Abstract
In the United States, the most prevalent mechanism of carbapenem resistance among Enterobacteriaceae is the production of a Klebsiella pneumoniae carbapenemase (KPC). KPC-producing isolates often exhibit a range of carbapenem MICs. To better understand the factors that contribute to overall carbapenem resistance, we analyzed 27 KPC-producing K. pneumoniae isolates with different levels of carbapenem resistance, 11 with low-level (i.e., meropenem or imipenem MIC ≤ 4 μg/ml), 2 with intermediate-level (i.e., meropenem and imipenem MIC = 8 μg/ml), and 14 with high-level (i.e., imipenem or meropenem MIC ≥ 16 μg/ml) carbapenem resistance, that were received from throughout the United States. Among 14 isolates that exhibited high-level carbapenem resistance, Western blot analysis indicated that 10 produced an elevated amount of KPC. These isolates either contained an increased blaKPC gene copy number (n = 3) or had deletions directly upstream of the blaKPC gene (n = 7). Four additional isolates lacked elevated KPC production but had high-level carbapenem resistance. Porin sequencing analysis identified 22 isolates potentially lacking a functional OmpK35 and three isolates potentially lacking a functional OmpK36. The highest carbapenem MICs were found in two isolates that lacked both functioning porins and produced elevated amounts of KPC. The 11 isolates with low-level carbapenem resistance contained neither an upstream deletion nor increased blaKPC copy number. These results suggest that both blaKPC copy number and deletions in the upstream genetic environment affect the level of KPC production and may contribute to high-level carbapenem resistance in KPC-producing K. pneumoniae, particularly when coupled with OmpK36 porin loss.
The occurrence of Gram-negative bacterial infections that are resistant to extended-spectrum β-lactam antimicrobial agents forces clinicians to rely on carbapenems as a “last resort” to combat these resistant pathogens. However, as carbapenems are more frequently utilized, an increasing number of bacteria with various mechanisms of resistance to this class of antimicrobial agents are identified. The most widespread resistance mechanisms include the production of a carbapenemase and the combination of porin loss with the production of either an AmpC enzyme or an extended-spectrum β-lactamase (4, 15). Klebsiella pneumoniae carbapenemase (KPC), an Ambler class A β-lactamase that can hydrolyze most β-lactam agents, including carbapenems, is now the most prevalent carbapenemase found among clinical Gram-negative isolates in the United States (22).
KPC was first reported in a K. pneumoniae isolate from North Carolina in 1996 (28). However, recent reports indicate that KPC-producing Gram-negative isolates are being identified throughout the United States as well as parts of Europe, Asia, and South America (13, 20, 22). Although these β-lactamases occur most commonly in K. pneumoniae, they have also been identified in other members of the Enterobacteriaceae family and in Pseudomonas and Acinetobacter species (3, 21, 24, 26, 27). The blaKPC gene is plasmid mediated and is carried on a Tn3-based transposon, Tn4401 (17), which may account for the high mobility of this resistance mechanism.
KPC-producing isolates can exhibit a range of carbapenem MICs, thus making their detection a significant challenge for clinical laboratories. By using 2009 Clinical and Laboratory Standards Institute (CLSI) breakpoints and testing methods (1, 6), some KPC-producing isolates may be identified as susceptible to carbapenems. The clinical significance of carbapenem-susceptible isolates with elevated carbapenem MICs is unclear (6), and the cellular changes that may convert a susceptible KPC-producing isolate to one with MICs indicating resistance to carbapenem are not well described. From previous reports, we know that KPC production combined with porin loss can result in higher carbapenem MICs (10, 14, 29). This finding suggests that the KPC enzyme alone is not always sufficient to confer carbapenem resistance, as defined by the 2009 CLSI breakpoints.
Other factors likely result in higher carbapenem MICs for KPC-producing isolates. For example, isolates with an increased expression of blaKPC were previously shown to have increased rates of hydrolysis of imipenem and meropenem (14). Directly upstream of the blaKPC gene is a nonconserved region of the Tn4401 transposon, located between the istB and the blaKPC genes (17). Previous reports describe four isoforms in this variable region: Tn4401a contains a 100-bp deletion, Tn4401b contains no deletion (17), and isoforms with 215-bp (GenBank accession no. DQ989640) and 255-bp (13) deletions were recently reported. Additional studies of this variable region suggest that the 100-bp deletion may result in a different −35 promoter region of the blaKPC gene (11). Upstream deletions that affect the promoter may impact the level of blaKPC expression and thus would influence the overall level of carbapenem resistance. Also, KPC-producing isolates may contain different levels of blaKPC dosage based on the presence of multiple copies of a blaKPC-carrying plasmid, multiple blaKPC-carrying plasmids, or multiple copies of the blaKPC gene located within the same plasmid (11). Increasing the blaKPC gene copy number could result in increased enzyme production and higher carbapenem MICs. Understanding the impact of these factors may help to predict the potential for KPC-producing isolates susceptible to carbapenems to convert to isolates resistant to carbapenems.
In this study, we examined genetic factors that may enhance the level of carbapenem resistance. We selected 27 KPC-producing K. pneumoniae isolates that were obtained from clinical patients in different areas of the country and exhibited a range of carbapenem MICs. These isolates were characterized by determining the sequences of the two main porins, OmpK35 and OmpK36 (9), examining levels of KPC production by Western blot analysis, comparing relative blaKPC copy numbers using quantitative real-time PCR, and analyzing sequence variations in the genetic environment directly upstream of the blaKPC gene.
MATERIALS AND METHODS
Selection of bacterial strains.
K. pneumoniae isolates sent to the Centers for Disease Control and Prevention (CDC) for reference antimicrobial susceptibility testing were analyzed by PCR for blaKPC if the MIC was ≥2 μg/ml for any of the carbapenems (i.e., imipenem, meropenem, or ertapenem) or if the isolate tested positive by the modified Hodge test (1, 6). For this study, KPC-producing K. pneumoniae isolates (n = 27) were selected from diverse geographic locations and represented isolates with MICs that spanned the 2009 CLSI breakpoints for imipenem and meropenem (i.e., susceptible, ≤4 μg/ml; intermediate, 8 μg/ml; resistant, ≥16 μg/ml) (6). These isolates were recovered from patients located in 17 cities in 9 states representing each major geographic region of the continental United States. We defined 11 isolates as having “low-level carbapenem resistance” (i.e., MIC ≤ 4 μg/ml for either imipenem or meropenem) and 14 isolates as having “high-level carbapenem resistance” (i.e., MIC ≥ 16 μg/ml for either imipenem or meropenem). Also, two additional isolates that had an intermediate level of resistance (MIC = 8 μg/ml for both imipenem and meropenem) were selected.
BMD.
Carbapenem MICs were measured using broth microdilution (BMD) on panels made in-house according to CLSI guidelines (5, 6).
PFGE.
The CHEF Mapper electrophoresis system (Bio-Rad, Hercules, CA) was used to type KPC-positive K. pneumoniae isolates selected for this study by pulsed-field gel electrophoresis (PFGE) of XbaI-digested DNA as described for Escherichia coli (http://www.cdc.gov/pulsenet/protocols.htm) and compared to the CDC's KPC-producing K. pneumoniae PFGE database (n > 430).
Sequencing of the blaKPC gene.
The KPC subtypes of the 27 isolates were determined by amplification of a 1,011-bp PCR product and bidirectional DNA sequence analysis using previously described primers (23).
Western blot analysis of KPC production.
Rabbit polyclonal antibodies were raised against the KPC type 2 (KPC-2) β-lactamase in order to measure blaKPC expression. The blaKPC-2 gene was expressed from pBR322-catI-blaKPC-2 in E. coli DH10B. The KPC-2 β-lactamase was then isolated and purified as previously described (18). Anti-KPC-2 polyclonal antibodies were raised by Sigma-Genosys (The Woodlands, TX) and isolated from serum using protein G column purification (Sigma-Genosys) (19). KPC-producing K. pneumoniae study isolates were grown in Luria-Bertani broth to an optical density at 600 nm (OD600) of 0.8. Preparation of samples, immunoblotting, and recognition of KPC expression were performed as previously reported (19).
Relative blaKPC copy number.
The quantity of the blaKPC gene was measured relative to an internal K. pneumoniae housekeeping gene, rpoB, using real-time PCR on the 7500 Fast system (Applied Biosystems Inc., Carlsbad, CA). Cell lysates were prepared as previously described (7) except that 50 μl of heated lysate was neutralized with 18 μl of 0.5 M Tris-HCl, pH 8, diluted with 400 μl of cold, sterilized reagent grade water, and stored at −20°C. DNA concentrations of cell lysates were subsequently normalized using a Nanodrop spectrophotometer (Thermo Scientific, West Palm Beach, FL). PCRs were performed in triplicate using QuantiFast reagents (Qiagen, Valencia, CA), 1 ng/μl of template, 500 nM each primer, and 250 nM each probe (listed in Table 1). Cycling conditions included a 3-min enzyme activation step at 95°C, followed by 40 cycles of melting (95°C for 3 s) and annealing/extension (60°C for 20 s). Standard curves were generated for both the target (blaKPC) and the endogenous control (rpoB) using 10-fold dilutions of template DNA at known concentrations and by plotting the logarithm of initial quantity of template (along the x axis) versus the respective cycle threshold (CT) values (along the y axis) (12). Absolute quantification analysis of gene copy number was performed using the following equations derived from these standard curves (rearrangement of y = mx + b): quantity of blaKPC (ng/μl) = 10(CT − 22.43)/−3.4665 (r2 = 0.994) and quantity of rpoB (ng/μl) = 10(CT − 22.05)/−3.2561 (r2 = 0.996). The ratio of blaKPC copy number to rpoB copy number was calculated in order to determine the relative blaKPC gene dosage in each isolate. For each isolate, an average of these ratios and their standard deviation are shown in Table 2.
TABLE 1.
Sequences of primers and probes utilized for blaKPCgene dosage studies and analysis of Tn4401 deletions (located between the istB and blaKPC genes)
| Primer or probe | Sequence (5′-3′) (reference) |
|---|---|
| KPC forward primer | GGC CGC CGT GCA ATA C |
| KPC reverse primer | GCC GCC CAA CTC CTT CA |
| KPC probe (FAM)a | TG ATA ACG CCG CCG CCA ATT TGT |
| RPOB forward primer | CTG ATG CCT CAG GAT ATG ATC AAC |
| RPOB reverse primer | CTG GCT GGA ACC AAA GAA CTC T |
| RPOB probe (Cy5)a | CA AGC CGA TTT CCG CAG CAG TGA |
| Tn4401 forward primer | TGA CCC TGA GCG GCG AAA GC (17) |
| Tn4401 reverse primer | CAC AGC GGC AGC AAG AAA GC |
KPC probes were labeled with 6-carboxyfluorescein (FAM), and RPOB probes were labeled with Cy5 on their 5′ ends. Each contained a black hole quencher (BHQ) on its 3′ end.
TABLE 2.
Characteristics of KPC-producing Klebsiella pneumoniae isolates with low-, intermediate-, and high-level carbapenem resistance
| Carbapenem resistance category and isolatea | MIC (μg/ml) ofi: |
KPC variant | Western blot resultd | Gene dosagef | Tn4401 variable region | ||
|---|---|---|---|---|---|---|---|
| IPM | MEM | ETP | |||||
| Low | |||||||
| ART2008226b | <1 | <0.25 | <0.5 | 3 | + | NAg | NA |
| ART2008142b | <1 | 4 | 8 | 3 | + | 0.62 ± 0.07 | No deletion |
| AIS081058 | 1 | 4 | 2 | 2 | + | 0.05 ± 0.07 | No deletion |
| AIS070654b | 4 | 1 | 2 | 3 | + | 0.27 ± 0.02 | No deletion |
| AIS081072b | 2 | 2 | 8 | 3 | + | 0.35 ± 0.03 | No deletion |
| AIS080884 | 2 | 4 | 8 | 3 | ++ | 0.49 ± 0.05 | No deletion |
| ART2008140b | 4 | 2 | 8 | 3 | ++ | 0.51 ± 0.01 | No deletion |
| AIS070446b | 4 | 4 | 4 | 3 | ++ | 0.64 ± 0.11 | No deletion |
| HIP10924 | 4 | 4 | 4 | 3 | ++ | 0.45 ± 0.01 | No deletion |
| ART2008028 | 4 | 8 | 8 | 3 | ++ | 0.52 ± 0.09 | No deletion |
| ART2008137b | 4 | 8 | 16 | 3 | + | 0.57 ± 0.04 | No deletion |
| Intermediate | |||||||
| HIP14474b | 8 | 8 | 16 | 3 | + | 0.48 ± 0.02 | No deletion |
| ART2008136b | 8 | 8 | 16 | 2 | + | 0.05h | 100-bp deletion |
| High | |||||||
| ART2008022b | 8 | 16 | 32 | 3 | ++ | 0.64 ± 0.01 | No deletion |
| ART2008139b | 32 | 16 | 16 | 3 | +++++ | 0.37 ± 0.14 | 68-bp deletion |
| ART2008143b | 8 | 16 | 64 | 3 | ++++ | 0.38 ± 0.02 | 68-bp deletion |
| AIS080949b | 32 | 16 | 64 | 3 | ++++ | 2.03 ± 0.01 | No deletion |
| HIP14358b | 32 | 16 | 64 | 2 | +++++ | 0.12 ± 0.05 | 100-bp deletion |
| AIS080792b | 32 | 32 | 64 | 2 | +++++++ | 0.55 ± 0.18 | 100-bp deletion |
| ART2008138b | 32 | 32 | 128 | 3 | ++++ | 0.28 ± 0.19 | 68-bp deletion |
| ART2008024 | 32 | 64 | 128 | 2 | +++++++ | 0.18 ± 0.06 | 255-bp deletion |
| ART2008026b | 64 | 64 | 256 | 3 | ++ | 0.56 ± 0.29 | No deletion |
| AIS081042b | 64 | 128 | 256 | 3 | +++++ | 0.25 ± 0.09 | 100-bp deletion |
| AIS080571b | 128 | 128 | 256 | 3 | ++ | 0.59 ± 0.13 | No deletion |
| ART2008141b,c | 256 | 256 | 256 | 3 | ++ | 0.47 ± 0.08 | No deletion |
| ART2008135b,c | 512 | 256 | 1,024 | 3 | ++++ | 1.81 ± 0.46 | No deletion |
| ART2008027b,c | 512 | 512 | 1,024 | 3 | ++++e | 3.03 ± 0.29 | No deletion |
Isolates are arranged in order of increasing carbapenem resistance. Classification of isolates as having low-, intermediate-, and high-level carbapenem resistance was as defined in this study.
Isolate that potentially lacks a functional OmpK35 porin (Table 3).
Isolate that potentially lacks a functional OmpK36 porin (Table 3).
KPC production is interpreted with plus signs to represent relative intensity of bands from the Western blot analysis (Fig. 2).
The only result that varied between repeated Western blots as seen in Fig. 2.
Gene dosage results are shown as a ratio of quantity of blaKPC (ng/μl) to that of rpoB (ng/μl).
NA, not applicable because the isolate lost its blaKPC-carrying plasmid upon passage in vitro.
blaKPCgene dosage result from only the first experimental repetition due to loss of the blaKPC-carrying plasmid prior to subsequent repetitions.
IPM, imipenem; MEM, meropenem; ETP, ertapenem.
Analysis of upstream genetic environment.
Directly upstream of the blaKPC gene is a variable region of the encompassing Tn4401 transposon structure. PCR and subsequent bidirectional DNA sequence analysis were performed using primers shown in Table 1 in order to analyze the region located between the istB and blaKPC genes (17).
Sequencing of ompK35 and ompK36 genes.
PCR for ompK35 and ompK36 was performed using previously described primers and conditions (16). DNA sequence analysis was performed using Lasergene 7.2 (DNASTAR, Madison, WI). The final amino acid sequences were determined using the ExPASy proteomics server (http://ca.expasy.org) and compared with those previously described in GenBank (i.e., accession numbers FJ577672, AJ011501, ADG27478, FJ577673, FJ577675, BAH64431, and ACI07318) (see Table 3).
TABLE 3.
Sequencing profiles of ompK35 and ompK36
| Carbapenem resistance category and isolatef | MIC (μg/ml) of: |
OmpK35 |
OmpK36 |
||||
|---|---|---|---|---|---|---|---|
| IPM | MEM | ETP | Sequence accession no.a | Modificationb | Sequence accession no. | Modification(s) | |
| Low | |||||||
| ART2008226 | <1 | <0.25 | <0.5 | FJ577672c | — | FJ577673 | Thr192 del |
| ART2008142 | <1 | 4 | 8 | FJ577672c | — | FJ577673 | — |
| AIS081058 | 1 | 4 | 2 | AJ011501 | — | BAH64431 | — |
| AIS070654 | 4 | 1 | 2 | FJ577672c | — | FJ577673 | — |
| AIS081072 | 2 | 2 | 8 | FJ577672c | — | FJ577673 | — |
| AIS080884 | 2 | 4 | 8 | AJ011501 | Glu132Lys | BAH64431 | Ser255Thr |
| ART2008140 | 4 | 2 | 8 | FJ577672c | — | FJ577673 | — |
| AIS070446 | 4 | 4 | 4 | FJ577672c | — | FJ577673 | — |
| HIP10924 | 4 | 4 | 4 | ADG27478 | — | ACI07318 | Asp91Asn, Tyr198Phe, Leu311Ile |
| ART2008028 | 4 | 8 | 8 | AJ011501 | — | FJ577675 | Gly136 and Asp137 del, Ala305Pro |
| ART2008137 | 4 | 8 | 16 | FJ577672c | — | FJ577673 | — |
| Intermediate | |||||||
| HIP14474 | 8 | 8 | 16 | FJ577672c | — | FJ577673 | — |
| ART2008136 | 8 | 8 | 16 | FJ577672c | — | FJ577673 | — |
| High | |||||||
| ART2008022 | 8 | 16 | 32 | FJ577672c | — | FJ577673 | — |
| ART2008139 | 32 | 16 | 16 | FJ577672c | — | FJ577673 | — |
| ART2008143 | 8 | 16 | 64 | FJ577672c | — | FJ577673 | — |
| AIS080949 | 32 | 16 | 64 | FJ577672c | — | FJ577673 | — |
| HIP14358 | 32 | 16 | 64 | FJ577672c | — | FJ577673 | — |
| AIS080792 | 32 | 32 | 64 | FJ577672c | — | FJ577673 | — |
| ART2008138 | 32 | 32 | 128 | FJ577672c | — | NAd | NDe |
| ART2008024 | 32 | 64 | 128 | AJ011501 | — | BAH64431 | 135 and 136 ins Asp |
| ART2008026 | 64 | 64 | 256 | FJ577672c | — | FJ577673 | — |
| AIS081042 | 64 | 128 | 256 | FJ577672c | — | FJ577673 | 135 and 136 ins GlyAsp |
| AIS080571 | 128 | 128 | 256 | FJ577672c | — | NA | ND |
| ART2008141 | 256 | 256 | 256 | FJ577672c | — | FJ577673 | Thr261fsX |
| ART2008135 | 512 | 256 | 1,024 | FJ577672c | — | FJ577673 | Thr126fsX |
| ART2008027 | 512 | 512 | 1,024 | FJ577672c | — | FJ577673 | Glu78fsX |
Accession numbers indicate most similar reference sequences in GenBank.
Predicted translational modifications, based on nucleotide sequencing data, that deviate from the sequence of the listed accession number. —, no modification; del, deletion; ins, insertion of; fsX, frameshift resulting in a premature stop codon.
Previously described sequence with a frameshift that ultimately results in a stop codon after the amino acid 88 codon.
NA, unable to generate a quality PCR product for sequencing despite multiple attempts.
ND, not determined.
Listing of isolates is as for Table 2.
RESULTS AND DISCUSSION
Characterization of isolates.
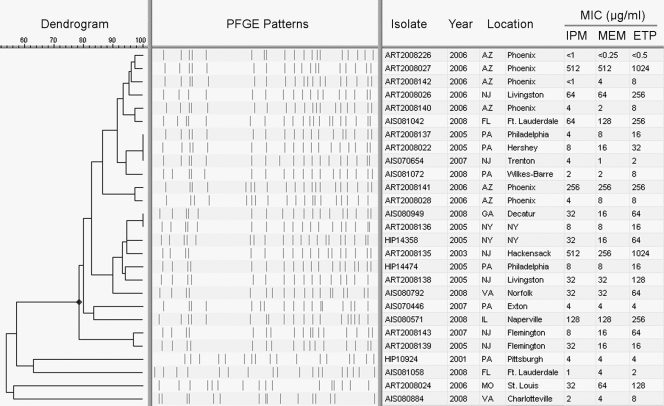
Isolates were selected to represent a diverse population of KPC-producing K. pneumoniae isolates seen in the United States. Notably, the majority of the isolates (n = 23) had ≥80% PFGE pattern similarity using the Dice coefficient and clustering by the unweighted-pair group method using average linkages (UPGMA) (Bionumerics 5.10; Applied Maths Inc., Austin, TX) (Fig. 1). Since six of these 23 isolates were previously confirmed to be multilocus sequence type 258 (ST 258), the dominant strain of KPC-producing K. pneumoniae in the United States (13), the remaining isolates whose PFGE patterns cluster with this group are also likely ST 258 (2). Sequencing the blaKPC genes for all 27 isolates showed that the majority (n = 22) produced KPC-3, while only five produced KPC-2 (Table 2). These are the most common KPC subtypes reported in the United States. The carbapenem MICs for these isolates are shown in Tables 2 and 3.
FIG. 1.
Dendrogram showing the relatedness of isolates based on PFGE patterns. Isolates were selected to represent a broad range of carbapenem MICs and geographic locations of isolation. The similar PFGE patterns observed for the majority of these isolates (sharing ≥80% similarity) correspond to a PFGE pattern associated with multilocus sequence type 258, the dominant strain of KPC-producing K. pneumoniae seen throughout the United States (13). IPM, imipenem; MEM, meropenem; ETP, ertapenem.
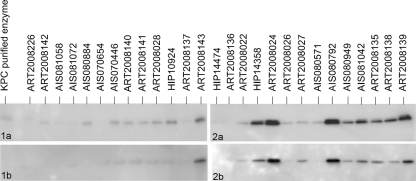
Levels of KPC production.
To evaluate the relative levels of steady-state KPC expression in the 27 isolates, Western blot analysis was performed. Immunoblotting experiments were conducted in duplicate and produced nearly identical results (Fig. 2). Only one isolate (ART2008027) had results that varied, demonstrating a relatively more intense band in the second blot (Fig. 2, blot 2b). All isolates exhibiting elevated KPC production (i.e., ≥4 pluses in Table 2) were associated with high-level carbapenem resistance. Conversely, only four of the isolates with high-level carbapenem resistance (ART2008022, ART2008026, AIS080571, and ART2008141) lacked elevated KPC production. All of the isolates with low and intermediate levels of carbapenem resistance (n = 13) exhibited lower KPC production (i.e., ≤2 pluses in Table 2). These results demonstrate that elevated KPC production is associated with high-level carbapenem resistance.
FIG. 2.
Western blot analysis of KPC production within each of the 27 isolates. Results from blots 1a and 2a were repeated (seen below as blots 1b and 2b, respectively). Comparison of each blot was crucial in determining the relative level of KPC production notated in Table 2.
Analysis of relative blaKPC copy number.
We hypothesized that the amount of KPC expression could be directly affected by blaKPC copy number. To analyze the relative blaKPC copy number, we used real-time PCR to measure the quantity of blaKPC DNA relative to that of an internal K. pneumoniae housekeeping gene, rpoB. Three of the isolates (AIS080949, ART2008135, and ART2008027) exhibited an elevated relative blaKPC copy number compared to the other isolates; all three of these isolates demonstrated elevated KPC production and high-level carbapenem resistance (Table 2). Of the remaining 24 isolates, 7 (ART2008143, HIP14358, ART2008024, AIS080792, AIS081042, ART2008138, and ART2008139) exhibited elevated KPC production but lacked an increased blaKPC copy number. This suggests that factors other than blaKPC copy number may contribute to the elevated KPC production and high-level carbapenem resistance observed in these isolates.
Analysis of the genetic environment upstream of the blaKPC gene.
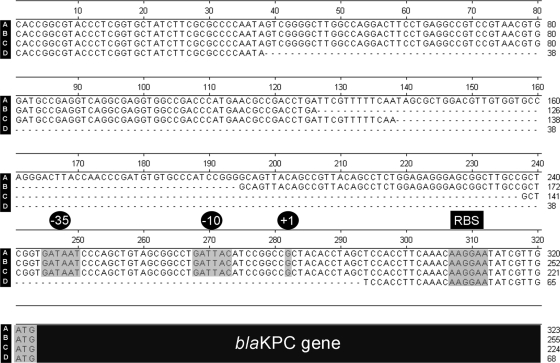
The upstream Tn4401 element was analyzed for the presence of deletions by PCR, and its sequence was analyzed for 26 of the 27 isolates. One isolate, ART2008226, did not produce a PCR product because its blaKPC-3-carrying plasmid was lost during passage of the isolate in vitro. Four different isoforms of Tn4401 were detected in this study, including sequences with no deletion (n = 18), a novel 68-bp deletion (n = 3), a 100-bp deletion (n = 4), and a 255-bp deletion (n = 1). Sequence analysis of these deletions indicated that the 68-bp and 100-bp deletions both end upstream of the −35 region of the putative promoter identified by Yigit et al. (28). However, the 255-bp deletion observed in this study eliminates the entire putative promoter region (28), including the transcription start site (Fig. 3).
FIG. 3.
Nucleotide sequence illustrating observed variations in the genetic environment directly upstream of the blaKPC gene (seen in black at the bottom). Highlighted regions include the putative −35 and −10 regions of the promoter, the transcription start site (marked as +1), the potential ribosome-binding site (RBS), and the blaKPC start codon (ATG), as previously reported by Yigit et al. (28). Sequences of isolates with no deletion (A), a 68-bp deletion (B), a 100-bp deletion (C), and a 255-bp deletion (D) were observed in this study.
Seven of the eight isolates with deletions demonstrated high-level carbapenem resistance and elevated KPC production (i.e., ≥4 pluses in Table 2), including ART2008024 (255-bp deletion), which produced one of the highest levels of KPC expression. One isolate with a 100-bp deletion, ART2008136, demonstrated an intermediate level of carbapenem resistance and did not exhibit elevated KPC production. The blaKPC-carrying plasmid in this isolate was likely unstable since the initial blaKPC gene dosage assay indicated a decreased amount of relative gene dosage (i.e., 0.05) (Table 2) and since in a repeat experiment blaKPC could not be detected. With the exception of this isolate, the observed upstream deletions corresponded with increased KPC production, suggesting that blaKPC expression is driven by a second upstream promoter.
Porin analysis.
Genes encoding the two major porins, ompK35 and ompK36, were sequenced. Most of the isolates (n = 22 out of 27) contained ompK35 genes that had a sequence similar to that of GenBank accession number FJ577672 (Table 3). This previously described ompK35 sequence has a G insertion after base pair 122 (in relation to the start of translation), resulting in an early frameshift and a premature stop codon following the amino acid 88 codon, producing a truncated protein that was nonfunctional (14). Specifically, genes with this sequence were found in 22 of the 23 ST 258-like isolates from this study, suggesting that this may be a common characteristic of ST 258 strains. Even though most isolates in this study appear to lack a functional OmpK35, previous reports suggest that OmpK36 likely plays a larger role in carbapenem resistance (8, 14).
DNA sequence analysis of ompK36 could not be performed in two isolates (ART2008138 and AIS080571) because quality PCR products could not be generated after multiple attempts; this may be attributed to unknown sequence modifications that interfered with primer binding. The majority of the other 25 isolates (n = 15) were found to possess ompK36 genes that were identical in sequence to a previously described ompK36 (GenBank accession no. FJ577673), which previously produced a functional protein (14). These 15 isolates had carbapenem MICs that spanned low, intermediate, and high levels of carbapenem resistance (Table 3). Four isolates with low-level carbapenem resistance and two isolates with high-level resistance also had OmpK36 sequences that contained modifications additional to those previously reported (e.g., single amino acid substitutions, insertions, or deletions) (Table 3). The significance of these modifications is unclear, and their impact on OmpK36 porin function is unknown. The three isolates that produced the highest carbapenem MICs (i.e., ≥256 μg/ml for each of the carbapenems) (ART2008141, ART2008135, and ART2008027) each contained ompK36 modifications that resulted in a frameshift (starting after amino acids 261, 126, and 78, respectively) and ultimately produced a premature stop codon (Table 3). The resulting truncated OmpK36 proteins would likely be nonfunctional (8, 25). The potential loss of functional OmpK35 and OmpK36 porins in these three isolates likely contributes in part to their high level of carbapenem resistance (Table 3).
Isolates with low-level carbapenem resistance.
Western blot analysis of the 11 isolates with low-level carbapenem resistance revealed relatively low-level KPC production for all of these isolates (i.e., ≤2 pluses in Table 2). Porin sequence analysis identified seven of these isolates with critically modified ompK35 genes (Table 3); however, all ompK36 genes in these isolates appear to lack significant modifications. One isolate with very low level carbapenem resistance, ART2008226, completely lost its blaKPC-3-carrying plasmid as described above. Evidence of unstable blaKPC-carrying plasmids may also be observed in other isolates with low-, intermediate-, and high-level carbapenem resistance (e.g., <0.2 relative blaKPC gene dosage) (Table 2). Gene dosage studies for each of the isolates with low-level carbapenem resistance did not detect any significant elevation in blaKPC copy number relative to those of the other study isolates (e.g., >0.8) (Table 2). Also, all isolates with low-level carbapenem resistance were found to have the Tn4401b isoform (containing no deletions upstream of the blaKPC gene). The lack of these two genetic factors in the 11 isolates with low-level carbapenem resistance supports our hypothesis that both elevated blaKPC copy number and deletions upstream of the blaKPC gene contribute to increased KPC production.
Isolates with high-level carbapenem resistance.
Ten of the 14 isolates with high-level carbapenem resistance exhibited elevated KPC production (i.e., ≥4 pluses in Table 2) and had either an elevated relative blaKPC copy number (n = 3) or deletions directly upstream of the blaKPC gene (n = 7). Seven of these 10 isolates with elevated KPC production (excluding ART2008138, ART2008135, and ART2008027) contained ompK36 genes that have either been previously shown to be functional (14) or appear to lack significant modifications (Table 3), suggesting that high-level carbapenem resistance can be achieved without OmpK36 porin loss. However, when critical ompK36 modifications were observed in isolates with elevated KPC production (i.e., ART2008135 and ART2008027), the level of carbapenem MICs incrementally increased. Four isolates with high-level carbapenem resistance (ART2008022, ART2008026, AIS080571, and ART2008141) did not exhibit an elevated blaKPC copy number, an upstream deletion, or increased KPC production (Table 2). The high-level carbapenem resistance in these four isolates may likely be attributable to identified porin modifications (Table 3) or other factors not examined in this study. For example, ART2008026 had high-level carbapenem resistance and contained an ompK36 sequence previously shown to encode a functional porin (GenBank accession no. FJ577673) but lacked upstream deletions, increased blaKPC copy number, and elevated KPC production.
In conclusion, our results suggest that both blaKPC copy number and deletions in the upstream genetic environment may affect the level of KPC production and thus contribute to high-level carbapenem resistance in KPC-producing K. pneumoniae. In addition, the combination of OmpK36 porin loss with elevated KPC production appears to contribute to incremental carbapenem MIC increases. These results indicate that KPC-positive isolates susceptible to carbapenems but with elevated carbapenem MICs may have multiple pathways to achieve high-level carbapenem resistance. Also, the identification of plasmid-based factors that elevate carbapenem MICs suggests that KPC-producing isolates with high-level carbapenem resistance may become more common as plasmids continue to disseminate within the Enterobacteriaceae population. Our analysis confirms the complexity of the carbapenem resistance phenotype and highlights the threat this continues to present to patients.
Acknowledgments
We acknowledge David R. Lonsway for his work on MIC panel preparation and testing, David Sue for his assistance with real-time PCR experiment design, and Anoop Pasupuleti for his assistance with ompK35 and ompK36 sequence analysis.
This work was supported in part by the Veterans Affairs Merit Review Program (R.A.B.), the National Institutes of Health (grant RO3-AI081036 to R.A.B.), and Geriatric Research Education and Clinical Center VISN 10 (R.A.B.).
The authors of this publication state that they do not have a commercial or other association that might be considered a conflict of interest regarding the information within this document.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 26 July 2010.
REFERENCES
- 1.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, T. J., P. Gerner-Smidt, and B. Swaminathan. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3:20-31. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, J. W., M. L. Herrera, J. S. Lewis II, B. W. Wickes, and J. H. Jorgensen. 2009. KPC-2-producing Enterobacter cloacae and Pseudomonas putida coinfection in a liver transplant recipient. Antimicrob. Agents Chemother. 53:292-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed. Approved standard M7-A8. CLSI, Wayne, PA.
- 6.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19. CLSI, Wayne, PA.
- 7.Conrad, S., M. Oethinger, K. Kaifel, G. Klotz, R. Marre, and W. V. Kern. 1996. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 38:443-455. [DOI] [PubMed] [Google Scholar]
- 8.Crowley, B., V. J. Benedí, and A. Doménech-Sánchez. 2002. Expression of SHV-2 β-lactamase and of reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob. Agents Chemother. 46:3679-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doumith, M., M. J. Ellington, D. M. Livermore, and N. Woodford. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659-667. [DOI] [PubMed] [Google Scholar]
- 10.Endimiani, A., J. M. Depasquale, S. Forero, F. Perez, A. M. Hujer, D. Roberts-Pollack, P. D. Fiorella, N. Pickens, B. Kitchel, A. E. Casiano-Colon, F. C. Tenover, and R. A. Bonomo. 2009. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J. Antimicrob. Chemother. 64:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootz, T. D., M. K. Lescoe, F. Dib-Hajj, B. A. Dougherty, W. He, P. Della-Latta, and R. C. Huard. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 13.Kitchel, B., J. K. Rasheed, J. B. Patel, A. Srinivasan, S. Navon-Venezia, Y. Carmeli, A. Brolund, and C. G. Giske. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landman, D., S. Bratu, and J. Quale. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J. Med. Microbiol. 58:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leavitt, A., I. Chmelnitsky, R. Colodner, I. Ofek, Y. Carmeli, and S. Navon-Venezia. 2009. Ertapenem resistance among extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 47:969-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomaestro, B. M., E. H. Tobin, W. Shang, and T. Gootz. 2006. The spread of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae to upstate New York. Clin. Infect. Dis. 43:e26-e28. [DOI] [PubMed] [Google Scholar]
- 17.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papp-Wallace, K. M., C. R. Bethel, A. M. Distler, C. Kasuboski, M. Taracila, and R. A. Bonomo. 2010. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob. Agents Chemother. 54:890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papp-Wallace, K. M., M. Taracila, J. M. Hornick, A. M. Hujer, K. M. Hujer, A. M. Distler, A. Endimiani, and R. A. Bonomo. 2010. Substrate selectivity and a novel role in inhibitor discrimination by position 237 in the KPC-2 β-lactamase. Antimicrob. Agents Chemother. 54:2867-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel, J. B., J. K. Rasheed, and B. Kitchel. 2009. Carbapenemases in Enterobacteriaceae: activity, epidemiology, and laboratory detection. Clin. Microbiol. Newsl. 31:55-62. [Google Scholar]
- 21.Poirel, L., P. Nordmann, E. Lagrutta, T. Cleary, and L. S. Munoz-Price. 2010. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 54:3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasheed, J. K., J. W. Biddle, K. F. Anderson, L. Washer, C. Chenoweth, J. Perrin, D. W. Newton, and J. B. Patel. 2008. Detection of the KPC-2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and Klebsiella oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robledo, I. E., E. E. Aquino, M. I. Sante, J. L. Santana, D. M. Otero, C. F. Leon, and G. J. Vazquez. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struyvé, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenyl-alanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 26.Villegas, M. V., K. Lolans, A. Corrrea, J. N. Kattan, J. A. Lopez, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 51:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther-Rasmussen, J., and N. Høiby. 2007. Class A carbapenemases. J. Antimicrob. Chemother. 60:470-482. [DOI] [PubMed] [Google Scholar]
- 28.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, R., L. Yang, J. C. Cai, H. W. Zhou, and G. X. Chen. 2008. High-level carbapenem resistance in a Citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J. Med. Microbiol. 57:332-337. [DOI] [PubMed] [Google Scholar]