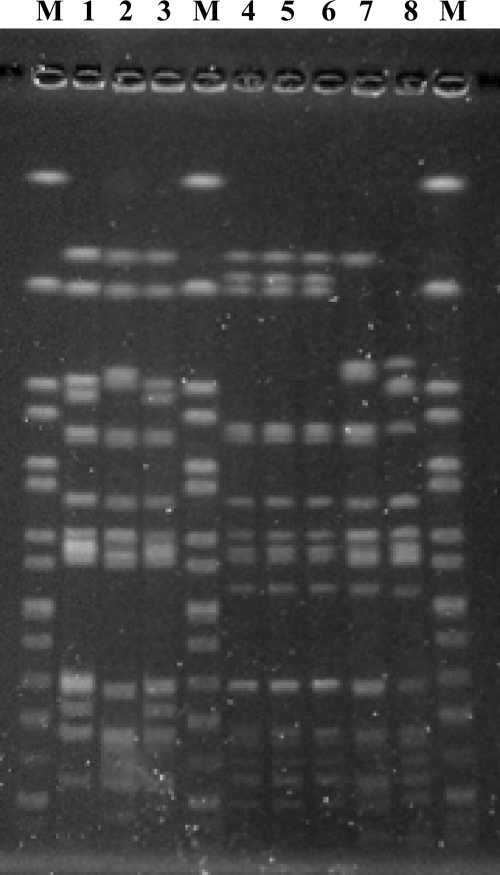
To assess the genetic diversity of the serovar Typhimurium strains, PulseNet standard pulsed-field gel electrophoresis (PFGE) of XbaI-digested chromosomal DNA was carried out (16). These strains showed four different PFGE profiles (Fig. 2 and Table 1). The XTYM-1 profile exhibited by both SGI1-carrying isolates is the most prevalent profile for DT104 strains in France (16).
FIG. 2.
XbaI-PFGE profiles of the S. enterica serovar Typhimurium (and monophasic variant) strains studied. M, XbaI-digested DNA from S. enterica serovar Braenderup strain H9812; lane 1, strain 05-9280; lane 2, strain 06-6550; lane 3, strain 08-843; lane 4, strain 08-1537; lane 5, strain 08-1745; lane 6, strain 08-2211; lane 7, strain 08-2712; lane 8, strain 09-1581 (unrelated to the study).
In conclusion, this study showed the spread of an IncI1 plasmid carrying the blaCTX-M-1 gene among S. enterica serovars Llandoff, London, Newport, and Typhimurium of animal and human origin. According to our plasmid analyses, this plasmid is likely the same as that found in E. coli from poultry in France. We thus suspect horizontal transfer events that can contribute to its spread between bacterial populations from animals and humans as well (14). The further spread of such plasmids in multidrug-resistant strains carrying SGI1 is of concern.
Acknowledgments
We thank Alessandra Carattoli for her kind assistance and suggestions in the characterization of the plasmids of this study. We are also grateful to Laurent Poirel, Jean-Yves Madec, and Danièle Meunier for providing the E. coli control plasmids of this study.
Footnotes
Published ahead of print on 19 July 2010.
REFERENCES
- 1.Bertrand, S., F. X. Weill, A. Cloeckaert, M. Vrints, E. Mairiaux, K. Praud, K. Dierick, C. Wildemauve, C. Godard, P. Butaye, H. Imberechts, P. A. D. Grimont, and J. M. Collard. 2006. Clonal emergence of extended-spectrum β-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 3.Cloeckaert, A., K. Praud, B. Doublet, A. Bertini, A. Carattoli, P. Butaye, H. Imberechts, S. Bertrand, J. M. Collard, G. Arlet, and F. X. Weill. 2007. Dissemination of an extended-spectrum-β-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 51:1872-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Fernandez, A., G. Chiaretto, A. Bertini, L. Villa, D. Fortini, A. Ricci, and A. Carattoli. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61:1229-1233. [DOI] [PubMed] [Google Scholar]
- 6.Garcia Fernandez, A., A. Cloeckaert, A. Bertini, K. Praud, B. Doublet, F. X. Weill, and A. Carattoli. 2007. Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51:4177-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girlich, D., L. Poirel, A. Carattoli, I. Kempf, M. F. Lartigue, A. Bertini, and P. Nordmann. 2007. Extended-spectrum β-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 73:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Sanz, R., S. Herrera-Leon, M. de la Fuente, M. Arroyo, and M. Aurora Echeita. 2009. Emergence of extended-spectrum β-lactamases and AmpC-type β-lactamases in human Salmonella isolated in Spain from 2001 to 2005. J. Antimicrob. Chemother. 64:1181-1186. [DOI] [PubMed] [Google Scholar]
- 9.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madec, J. Y., C. Lazizzera, P. Châtre, D. Meunier, S. Martin, G. Lepage, M. F. Ménard, P. Lebreton, and T. Rambaud. 2008. Prevalence of fecal carriage of acquired expanded-spectrum cephalosporin resistance in Enterobacteriaceae strains from cattle in France. J. Clin. Microbiol. 46:1566-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meunier, D., E. Jouy, C. Lazizzera, M. Kobisch, and J. Y. Madec. 2006. CTX-M-1 and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28:402-407. [DOI] [PubMed] [Google Scholar]
- 12.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915-1922. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez, I., W. Barownick, R. Helmuth, M. Carmen Mendoza, M. Rosario Rodicio, A. Schroeter, and B. Guerra. 2009. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 64:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Smet, A., A. Martel, D. Persoons, J. Dewulf, M. Heyndrickx, A. Cloeckaert, K. Praud, G. Claeys, B. Catry, L. Herman, F. Haesebrouck, and P. Butaye. 2009. Comparative analysis of extended-spectrum-β-lactamase-carrying plasmids from different members of Enterobacteriaceae isolated from poultry, pigs and humans: evidence for a shared β-lactam resistance gene pool? J. Antimicrob. Chemother. 63:1286-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weill, F. X., M. Demartin, L. Fabre, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (TEM-52)-producing strains of Salmonella enterica of various serotypes isolated in France. J. Clin. Microbiol. 42:3359-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weill, F. X., F. Guesnier, V. Guibert, M. Timinouni, M. Demartin, L. Polomack, and P. A. D. Grimont. 2006. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weill, F. X., R. Lailler, K. Praud, A. Kérouanton, L. Fabre, A. Brisabois, P. A. D. Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]