Summary
The determinant of verapamil-reversible chloroquine resistance (CQR) in a Plasmodium falciparum genetic cross maps to a 36 kb segment of chromosome 7. This segment harbors a 13-exon gene, pfcrt, having point mutations that associate completely with CQR in parasite lines from Asia, Africa, and South America. These data, transfection results, and selection of a CQR line harboring a novel K76I mutation point to a central role for the PfCRT protein in CQR. This transmembrane protein localizes to the parasite digestive vacuole (DV), the site of CQ action, where increased compartment acidification associates with PfCRT point mutations. Mutations in PfCRT may result in altered chloroquine flux or reduced drug binding to hematin through an effect on DV pH.
Introduction
The increasing failure of chloroquine (CQ) against P. falciparum malaria has produced severe public health setbacks in recent years, especially among pediatric populations of sub-Saharan Africa, where malaria is a leading cause of morbidity and mortality (Marsh, 1998; Trape et al., 1998). No alternative drug now exists with the affordability, low toxicity, and efficacy that once characterized CQ. Understanding the molecular basis of CQR should assist design of new antimalarials that can replace or be used in adjunct with CQ.
The antimalarial activity of CQ is thought to occur in the DV of the haploid intraerythrocytic stages of P. falciparum, where the drug interferes with the polymerization and detoxification of heme molecules released by digestion of hemoglobin (Goldberg et al., 1990). Two characteristics that distinguish CQR parasites are diminished accumulation of CQ (Fitch, 1970) and chemosensitization (“reversal”) by verapamil (VP) and diverse tricyclic antidepressants and antihistaminics (Martin et al., 1987; Bitonti et al., 1988; Peters et al., 1989). Proposals for the CQR mechanism include (1) altered CQ uptake or efflux at the cytoplasmic membrane of the intraerythrocytic parasite, (2) altered H+ flux or CQ uptake at the parasite DV membrane, (3) reduced CQ access to its receptor, hematin (formed in the DV upon hemoglobin digestion), and (4) increased glutathione-mediated detoxification of CQ-hematin complexes. Molecules invoked in these proposals include P-glycoproteins, a CL− channel regulator, a Na+/H+ exchanger, a DV H+ pump, a factor that reduces CQ access to hematin, and glutathione S-transferase or related molecule involved in heme detoxification (Martin et al., 1987; Martiney et al., 1995; Bray et al., 1998; Ginsburg et al., 1998; Wunsch et al., 1998).
The VP-reversible CQR phenotype segregated as a Mendelian trait in a P. falciparum genetic cross between a CQR clone (Dd2) and a chloroquine-sensitive (CQS) clone (HB3) (Wellems et al., 1990). Linkage mapping localized the CQR determinant to a 36 kb segment of P. falciparum chromosome 7; further, the analysis provided evidence against a determining role for the pfmdr1 gene whose P glycoprotein–like product may be involved in modulating the degree of in vitro CQR in parasites already resistant to CQ (Wellems et al., 1991; Su et al., 1997; Reed et al., 2000). DNA sequencing and searches for open reading frames (ORFs) encoding >100 amino acids identified eight genes within this 36 kb segment, of which two, cg2 and cg1, displayed multiple codon changes that associated closely with CQR in a set of isolates from Asia and Africa (Su et al., 1997). An exception was found, however, in the 106/1 Sudan clone, which contained the Dd2-type cg2 allele but was CQS (Su et al., 1997). Genetic modifications of cg2 and cg1 in CQR parasites recently demonstrated that the Dd2 complement of polymorphisms in these genes is not necessary for VP-reversible CQR (Fidock et al., 2000).
We now report a highly interrupted and previously undetected gene, pfcrt, in the 36 kb segment. Several lines of evidence implicate pfcrt mutations in the VP-reversible CQR phenotype.
Results
A Novel Transmembrane Protein Is Encoded by a 13-exon P. falciparum Gene within the CQR-Associated 36 kb Segment
To test for previously unidentified transcripts in the 36 kb region, we designed primer pairs from small putative ORFs (<100 codons). Upon screening of cDNA libraries by PCR, 3 of 21 pairs yielded products indicative of a transcript produced from small exons located less than 10 kb from cg1 and cg2. Sequence extension by PCR from cDNA revealed that this transcript was from a gene, pfcrt, whose coding region spans 3.1 kb in 13 exons ranging in size from 45–269 base pairs. RT-PCR analysis identified no additional transcribed sequences between pfcrt and the neighboring cg3 and cg9 genes.
Translation of the Dd2 pfcrt coding region predicted a 424 amino acid, 48.6 kDa protein (PfCRT; Figure 1). We evaluated PfCRT together with ortholog sequences from P. vivax and P. knowlesi and a more distant homolog from Dictyostelium discoideum (T. N., J. C. W., and T. E. W., unpublished data). Database searches, alignments, and prediction algorithms indicated that PfCRT belongs to a previously undescribed family of putative transporters or channels with ten transmembrane segments. These analyses showed no evidence of a typical signal sequence or other recognizable features such as ATP binding motifs.
Figure 1. The Dd2 pfcrt Gene Predicts a 424 Amino Acid Transmembrane Protein.
The ten predicted transmembrane segments are shaded, and positions of eight amino acid substitutions are indicated that distinguish CQR and CQS parasites in the HB3 × Dd2 genetic cross. Inverted arrows indicate the intron splice sites in the corresponding nucleotide sequence. A map illustrating the exon-intron organization of pfcrt and its position relative to neighboring genes of chromosome 7 is available as supplemental data at www.molecule.org/cgi/content/full/6/4/861/DC1.
pfcrt Mutations and the CQR Phenotype Are Perfectly Associated in a Set of Old World and New World Isolates
We identified pfcrt polymorphisms in the genetic cross by sequencing cDNA transcripts of Dd2, HB3, and two progeny, SC-01 (CQR) and GC-03 (CQS). Sequence comparisons revealed eight codon differences between the CQR and CQS clones at positions (amino acid numbering) 74, 75, 76, 220, 271, 326, 356, and 371. These amino acid substitutions localized within or near predicted transmembrane segments, and six involved changes in charge or hydrophilicity (Figure 1).
Sequence analysis of DNA from laboratory-adapted Old World isolates demonstrated a clear association between pfcrt codon mutations and CQR. CQR isolates from Asia and Africa consistently showed seven of the mutations found in codons of the Dd2 allele (M74I, N75E, K76T, A220S, Q271E, N326S, and R371I) with or without the eighth Dd2 mutation, I356T (Table 1). Of the 16 CQS parasites tested, 15 displayed the HB3 canonical pfcrt sequence. The remaining clone, 106/1, a previous exception to complete association between cg2 and CQR (Su et al., 1997), was particularly informative. By micro-satellite analysis (X-z. S., unpublished data), the genome of this clone is very closely related to an Old World CQR line designated FCB. Yet 106/1 is unambiguously of the CQS phenotype not chemosensitized by VP (Su et al., 1997). The 106/1 pfcrt allele shows six of the seven mutations consistently found in Old World CQR parasites but differs in having a K76 codon characteristic of CQS isolates. We have also identified a Malian isolate, S35CQ, which is CQR but contains a non-Dd2 type cg2 allele. The pfcrt sequence of S35CQ contains the same set of seven mutations observed for other CQR parasites from Africa (Table 1).
Table 1.
PfCRT Mutations and CQ Response Phenotypes in P. falciparum Lines from Different Geographical Regions
| Region | Line | 72 | 74 | 75 | 76 | 97 | 220 | 271 | 326 | 356 | 371 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chloroquine-sensitive lines | PNG | D10 | C | M | N | K | H | A | Q | N | I | R |
| All regions | Thailand | T2/C6 | C | M | N | K | H | A | Q | N | I | R |
| Malaysia | Camp/A1 | C | M | N | K | H | A | Q | N | I | R | |
| Sudan | 105/7a | C | M | N | K | H | A | Q | N | I | R | |
| Sudan | 106/1a | C | I | E | K | H | S | E | S | I | I | |
| Sudan | REN | C | M | N | K | H | A | Q | N | I | R | |
| South Africa | FAB9 | C | M | N | K | H | A | Q | N | I | R | |
| Kenya | K39a | C | M | N | K | H | A | Q | N | I | R | |
| Kenya | M24a | C | M | N | K | H | A | Q | N | I | R | |
| Liberia | LF4/1 | C | M | N | K | H | A | Q | N | I | R | |
| The Netherlands | NF54a | C | M | N | K | H | A | Q | N | I | R | |
| Sierra Leone | SL/D6 | C | M | N | K | H | A | Q | N | I | R | |
| Mali | BC5 | C | M | N | K | H | A | Q | N | I | R | |
| Mali | M5 | C | M | N | K | H | A | Q | N | I | R | |
| Haiti | Haiti | C | M | N | K | H | A | Q | N | I | R | |
| Honduras | HB3a | C | M | N | K | H | A | Q | N | I | R | |
| Chloroquine-resistant lines | Indochina | Dd2a | C | I | E | T | H | S | E | S | T | I |
| Southeast Asia and Africa | Vietnam | V1/S | C | I | E | T | H | S | E | S | T | I |
| Thailand | TM284 | C | I | E | T | H | S | E | S | T | I | |
| SE Asia | ItG2F6 | C | I | E | T | H | S | E | S | I | I | |
| SE Asia | FCB | C | I | E | T | H | S | E | S | I | I | |
| SE Asia | R29 | C | I | E | T | H | S | E | S | I | I | |
| Cambodia | Jcl | C | I | E | T | H | S | E | S | T | I | |
| Sudan | 102/1a | C | I | E | T | H | S | E | S | T | I | |
| South Africa | RB8 | C | I | E | T | H | S | E | S | I | I | |
| South Africa | RB20 | C | I | E | T | H | S | E | S | I | I | |
| Uganda | PARa | C | I | E | T | H | S | E | S | I | I | |
| Kenya | KMWII | C | I | E | T | H | S | E | S | I | I | |
| Mali | M2a | C | I | E | T | H | S | E | S | I | I | |
| Mali | S35CQa | C | I | E | T | H | S | E | S | I | I | |
| Saõ Tomé | Cai | C | I | E | T | H | S | E | S | I | I | |
| South America | Brazil | 7G8a | S | M | N | T | H | S | Q | D | L | R |
| Brazil | DIV30a | S | M | N | T | H | S | Q | D | L | R | |
| Brazil | ICSN66 | S | M | N | T | H | S | Q | D | L | R | |
| Brazil | ECPa | S | M | N | T | H | S | Q | D | L | R | |
| Brazil | PAD | S | M | N | T | H | S | Q | D | L | R | |
| Brazil | ItD12 | S | M | N | T | H | S | Q | D | L | R | |
| Ecuador | Ecu1110 | C | M | N | T | H | S | Q | D | L | R | |
| Colombia | Java | C | M | E | T | Q | S | Q | N | I | T | |
| Colombia | IAJa | C | M | E | T | Q | S | Q | N | I | T |
Complete pfcrt coding sequences for these lines were determined directly from RT-PCR amplified products. For all other lines, point mutations were determined from genomic DNA PCR products.
Sequences of pfcrt were also examined from South America, where CQR arose independently from Southeast Asia (Payne, 1987). Patterns of mutation in nine CQR isolates from Brazil, Ecuador, and Colombia differed from those in the Old World parasites and showed three distinct PfCRT sequences, which in all cases included K76T and A220S. These South American isolates revealed different mutations at codon positions 326, 356, and 371 as well as additional mutations at positions 72 and 97, relative to the Old World CQR sequences (Table 1). Amino acids 74 and 271 of the South American lines were identical to those in the canonical CQS (HB3) sequence.
CQS Parasites Acquire Features of CQR upon Episomal Transformation with the Dd2 pfcrt Sequence
We transfected the CQS clone 106/1 with the plasmid pNHSC, designed to express the Dd2-type pfcrt ORF under the regulatory control of hrp3 5′ and hrp2 3′ elements. Parasites were detected 46 days post-electroporation in a population (34-1) that had been selected with 46–92 nM CQ and maintained at 46 nM (line 34-1/A; results were reproduced with a second pNHSC-transformed line, 34-2, data not shown). Using the number of days required to obtain a parasitemia of 2% (or measurements of proliferation rates, see below), we found the efficiencies of pNHSC transformations were within the range of those observed for pHD22Y and pDC plasmids that employ human dhfr as a selectable maker (Table 2; Fidock et al., 1998). No CQR parasites were obtained when 106/1 parasites were transfected with pNHGC, designed to express the canonical CQS pfcrt ORF from GC-03, and kept in culture for 67 days in the presence of 46 nM CQ. Similarly, no CQR parasites were obtained on three occasions when 106/1 parasites (3–6 × 107, the range used for electroporation) were maintained in culture with 36–56 nM CQ for 54–69 days.
Table 2.
Proliferation Rates of Parasite Lines and CQ Inhibition Concentrations from [3H]hypoxanthine Uptake Assays
| Line | Parent | Plasmid | Selection Agent | No. Days to Detect Parasitese | No. Days to Attain 2% par. | Observed Proliferation Rate |
Uptake Assay, CQh |
Uptake Assay, CQ+VPh |
Fractional [3H]hypoxanthine Uptake at Selection Agent Concentrationi | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Per 48 hr Cyclef | Relative to Parentg | IC50 | IC90 | IC50 | IC90 | |||||||
| 106/1 | none | 3.6 | 1.00 | 32 ± 5 | 42 ± 6 | 32 ± 8 | 43 ± 11 | |||||
| 34-1/Aa,b | 106/1 | pNHSC | 46 nM CQ | 46 | 52 | 2.2 | 0.61 | 31 ± 3 | 78 ± 18 | 28 ± 2 | 42 ± 7 | 0.37 |
| 34-1/Arm | 106/1 | removed | 3.5 | 0.97 | 23 ± 3 | 28 ± 7 | 28 ± 2 | 39 ± 3 | ||||
| 34-1/Ba | 106/1 | pNHSC | 92 nM CQ | 1.9 | 0.53 | 30 ± 4 | 114 ± 30 | 22 ± 3 | 41 ± 9 | 0.20 | ||
| 34-1/Ca | 106/1 | pNHSC | 153 nM CQ | 1.7 | 0.47 | 39 ± 8 | 201 ± 72 | 23 ± 3 | 55 ± 10 | 0.22 | ||
| 34-1/Da | 106/1 | pNHSC | 192 nM CQ | 1.7 | 0.47 | 37 ± 12 | 180 ± 92 | 20 ± 4 | 63 ± 6 | 0.06 | ||
| 34-1/E | 106/1 | 230 nM CQ | 4.1 | 1.14 | 409 ± 80 | 597 ± 67 | 88 ± 6 | 161 ± 24 | 0.72 | |||
| 34-1/Erm | 106/1 | removed | 3.7 | 1.03 | 364 ± 121 | 583 ± 192 | 81 ± 7 | 142 ± 19 | ||||
| Dd2 | none | 4.4 | 1.00 | 278 ± 27 | 385 ± 52 | 77 ± 10 | 124 ± 22 | |||||
| FCB | none | 6.2 | 1.00 | 291 ± 30 | 398 ± 32 | 73 ± 16 | 119 ± 12 | |||||
| 33-5a | 106/1 | pHD22Y | 5 nM WR99210 | 29 | 34 | 2.7 | 0.72 | 20 ± 4 | 37 ± 5 | 20 ± 1 | 38 ± 6 | |
| 30-3a | 106/1 | pHD22Y | 5 nM WR99210 | 41 | 64 | nd | nd | nd | nd | nd | ||
| GC-03 | none | 4.0 | 1.00 | 31 ± 3 | 40 ± 3 | 30 ± 2 | 39 ± 3 | |||||
| 24-5a | GC-03 | pDC/CRT-Dd2/trunc. | 5 nM WR99210 | 17 | 28 | 2.5 | 0.63 | 53 ± 11 | 87 ± 18 | 48 ± 4 | 58 ± 5 | |
| 25-3a | GC-03 | pDC/CRT-HB3/all | 5 nM WR99210 | 25 | 37 | 2.5 | 0.63 | 32 ± 2 | 47 ± 2 | nd | nd | |
| 24-6a,c | GC-03 | pDC/CRT-frameshift | 5 nM WR99210 | 46 | 58 | 2.6 | 0.65 | 36 | 44 | 27 | 42 | |
| 25-7a,c | GC-03 | pDC/CRT/T76K | 5 nM WR99210 | 25 | 48 | 1.6 | 0.40 | 30 | 45 | nd | nd | |
| 25-4a,c | GC-03 | pDC/cg6 | 5 nM WR99210 | 25 | 37 | 2.5 | 0.63 | 31 | 45 | nd | nd | |
| 25-ga | GC-03 | pDC/CAT | 5 nM WR99210 | 15 | 16 | nd | ||||||
| 22-3a | GC-03 | pDC | 5 nM WR99210 | 43 | 49 | 2.8 | 0.70 | |||||
| 25-8a | GC-03 | pHD22Y | 5 nM WR99210 | 15 | 23 | nd | ||||||
| 106/1 | none | 3.6 | 1.00 | 33 ± 4 | 41 ± 5 | 31 ± 3 | 40 ± 6 | |||||
| 45-1a,b,d | 106/1 | pNH76l | 46 nM CQ | 34 | 38 | 2.5 | 0.69 | 43 ± 5 | 128 ± 26 | 39 ± 3 | 61 ± 5 | 0.48 |
| GC-03 | none | 3.9 | 1.00 | 30 ± 1 | 34 ± 2 | 28 ± 2 | 33 ± 1 | |||||
| 46-4a,b,d | GC-03 | pNH76l | 52 nM CQ | 31 | 42 | 2.8 | 0.72 | 51 ± 14 | 79 ± 13 | 29 ± 6 | 43 ± 9 | 0.49 |
| 3D7 | none | 3.6 | 1.00 | 27 ± 4 | 33 ± 5 | 27 ± 3 | 34 ± 4 | |||||
| 46-2a,b,d | 3D7 | pNH76l | 26 nM CQ | 44 | 55 | 2.6 | 0.72 | 26 ± 5 | 66 ± 21 | 20 ± 4 | 32 ± 4 | 0.50 |
Episomal transformation of these lines was confirmed using plasmid rescue.
When cured of episomes by removal of CQ selection, these lines demonstrated complete reversion to the CQS phenotype with no residual increase in CQ IC90.
These lines were tested on only a single occasion in duplicate; the other transformed lines were tested on at least three occasions.
These pNH76I-transformed lines have subsequently been maintained at 70–100 nM CQ.
Listed are the number of days post-electroporation when transformed parasites were first microscopically detected. The number of days required to attain a 2% parasitemia is considered a more accurate benchmark of transformation efficiency.
Each observed proliferation rate indicates the factor by which the parasite number increased per 48 hr cycle at the indicated selection agent concentration. Values were calculated from parasite counts of Giemsa-stained thin blood smears (0.5%–5% parasitemia) and represent geometric means of 3–8 sets of measurements collected together throughout different weeks of this work.
Proliferation rates are listed relative to that of the untransformed parental line cultured in medium without selection agent.
Inhibition values are in nM. Verapamil was included at a concentration of 0.8 μM; nd, not determined. Extrapolation of drug assay IC values consistently underestimated the rate of proliferation of the parasite lines episomally transformed with mutant pfcrt and cultivated in the presence of CQ concentrations lethal to the nontransformed parents.
Each fractional value was determined relative to [3H]hypoxanthine uptake by the same line in the absence of selection agent. Parasites were washed free of selection agent and maintained in culture for 6–8 hrs prior to initiation of the assays.
Lines from the 34-1 population were split and grown for three weeks in CQ at 92 nM (line 34-1/B) and 153 nM (line 34-1/C), a level that approaches the tolerance limit of native CQR parasites subjected to continuous CQ pressure in vitro. Presence of episomes was confirmed by patterns of DpnI digestion and plasmid rescue (Wu et al., 1996). Transcription levels from the episomal pfcrt expression cassette and from the chromosomal pfcrt gene were found to be comparable by RT-PCR (data not shown).
Proliferation rates of these lines were measured by microscopic analysis of Giemsa-stained thin blood smears made throughout the course of the experiments (Table 2). pNHSC-transformed 106/1 parasites showed a proliferation rate corresponding to a 2.2-fold increase in parasite numbers per 48 hr cycle in 46 nM CQ. Proliferation rates decreased to 1.9–1.7 in 92 nM and 153 nM CQ, compared to a rate of 3.6 for untransformed 106/1 parasites in CQ-free culture.
In parallel with microscopic analysis, we performed [3H]hypoxanthine drug assays of the episomally transformed lines that had been maintained in 46, 92, and 153 nM CQ (Table 2). These pNHSC transformants showed IC90 values of 114–201 nM CQ, significantly above the IC90 values of 37–42 nM obtained with the 106/1 parent or with a 106/1 line (33-5) episomally transformed with the pHD22Y plasmid that expresses human dihydrofolate reductase (dhfr) instead of pfcrt under the control of the same regulatory elements (Table 2).
Analysis of the pfcrt transformants revealed an apparent paradox in that IC50 and IC90 values obtained from [3H]hypoxanthine assays were routinely lower than those observed by microscopy. This finding contrasted with the close agreement between these two methods in assays of untransformed CQS and CQR lines (Su et al., 1997; unpublished observations). For the pfcrt episomally transformed lines, extrapolation of the [3H]hypoxanthine assay curves also consistently underestimated the observed rates of proliferation under continuous CQ pressure (Table 2). For example, in 192 nM CQ, the 34-1/D line maintained a proliferation rate of 1.7 compared to 3.6 for the untransformed 106/1 parent. This contrasted with a fractional [3H]hypoxanthine uptake at this CQ concentration that was only 0.06× the uptake by this line in the absence of CQ (Table 2).
We also compared the [3H]hypoxanthine uptake data against lactate dehydrogenase (LDH) immunocapture assays that measure parasite protein production. Equivalent response profiles were obtained from the two assays. Comparisons of these results and microscopic observations showed that reductions in [3H]hypoxanthine uptake and LDH production were accompanied by reductions in schizont size in the elevated CQ concentrations at which the transformants remained viable. This morphologic picture was in clear contrast to the effect of CQ on untransformed parasite lines that, in the IC50 range, produces large percentages of pyknotic, nonviable parasites within a single generation and, at higher concentrations or longer exposure times, clears the culture of intraerythrocytic forms.
Two additional findings confirmed that pNHSC conferred features of CQR to 106/1. The first was the complete return to CQS of parasites that had lost their episomes by proliferation in the absence of CQ for one month. Proliferation rates and drug response curves of these lines (e.g., line 34-1/Arm, Table 2) reverted to that of the 106/1 control, with no residual increase in IC90 value. The second finding was the reversal of the IC90 shift in drug response curves by VP in pNHSC-transformed lines, an effect characteristic of CQR but not CQS parasites (Martin et al., 1987). VP alone (0.8 μM) also proved partially inhibitory to growth of the episomally transformed lines, as we have observed with the CQR parasites FCB and Dd2 but not with the CQS parasites HB3 and 106/1.
Transformation with Mutant pfcrt Alleles Expressed in trans from a Plasmid Containing a Human dhfr Selectable Marker Can Also Confer Features of CQR to CQS Parasites
To assess the consequence of pfcrt coexpression in the absence of long-term CQ pressure, we developed the pDC vector that has a selectable human dhfr cassette conferring resistance to the antifolate WR99210. This vector ultimately enabled stable cloning of pfcrt expression cassettes under the control of the P. falciparum calmodulin promoter and allowed transfection of the CQS GC-03 clone with a pDC construct that expresses a truncated Dd2 pfcrt allele encoding all but the last ten amino acids of PfCRT (transfection attempts with full-length Dd2-type pfcrt were not successful). These resulting transformants (line 24-5) exhibited 1.5- to 2.1-fold increases in the CQ IC50 and IC90 values by [3H]hypoxanthine uptake assays, with dose response curves similar to those observed in pNHSC and pNH76I transformants (see below). VP reversibility was also evident in these transformants (Table 2). No change in CQ response was detected in GC-03 lines transformed with alternative pDC constructs expressing either the full-length canonical CQS pfcrt allele, a PCR-amplified pfcrt sequence from Dd2 that contained a frameshift immediately after the transmembrane domain II region (introducing a stop codon after the first 116 codons), full-length pfcrt from Dd2 mutated to encode a lysine at position 76 (analogous to the 106/1 allele), or the full-length neighboring cg6 gene (Table 2).
Continuous Selection of pNHSC-Transformed 106/1 Parasites Produces Stable, Highly CQR Lines
With continued application of CQ pressure to the pNHSC-transformed 34-1 parasites, a line (34-1/E) was obtained that demonstrated markedly increased growth rates comparable to untransformed 106/1 as well as a very high level of CQR (Table 2). IC50 and IC90 values from [3H]hypoxanthine assays with this line were 409 nM and 597 nM, compared to 278 and 385 for Dd2 parasites. The 34-1/E line maintained its CQR phenotype long after removal of CQ pressure (line 34-1/Erm), in contrast to the observed return to sensitivity by the 34-1/A line. CQ IC50 and IC90 values of the 34-1/Erm line were 1.2- to 1.5-fold and 1.2- to 1.9-fold the values of FCB and Dd2, as determined from nine assays performed over three to four months. Microsatellite analysis and DNA fingerprinting (Su et al., 1998, 1999) of the 34-1/Erm, 34-1/E, and 106/1 lines confirmed the related backgrounds and lack of contamination in these lineages.
The Stable, Highly CQR Lines Selected from 106/1 Transformants Lack Episomal DNA and Harbor a Novel pfcrt Mutation Encoding K76I Instead of K76T
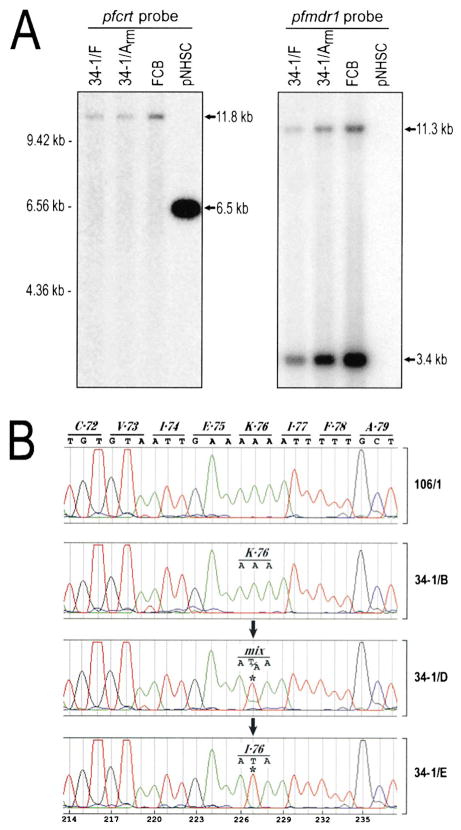
Plasmid rescue experiments with the set of 34-1 transformants detected episomes in lines 34-1/A to 34-1/D but not in lines 34-1/Erm, 34-1/E, and 34-1/F (derived from 34-1/E and grown at 375 nM CQ). PCR analyses revealed no integration of pNHSC into the pfcrt, hrp3, or hrp2 chromosomal regions that share sequences with the plasmid. Hybridization of a pfcrt probe against BamHI-restricted genomic DNA from 34-1/F, 34-1/Arm, and FCB parasites detected the single 11.8 kb band predicted from the known chromosomal sequence (Figure 2A) but no 6.5 kb band from pNHSC (this 6.5 kb band was detected as expected in the episomally transformed line 34-1/A; data not shown).
Figure 2. The Mutant CQR 106/1 Line 34-1/E Does Not Show Size Modifications in the pfcrt or pfmdr1 Genes but Instead Has Acquired a Novel K76I Codon in the pfcrt Sequence.
(A) Southern blot analysis of BamHI-digested P. falciparum genomic DNA (from CQR lines 34-1/F and FCB and the CQS line 34-1/Arm) or pNHSC plasmid DNA hybridized with sequences from pfcrt (left) or pfmdr1 (right). The pfcrt probe can hybridize to both an 11.8 kb BamHI chromosomal fragment including all the pfcrt coding regions and the pfcrt cDNA sequence present in linearized 6.5 kb pNHSC. The pfmdr1 probe is a mixture of 5′ and 3′ sequences that hybridize to 11.3 kb and 3.4 kb BamHI chromosomal fragments. The pfcrt and pfmdr1 hybridization results were the same for FCB and 106/1 as well as for 34-1/F and 34-1/E (data not shown).
(B) The mutant CQR 106/1 line 34-1/E acquired a novel pfcrt mutation encoding K76I. Electropherograms show pfcrt sequences spanning codon 76 in lines 106/1, 34-1/B, 34-1/D, and 34-1/E (DNA prepared on days 74, 112, and 111 post-electroporation, respectively). The 34-1/E, 34-1/Erm, and 34-1/F lines carried only the I76 codon (34-1/E panel; other data not shown). This mutation represented about 65% of the mixed signal from line 34-1/D. Only the original K76 codon was found in the episomally transformed 34-1/B line as well as the CQS lines 34-1/Arm and 106/1.
To explore the basis of high-level resistance in the lines that had lost pNHSC, we determined the chromosomally transcribed pfcrt sequences in the 34-1 series. This analysis revealed a novel point mutation at codon 76 in the pfcrt allele of 34-1/E and its derivative lines (Figure 2B). Remarkably, this mutation was not the K76T change characteristic of other CQR parasites; a novel mutation was instead found in which the hydrophobic isoleucine residue replaced the charged lysine (K76I). This mutation had occurred on the background of the other pfcrt mutations already described for the allele of the 106/1 clone used in these transformation experiments (Table 1). The K76I mutation was also detected along with the original K76 codon in the 34-1/D line, whereas only the original K76 codon was detected in the 34-1/B and 106/1 lines.
We also analyzed the 34-1 lines for possible changes in the pfmdr1 gene. BamHI-restricted DNA probed with 5′ and 3′ pfmdr1 coding sequences did not show alterations in the sizes of the 11.3 and 3.4 kb bands, indicating no intragenic deletions or insertions (Figure 2A). Comparison of the nucleotide sequences of 106/1, 34-1/E, and 34-1/F at known polymorphic positions (Reed et al., 2000) demonstrated that these lines all carried the same pfmdr1 codons for 86Y, 184Y, 1034S, 1042N, and 1246D.
Episomal Expression of a Mutant Form of pfcrt Encoding the K76I Mutation Confers Features of CQR to Diverse CQS Parasites
To test for a causal relationship between CQR and the K76I mutation selected on the 106/1 pfcrt background, we modified the original pfcrt sequence from Dd2 in the pNHSC plasmid to encode the K76I mutation. The modified plasmid, pNH76I, was transfected into the original 106/1 stock and selection was performed with the same CQ treatment as for pNHSC transformation. On three separate occasions, resistant parasites were detected after 20–38 days, attaining 2% parasitemia within 26–52 days. Episomal transformation was confirmed by plasmid rescue and Southern analysis. These lines proliferated in 46 nM CQ at rates 0.67–0.72 that of parental 106/1 and showed CQ IC90 values of 110–141nM, comparable to the values observed with the pNHSC-transformed 34-1/B and 34-1/C lines (Table 2; data shown for the representative line 45-1). VP reversibility was also observed, as was increased sensitivity to VP alone. These pNH76I transformants also readily proliferated at 112 nM CQ (proliferation rates of 1.6–1.9 every 48 hr).
Successful transformations were also achieved with other CQS lines of distinct genetic backgrounds: GC-03 (a progeny clone from the HB3 × Dd2 cross) and the benchmark 3D7 clone. On two separate occasions, transfection of these lines with pNH76I yielded microscopically evident parasites after 31–44 days (Table 2 lists values for one line of each genetic background). These transformants again showed responses in [3H]hypoxanthine uptake assays not representative of the actual rates of proliferation in CQ. Loss of episomes from these lines resulted in complete reversion to the CQS phenotype of the untransformed parental lines (data not shown).
K76T or K76I in Conjunction with Other PfCRT Mutations Is Associated with Increased DV Acidification
Table 3 shows the results of single-cell cytosolic and DV pH (pHcyt, pHvac) determinations from fluorophotometric measurements of individual parasites perfused with BCECF or AO probes. BCECF results indicated that pHcyt values of HB3, 106/1, 34-1/Erm, and FCB were in the range of 7.23–7.44 and showed no association with CQR. In contrast, CQR was associated with significant increases in AO accumulation, yielding more acid pHvac values of 5.08 and 5.23 for the CQR lines versus 5.54 and 5.60 for the CQS lines. A reduction of 0.46 units was determined for pHvac in 34-1/Erm relative to 106/1. This was accompanied by a change in pHcyt to 7.23, resulting in a DV membrane pH gradient for 34-1/Erm comparable to that in FCB and increased relative to the CQS lines HB3 and 106/1.
Table 3.
Compartmental pH in CQR versus CQS Parasites
| HB3 | 106/1 | 34-1/Erm | FCB | |
|---|---|---|---|---|
| pHcyt | 7.31 ± 0.03 | 7.44 ± 0.02 | 7.23 ± 0.03 | 7.41 ± 0.03 |
| pHvac | 5.60 ± 0.03 | 5.54 ± 0.04 | 5.08 ± 0.02 | 5.23 ± 0.03 |
| ΔpH (mV) | 1.71 (101) | 1.90 (112) | 2.15 (127) | 2.18 (129) |
BCECF-based pHcyt (Wunsch et al., 1998; Martiney et al., 1999) and AO-based pHvac (Dzekunov et al., 2000) are shown for the CQS lines HB3 and 106/1 and the CQR lines FCB and 34-1/Erm. Also shown are the calculated ΔpH (vacuole to cytosol) values. Differences in pHcyt between 106/1 and FCB or between pHvac of HB3 and 106/1 are not statistically significant, but other differences are (p <0.001 in each pairwise comparison). pHcyt and pHvac were determined for each line from 35–103 individual measurements from PE. mV = −(2.3 × RT/F) × (pHvac − pHcyt), where R is the gas constant, T is temperature, and F is the Faraday constant.
PfCRT Localizes to the Parasite DV
The subcellular location of PfCRT was initially investigated using the pDC vector modified to express the canonical CQS pfcrt ORF plus the FLAG epitope appended at the carboxyl terminus. FCB parasitized erythrocytes electroporated with this construct (pDC/CRT-FLAG) were placed under selection with 2.5 nM WR99210, yielding the episomally transformed population 33-1/FLAG (first detected at day 41 post-electroporation). M2 monoclonal antibody (mAb) specific to the FLAG epitope detected a Mr 45K band in 33-1/FLAG trophozoites but not untransformed FCB trophozoites (Figure 3A). This Mr 45K band was also detected in 33-1/FLAG DV extracts purified by subcellular fractionation.
Figure 3. The Transmembrane Protein PfCRT Migrates as an Mr 45K Band and Localizes to the Digestive Vacuole.
(A) FLAG-specific M2 mAb recognizes epitope-tagged PfCRT as a Mr 45K protein in transformed 33-1/FLAG trophozoites. Rabbit IgG specific to the PfCRT-K peptide also recognize a protein of the same Mr in trophozoites from the 34-1/Erm line and in DV from the 34-1/Erm and 33-1/FLAG lines.
(B–D) Rabbit anti-PfCRT-K IgG specifically stain the DV in both 34-1/Erm (B and C) and 33-1/FLAG (D) parasites. Parasite location within the erythrocytes is evident in the differential interference contrast images (upper panels). Arrows indicate margins of the imaged intraerythrocytic parasites. Green fluorescence localizes PfCRT to the DV around the hemozoin pigment.
We also localized PfCRT using purified IgG from rabbit antisera raised against the synthetic peptide PfCRT-K. These antibodies showed a strong signal against a 51 kDa GST–PfCRT–His6 fusion protein expressed in E. coli (C2exp; Figure 3A). In extracts of 33-1/FLAG trophozoites, rabbit anti-PfCRT-K IgG recognized a Mr 45K band of the same size as that recognized by the M2 mAb (Figure 3A). These IgG also strongly detected this band in purified DV from both 33-1/FLAG and 34-1/Erm parasites (Figure 3A) but not in Percoll/sucrose subcellular preparation upper layer extracts that contained parasite membranous material depleted of intact DV. No bands were detected in the supernatants corresponding to cytoplasmic fractions obtained after hypotonic lysis of saponin-released parasites.
Immunofluorescence images of 34-1/Erm parasites stained with anti-PfCRT-K IgG localized PfCRT to the DV of trophozoites and schizonts (Figures 3B and 3C). Similar results were also obtained from 33-1/FLAG trophozoites expressing the epitope-tagged PfCRT protein (Figure 3D).
Discussion
Historical records indicate that CQR arose independently in the Old and New World after many years of extensive CQ use (Payne, 1987). This slow genesis of resistance and its steady spread from the initial foci argues for significantly greater genetic complexity than is present in some other forms of resistance, e.g., the single DHFR S108N point mutation that produces P. falciparum resistance to pyrimethamine (Wu et al., 1996). Our evidence suggests that this involves a requirement for multiple mutations in PfCRT that create important DV physiological changes necessary for CQR. All CQR strains from Asia and Africa in our analysis show one of two closely related pfcrt alleles that differ from the canonical CQS allele by seven or eight point mutations. In South America, other distinct pfcrt alleles with multiple mutations are present, in agreement with the independent genesis of CQR in the New World. All Old and New World pfcrt alleles in CQR parasites consistently include mutations for K76T and A220S accompanied by changes in two to six other codons. These observations are consistent with findings that CQR parasites from all regions have comparable verapamil-reversible phenotypes. The significance of K76T and A220S in CQR has recently been reinforced by a CQ trial in West Africa, where these mutations were found in 100% of persisting post-treatment infections versus 40% of randomly sampled pretreatment infections (Djimde et al., submitted).
The transformations reported here represent the successful production of CQR lines from CQS parasites in vitro. These transformations were repeated with three CQS clones distinct in both geographical origin and genetic backgrounds, thereby supporting the genetic cross and population survey results that associate pfcrt mutations with CQR. Transfections with each of two different pfcrt alleles from CQR parasites produced episomal transformants capable of surviving the same CQ concentrations as naturally resistant parasites. While the use of CQ selection in these experiments has the concern that it may have produced point mutations in other genes hypothetically necessary for CQR, several observations weigh against this possibility: (1) removal of CQ pressure from parasites transformed with CQ-selected episomes resulted in loss of the episomes and complete reversion of the parasites to the CQS state; (2) CQS parasites transfected with mutant pfcrt alleles and selected with CQ yielded drug resistant populations within time periods similar to those observed in dhfr transformations that do not require additional point mutations for drug resistance (Table 2; Wu et al., 1996); (3) elevated CQ IC90 values were observed in lines episomally transformed with plasmids that employ human dhfr as a selectable marker and express pfcrt in trans; (4) transfection of CQS lines with control plasmids carrying a pfcrt sequence predictive of CQS, or no pfcrt sequence, did not result in parasites with increased CQ IC90 values; and (5) VP chemosensitization, which perfectly associates with pfcrt and CQR in the HB3 × Dd2 cross, produced reversal of the CQR phenotype in the mutant pfcrt transformed lines selected with either CQ or WR99210.
The [3H]hypoxanthine uptake (or LDH production) profiles from drug assays of the episomal CQR transformants differ from those of naturally CQR parasites. Measurements of the actual proliferation rates by parasite counts from Giemsa-stained blood smears showed that the [3H]hypoxanthine drug assays did not accurately reflect the ability of these transformants to proliferate in high CQ concentrations. These unusual results may be a physiologic consequence of the coexpression of the episomal CQR and endogenous chromosomal CQS pfcrt sequences under the different regulatory controls. Other effects may also be present from variable copy number and irregular parsing of the episomes in the transformed populations. We note that production of CQR by coexpression of two different functional pfcrt sequences differs fundamentally from systems of dhfr transformation, in which there is a functional knock out of the endogenous enzyme in the presence of drug so that expression of the transfected gene provides the only source of active enzyme (Fidock and Wellems, 1997).
The CQS 106/1 P. falciparum clone carries all the pfcrt mutations of an Old World CQR allele except the mutation resulting in the critical K76T codon change. Following transformation of 106/1 parasites with plasmid constructs expressing a CQR pfcrt sequence with this codon change, continued CQ pressure produced a highly CQR line (34-1/Erm) that had lost the transfected DNA and had acquired a novel mutation encoding K76I in the chromosomal pfcrt gene. Our data do not explicitly rule out the possible involvement of a second gene in acquisition of this CQR phenotype. Nevertheless, four observations indicate that the PfCRT K76I mutation in the 106/1 sequence directly contributed to the change in CQ phenotype: (1) the point mutation occurred exactly in the position identified as critical by preceding genetic analyses; (2) the CQR phenotype was VP-reversible, as found with the pfcrt episomal transformants; (3) the K76I mutation in the 106/1 allele appeared after a period similar to that reported for the appearance of single point mutations in other studies (Tanaka et al., 1990; Thaithong et al., 1992); and (4) independent transformations with the pfcrt K76I expression plasmid pNH76I yielded parasites that survived CQ concentrations lethal to the original CQS lines 106/1, GC-03, and 3D7.
Subcellular fractionation and immunofluorescence microscopy place the PfCRT transmembrane protein at the DV. The finding of more acidic pHvac in CQR parasites suggests PfCRT mutations affect the magnitude of the H+ gradient across this membrane. While the 34-1/Erm line carrying K76I also exhibits a more acid pHcyt than the CQR FCB line carrying K76T, both lines possess a similarly increased ΔpH across the DV membrane relative to the CQS HB3 and 106/1 lines, consistent with findings reported by Dzekunov et al. (2000) and Ursos et al. (2000) for CQR parasites. These mutations at position 76 have in common the removal of a positive charge from the first PfCRT transmembrane segment, which may affect ion transport across the DV membrane.
Models of CQR can be envisaged that incorporate the effects of PfCRT mutations in different ways: (1) the 0.3–0.5 unit decrease in pHvac associated with PfCRT mutations reduces drug–heme interactions responsible for toxicity; and (2) drug flux across the DV membrane is directly altered by a structural change in the PfCRT molecule itself or by an effect of PfCRT on the function of other molecules involved in DV physiology. The first model would appear paradoxical if CQ accumulation in the DV were explained only by the weak-base trapping of diprotic CQ predicted by Henderson-Hasselbach equilibrium (Yayon et al., 1984; Krogstad et al., 1985). CQ accumulation in the DV, however, is also driven by binding to soluble heme that acts as a receptor (Fitch, 1970; Bray et al., 1998). Changes in pH are reported to have a steep effect on the conversion of soluble heme to insoluble aggregates (Dorn et al., 1998; Dzekunov et al., 2000). The pH midpoint of this conversion is strikingly close to the pHvac values of CQS parasites, and formation of insoluble heme is much more efficient at the more acid pHvac values of CQR parasites. DV acidification would consequently leave significantly less free heme available for the formation of toxic complexes with CQ, an effect that is consistent with experimental data showing a reduction in high-affinity drug receptor sites (Fitch, 1970; Bray et al., 1998). Such a pH effect, however, may not adequately explain the reported efficacy of certain CQ side chain derivatives on CQR as well as CQS parasites (De et al., 1996; Ridley et al., 1996). This raises consideration of the second model in which PfCRT mutations may directly or indirectly alter a structurally specific drug interaction affecting CQ flux across the DV membrane.
PfCRT mutations may be accompanied by adaptive changes in activities or expression levels of other proteins that affect parasite fitness. Such adaptive changes would be consistent with results from drug-resistant pathogens that show genome-wide changes in response to drug pressure and mutations in drug resistance determinants (Srivastava et al., 1999; Wilson et al., 1999). Levin et al. (2000) report that drug-resistant microorganisms can reduce the physiological “cost of resistance” associated with resistance-conferring mutations by compensatory genetic changes. Our finding that PfCRT mutations associate with large changes in DV pH does raise the possibility that P. falciparum CQR parasites may undergo adaptive adjustments in the levels or activities of additional proteins important to DV physiology. Adjustments associated with CQR may also involve mutations in proteins other than PfCRT. In this regard, we note that the pfmdr1-encoded Pgh-1 molecule has been localized to the DV (Cowman et al., 1991). Though not the determinant of CQR in P. falciparum, Pgh-1 provides an example in which mutations or changes of expression levels may affect the IC50 responses of parasites already resistant to CQ and influence susceptibility to other drugs such as quinine and mefloquine (Reed et al., 2000).
Identification of PfCRT will facilitate molecular investigations into the mechanism of P. falciparum CQR. Assays for PfCRT mutations will also serve for epidemiological surveys of CQR strains in malarious regions.
Experimental Procedures
cDNA Screening
PCR screening of the NF54 cDNA library was performed at 62°C extension temperature with 25 ng DNA and the three primer pairs 5′-AGATGGCTCACGTTTAGGTGGAGG + 5′-TTGTCATGTTTGAAAAGCATACAGG, 5′-CCTTTTTAGGAACGACACCGAAG + 5′-CCCAA GAATAAACATGCGAAACC, and 5′-GTGATGATTGTGACGGAGCA TGG + 5′-CGACGTTGGTTAATTCTCCTTCGG. Complete pfcrt cDNA ORF sequences were obtained from NF54 and Dd2 cDNA libraries and from oligo (dT) primed asexual blood stage RNA by PCR amplification with primers 5′-CACCGCTCGAGTAATTTCTTACATATAACA AAATGAAATTCGCAAG + 5′-CACCGCTCGAGTTATTGTGTAATAA TTGAATCGACG (underlined XhoI sites were used for subcloning).
Mutational Surveys
Cultured-adapted lines were obtained from regions of Asia, Africa, and South America (Su et al., 1997). cDNA pfcrt sequences were determined from overlapping products amplified using primers 5′-CCGTTAATAATAAATACACGCAG + 5′-GTTCTTGTAAGACCTATGAAGGCC, 5′-ATCCATGTTAGATGCCTGTTCAGTC + 5′-CCCAAGA ATAAACATGCGAAACC, 5′-GCTTTTCAAACATGACAAGGG + 5′-CGA CGTTGGTTAATTCTCCTTCGG, and 5′-GTCTTATATTACCTGTAT ACACCC + 5′-CCTTATAAAGTGTAATGCGATAGC. pfcrt genomic DNA sequences were from products amplified using primers 5′-CTTTCCCAAGTTGTACTGCTTC + 5′-CCTTATAAAGTGTAATGCGATAGC or 5′-TGACTTTCGATTGTTCATTCTG + 5′-ATTGGACAA AATTGTTTGCTCC.
Transfection Constructs
The full-length pfcrt ORF from SC-01 (an HB3 × Dd2 progeny clone with the same pfcrt gene as Dd2) was amplified from cDNA with primers 5′-CCTTTTTATGCATTTCGCAAGTAAAAAAAATAATCAAAA AAATTCAAGC + 5′-AATTTCAAGCTTATTGTGTAATAATTGAATCGACGTTG containing NsiI and HindIII sites (underlined). Insertion of the amplified product in place of luc between these sites in pHLH-1 (Wu et al., 1995) yielded the plasmid pNHSC. pNH76I, carrying a pfcrt allele from SC-01 modified to encode 76I, was derived from pNHSC using the primers 5′-GTGTATGTGTAATTGAAATAATTTTTGCTAAAAGAAC + 5′-GTTCTTTTAGCAAAAATTATTTCAATTACACATACAC and the QuikChange mutagenesis system (Stratagene).
To generate the 6.0 kb pDC transgene expression plasmid, we first modified the 7.2 kb plasmid pHC-1 (Crabb et al., 1997) by replacing the BamHI site at the start of the Toxoplasma gondii dhfr-ts gene with a NcoI site. The human dhfr cDNA sequence was then amplified from pHDWT (Fidock et al., 1998) with primers 5′-GCCATGCCATGGTTGGTTCGCTAAACTGCATCG and 5′-TTTGGGTACCCCGCGGTTAATCATTCTTCTCATATACTTC that contain NcoI and KpnI sites (underlined). The amplified product was substituted for the T. gondii dhfr-ts sequence between these sites in the plasmid, yielding pDC. pDC/CRT-GC03 was generated by amplifying and cloning the pfcrt cDNA ORF from the CQS GC-03 parasite into the unique XhoI site of pDC. The pfcrt sequence with the appended C-terminal FLAG epitope coding region was generated from this intermediate with primers 5′-CACCGCTCGAGTAATTTCTTACA TATAACAAAATGAAATTCGCAAG and 5′-CACCGCTCGAGTTAAG ATCTCTTGTCATCGTCGTCCTTGTAGTCACTAGTTTGTGTAATAAT TGAATCGACG (XhoI sites are underlined; FLAG sequence is italicized). The final tagged pfcrt sequence was cloned as a XhoI fragment into pDC, yielding pDC/CRT-FLAG.
Maps of plasmids pNHSC and pDC/CRT-FLAG are available as supplemental data at www.molecule.org/cgi/content/full/6/4/861/DC1.
P. falciparum Transfection
Culturing and transfection of P. falciparum asexual blood stages were performed as described (Fidock and Wellems, 1997; Fidock et al., 1998). 106/1 parasites transfected with pNHSC or pNH76I were treated with 46 nM CQ during days 2–5 and 92 nM CQ during days 6–8 post-electroporation and thereafter selected at 46 nM CQ. Lines were initiated on the following days post-electroporation: 34-1/A, day 0; 34-1/B, day 48; 34-1/C, day 67; 34-1/D, day 88; 34-1/E, day 98; 34-1/Erm and 34-1/F, day 101. Lines GC-03 and 3D7 transfected with pNH76I were selected with 36 nM CQ on days 2–8 and thereafter with 52 and 26 nM CQ, respectively.
Plasmid rescue was by electroporation of 100 ng P. falciparum DNA into E. coli and plating on ampicillin plates (Fidock and Wellems, 1997). Probes for Southern analysis were amplified from the pfcrt gene with the primers 5′-GGTTTCGCATGTTTATTCTTGGG + 5′-TTGCTGGACCTTGTATACAACTAAC (0.62 kb; spanning pfcrt exons 7–10) or from the pfmdr1 gene as a mixture of two 0.55 kb fragments (nt 1735–2280 and 3976–4527; GenBank accession number M29154) with primers 5′-GT(T/A)GG(T/A)TC(T/A)TC(T/A)GG(T/A)TG(T/C)GG(T/A)AAATC(T/A)AC + 5′-ATCTAA(A/T)GC(A/T)GA(A/T)GT(A/T)GCTTCATC (Wellems et al., 1990).
Drug Response Determinations
Parasite proliferation rates were determined from microscopic evaluation of Giemsa-stained culture thin smears every 2–4 days. Drug response determinations using 72 hr [3H]hypoxanthine uptake assays were as described (Fidock et al., 1998). Assays included 3–6 wells of uninfected erythrocytes to adjust for background cpm. Percentage reduction in [3H]hypoxanthine uptake, a surrogate marker of growth inhibition, was calculated by the equation 100 × ([geometric mean cpm of wells without drug] − [mean cpm of test wells])/(geometric mean cpm of wells without drug). Lactate dehydrogenase immunocapture assays were performed as described by Piper et al. (1999).
Vacuolar and Cytosolic pH Determinations by Single-Cell Photometry
Nigericin, acridine orange (AO), and 2′, 7′-bis-(2-carboxyethyl)-5 (and 6)-carboxyfluorescein, acetoxymethyl ester (BCECF, AM) were from Molecular Probes, Inc. Parasitized erythrocytes (PE) were attached to polylysine-treated glass coverslips and incubated for 10 min with probe and 10 units of streptolysin-O (Sigma). The attached PE were continuously perfused with NaHCO3/5% CO2-equilibrated buffers and gassed with 5% O2/5% CO2/90% N2. AO and BCECF exposures in these experiments were determined to be nontoxic via pulse cytotoxicity experiments (Hoffman et al., 1996). pHcyt determinations were made after preincubation in 20 μM BCECF-AM for 30 min. pHvac determinations were made from PE perfused with buffers containing stepwise decreasing concentrations of 1000 nM, 500 nM, 250 nM, and 0 nM AO in 10 min intervals (Luz et al., 1994; Hoffman et al., 1996; Dzekunov et al., 2000). In vivo pH calibration of converted BCECF was via the K+/nigericin method (Luz et al., 1994; Hoffman et al., 1996). Calibration of AO response was via the thin layer method developed for analyzing intact intraerythrocytic P. falciparum (Dzekunov et al., 2000). Localization of AO to the DV and of BCECF to the cytosol was confirmed by comparison of bright-field and fluorescence images of the same PE along with laser confocal microscopy analysis of simultaneous Z-axis series. Single-cell photometry was performed with a modified InCyt Im2 dual wavelength imaging system (Intracellular Imaging Inc.) and 640 × 480 pixel images were captured by an integrating Cohu CCD camera. Data recorded at similar settings from unstained PE and stained but unparasitized erythrocytes were used to correct for nonspecific light scattering.
PfCRT Recombinant Protein Expression, Ab Production, and Protein Analysis
Antisera to the synthetic peptide PfCRT-K (KKMRNEENEDSEG ELTNVDC, corresponding to PfCRT amino acids 401–419 plus an added carboxyterminal cysteine) were raised in rabbits (Spring Valley Laboratories). IgG were purified using protein G–conjugated Sepharose (Amersham Pharmacia).
To generate recombinant PfCRT, the coding region corresponding to amino acids 217–424 was amplified from pDC/CRT-GC03 using the primers 5′-CTGGGATCCTTAATTAGTGCCTTAATTCCT + 5′-CATGAATTCTTAGTGATGGTGGTGATGGTGTTGTGTAATAATTGAATC GAC and ligated as a BamHI + EcoRI (sites underlined) fragment with a His6 tag (italicized) into pGEX-6P1 (Amersham Pharmacia), yielding C2exp. This was transformed into E. coli BL21 (Stratagene), and expression of the resulting 51 kDa GST-PfCRT-His6 polypeptide was induced with 1 mM IPTG.
Parasite trophozoite lysates for SDS–PAGE electrophoresis were prepared from saponin-lysed PE cultures synchronized by sorbitol exposure. DV were isolated using Percoll-sucrose gradient centrifugation as described (Goldberg et al., 1990; Saliba et al., 1998). After SDS–polyacrylamide gel electrophoresis, proteins were transferred to PVDF membranes and detected by chemiluminescence (Amersham Pharmacia) after incubation with rabbit anti-PfCRT-K IgG (1:200 dilution) followed by HRP-conjugated goat anti-rabbit IgG (Jackson Immunoresearch). Loadings were adjusted so that each lane contained protein from similar numbers of parasites.
For immunofluorescence microscopy, PE were deposited onto toxoplasmosis slides (Bellco Glass), air dried, placed in 100% methanol at −70°C for 15 min, air dried again, and blocked in 3% skim milk in PBS for 30 min. Rabbit anti-PfCRT-K IgG (1:400 dilution) were used as primary Ab overnight, followed by Alexa Fluor 488–conjugated goat anti-rabbit IgG (Molecular Probes). Samples were mounted in Vectashield medium (Vector Laboratories). Images were collected on a Leica TSC-NT/SP confocal microscope.
Supplementary Material
Acknowledgments
We are grateful to Owen Schwartz of the NIAID Biological Imaging Facility for confocal microscopy and Brenda Rae Marshall for editorial assistance. P. D. R. thanks the Burroughs Wellcome Fund, and A. K. T. thanks the Howard Hughes Medical Institute Research Scholars Program for support.
Footnotes
GenBank Accession Numbers
Sequences reported in this paper have GenBank accession numbers AF030694 and AF233064-AF233068.
References
- Bitonti AJ, Sjoerdsma A, McCann PP, Kyle DE, Oduola AM, Rossan RN, Milhous WK, Davidson D., Jr Reversal of chloroquine resistance in malaria parasite Plasmodium falciparum by desipramine. Science. 1988;242:1301–1303. doi: 10.1126/science.3057629. [DOI] [PubMed] [Google Scholar]
- Bray PG, Mungthin M, Ridley RG, Ward SA. Access to hematin: the basis of chloroquine resistance. Mol Pharmacol. 1998;54:170–179. doi: 10.1124/mol.54.1.170. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Karcz S, Galatis D, Culvenor JG. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J Cell Biol. 1991;113:1033–1042. doi: 10.1083/jcb.113.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- De D, Krogstad FM, Cogswell FB, Krogstad DJ. Aminoquinolines that circumvent resistance in Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1996;55:579–583. doi: 10.4269/ajtmh.1996.55.579. [DOI] [PubMed] [Google Scholar]
- Dorn A, Vippagunta SR, Matile H, Jaquet C, Vennerstrom JL, Ridley RG. An assessment of drug-haematin binding as a mechanism for inhibition of haematin polymerisation by quinoline antimalarials. Biochem Pharmacol. 1998;55:727–736. doi: 10.1016/s0006-2952(97)00510-8. [DOI] [PubMed] [Google Scholar]
- Dzekunov SM, Ursos LMB, Roepe PD. Digestive vacuolar pH of intact intraerythrocytic P. falciparum either sensitive or resistant to chloroquine. Mol Biochem Parasitol. 2000;110:107–124. doi: 10.1016/s0166-6851(00)00261-9. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Cooper RA, Su X-z, Talley AK, Wellems TE. Allelic modifications of the cg2 and cg1 genes do not alter the chloroquine response of drug-resistant Plasmodium falciparum. Mol Biochem Parasitol. 2000;110:1–10. doi: 10.1016/s0166-6851(00)00249-8. [DOI] [PubMed] [Google Scholar]
- Fitch CD. Plasmodium falciparum in owl monkeys: drug resistance and chloroquine binding capacity. Science. 1970;169:289–290. doi: 10.1126/science.169.3942.289. [DOI] [PubMed] [Google Scholar]
- Ginsburg H, Famin O, Zhang J, Krugliak M. Inhibition of glutathione-dependent degradation of heme by chloroquine and amodiaquine as a possible basis for their antimalarial mode of action. Biochem Pharmacol. 1998;56:1305–1313. doi: 10.1016/s0006-2952(98)00184-1. [DOI] [PubMed] [Google Scholar]
- Goldberg DE, Slater AF, Cerami A, Henderson GB. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc Natl Acad Sci USA. 1990;87:2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Wei LY, Roepe PD. Are altered pHi and membrane potential in hu MDR 1 transfectants sufficient to cause MDR protein-mediated multidrug resistance? J Gen Physiol. 1996;108:295–313. doi: 10.1085/jgp.108.4.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogstad DJ, Schlesinger PH, Gluzman IY. Antimalarials increase vesicle pH in Plasmodium falciparum. J Cell Biol. 1985;101:2302–2309. doi: 10.1083/jcb.101.6.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz JG, Wei LY, Basu S, Roepe PD. Transfection of mu MDR 1 inhibits Na(+)-independent Cl−/−HCO3 exchange in Chinese hamster ovary cells. Biochemistry. 1994;33:7239–7249. doi: 10.1021/bi00189a028. [DOI] [PubMed] [Google Scholar]
- Marsh K. Malaria disaster in Africa. Lancet. 1998;352:924. doi: 10.1016/S0140-6736(05)61510-3. [DOI] [PubMed] [Google Scholar]
- Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- Martiney JA, Cerami A, Slater AF. Verapamil reversal of chloroquine resistance in the malaria parasite Plasmodium falciparum is specific for resistant parasites and independent of the weak base effect. J Biol Chem. 1995;270:22393–22398. doi: 10.1074/jbc.270.38.22393. [DOI] [PubMed] [Google Scholar]
- Martiney JA, Ferrer AS, Cerami A, Dzekunov S, Roepe PD. Chloroquine uptake, altered partitioning and the basis of drug resistance: evidence for a role of chloride-dependent ionic regulation. Novartis Foundation Symposium 226: Transport and Trafficking in the Malaria-Infected Erythrocyte; Chichester: John Wiley and Sons; 1999. pp. 265–274. [DOI] [PubMed] [Google Scholar]
- Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- Peters W, Ekong R, Robinson BL, Warhust DC. Antihistaminic drugs that reverse chloroquine resistance in Plasmodium falciparum. Lancet. 1989;2:334–335. doi: 10.1016/s0140-6736(89)90522-9. [DOI] [PubMed] [Google Scholar]
- Piper R, Lebras J, Wentworth L, Hunt-Cooke A, Houze S, Chiodini P, Makler M. Immunocapture diagnostic assays for malaria using Plasmodium lactate dehydrogenase (pLDH) Am J Trop Med Hyg. 1999;60:109–118. doi: 10.4269/ajtmh.1999.60.109. [DOI] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman A. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Ridley RG, Hofheinz W, Matile H, Jaquet C, Dorn A, Masciadri R, Jolidon S, Richter WF, Guenzi A, Girometta MA, et al. 4-aminoquinoline analogs of chloroquine with shortened side chains retain activity against chloroquine-resistant Plasmodium falciparum. Antimicrob Agents Chemother. 1996;40:1846–1854. doi: 10.1128/aac.40.8.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba KJ, Folb PI, Smith PJ. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem Pharmacol. 1998;56:313–320. doi: 10.1016/s0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Eidelman O, Pollard HB. Pharmacogenomics of the cystic fibrosis transmembrane conductance regulator (CFTR) and the cystic fibrosis drug CPX using genome microarray analysis. Mol Med. 1999;5:753–767. [PMC free article] [PubMed] [Google Scholar]
- Su X-z, Kirkman LS, Wellems TE. Complex polymorphisms in a ~330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- Su X-z, Carucci DJ, Wellems TE. Plasmodium falciparum: parasite typing by using a multicopy microsatellite marker, PfRRM. Exp Parasitol. 1998;89:262–265. doi: 10.1006/expr.1998.4299. [DOI] [PubMed] [Google Scholar]
- Su X-z, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gu HM, Bzik DJ, Li WB, Inselburg J. Mutant dihydrofolate reductase-thymidylate synthase genes in pyrimethamine-resistant Plasmodium falciparum with polymorphic chromosome duplications. Mol Biochem Parasitol. 1990;42:83–91. doi: 10.1016/0166-6851(90)90115-3. [DOI] [PubMed] [Google Scholar]
- Thaithong S, Chan SW, Songsomboon S, Wilairat P, Seesod N, Sueblinwong T, Goman M, Ridley R, Beale G. Pyrimethamine resistant mutations in Plasmodium falciparum. Mol Biochem Parasitol. 1992;52:149–157. doi: 10.1016/0166-6851(92)90047-n. [DOI] [PubMed] [Google Scholar]
- Trape JF, Pison G, Preziosi MP, Enel C, Desgrees du Lou A, Delaunay V, Samb B, Lagarde E, Molez JF, Simondon F. Impact of chloroquine resistance on malaria mortality. C R Acad Sci III. 1998;321:689–697. doi: 10.1016/s0764-4469(98)80009-7. [DOI] [PubMed] [Google Scholar]
- Ursos LMB, Dzekunov SM, Roepe PD. The effects of chloroquine and verapamil on digestive vacuolar pH of P. falciparum either sensitive of resistant to chloroquine. Mol Biochem Parasitol. 2000;110:125–134. doi: 10.1016/s0166-6851(00)00262-0. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Schoolnik GK. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci USA. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kirkman LA, Wellems TE. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc Natl Acad Sci USA. 1996;93:1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunsch S, Sanchez CP, Gekle M, Grosse-Wortmann L, Wiesner J, Lanzer M. Differential stimulation of the Na+/H+ exchanger determines chloroquine uptake in Plasmodium falciparum. J Cell Biol. 1998;140:335–345. doi: 10.1083/jcb.140.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayon A, Cabantchik ZI, Ginsburg H. Identification of the acidic compartment of Plasmodium falciparum-infected human erythrocytes as the target of the antimalarial drug chloroquine. EMBO J. 1984;3:2695–2700. doi: 10.1002/j.1460-2075.1984.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.