Abstract
This paper reports a general in situ method to grow a polymer conjugate solely from the C terminus of a recombinant protein. GFP was fused at its C terminus with an intein; cleavage of the intein provided a unique thioester moiety at the C terminus of GFP that was used to install an atom transfer radical polymerization (ATRP) initiator. Subsequent in situ ATRP of oligo(ethylene glycol) methyl ether methacrylate (OEGMA) yielded a site-specific (C-terminal) and stoichiometric conjugate with high yield and good retention of protein activity. A GFP-C-poly(OEGMA) conjugate (hydrodynamic radius (Rh): 21 nm) showed a 15-fold increase in its blood exposure compared to the protein (Rh: 3.0 nm) after intravenous administration to mice. This conjugate also showed a 50-fold increase in tumor accumulation, 24 h after intravenous administration to tumor-bearing mice, compared to the unmodified protein. This approach for in situ C-terminal polymer modification of a recombinant protein is applicable to a large subset of recombinant protein and peptide drugs and provides a general methodology for improvement of their pharmacological profiles.
Keywords: drug delivery, polymer bioconjugate, protein engineering
Polymer modification of therapeutically relevant proteins is important because it can improve their stability, increase their solubility, enhance their systemic circulation, and also potentially reduce their immunogenicity and antigenicity (1–4). Traditionally, synthetic polymers have been conjugated to proteins by reaction of the polymer with the reactive side chains of protein residues such as lysine or cysteine (5–10). Attachment of PEG to proteins, termed PEGylation, is the most widely used polymer conjugation methodology to improve the pharmacological profiles of proteins (11–14). Recently, in situ polymerization directly from the surface of a protein, in which a polymerization initiator is conjugated to the protein, followed by in situ growth of the polymer has emerged as a promising alternative to postpolymerization conjugation to synthesize protein-polymer conjugates with high yield (15–20). However the chemically reactive residues in most proteins are typically promiscuously distributed on their surface, which can result in the growth of polymers from many sites from the protein, resulting in heterogeneous protein-polymer conjugates with poorly controlled stoichiometry and decreased biological activity compared to the protein, thereby limiting the utility of these methods. Hence, the major challenge in protein-polymer conjugation is to create: (i) site-specific, (ii) stoichiometric protein-polymer conjugates, with (iii) high yield, (iv) good retention of protein activity, and (v) significantly improved pharmacological properties over the native protein.
Recently, we demonstrated a general strategy for the in situ growth of a stoichiometric, PEG-like polymer from the N terminus of a protein by a N-terminal transamination reaction followed by a chemoselective aldehyde-hydroxylamine reaction to install an atom transfer radical polymerization (ATRP) initiator at the N terminus and subsequent in situ growth of a PEG-like polymer -poly(oligo(ethylene glycol) methyl ether methacrylate) [poly(OEGMA)]- from the N terminus of the protein (21). Although this strategy is useful for a subset of proteins and peptide drugs to improve their pharmacological profiles, many proteins and peptide drugs -interferon alpha, glucagon-like peptide-1 (GLP-1), and parathyroid hormone are three examples- cannot be modified at their N terminus because their active site is at or near the N terminus of the molecule (22–24). Given these constraints on the modification of the N terminus of many pharmacologically relevant protein and peptide drugs, and the need for a toolbox of complementary and widely applicable methods for site-specific and stoichiometric polymer conjugation, we turned our attention to the C terminus, because it is the only other ubiquitous and universal site on a proteins’ scaffold. However, the C-terminal carboxylic acid has a similar pKa (∼3.0) to the carboxylic acid group present on the side chains of aspartic acid (pKa ∼ 3.7) and glutamic acid (pKa ∼ 4.0) that are typically distributed on the protein surface, and thus cannot be chemoselectively modified, unlike the N-terminal amine.
To address this challenge, we introduce a unique and potentially general methodology to directly grow a polymer from a recombinant protein at its C terminus; this strategy uses intein-mediated ligation to install an ATRP initiator solely at the C terminus of a protein. An intein is a protein sequence embedded in-frame within a precursor protein sequence that excises itself out, ligating the flanking sequences together through a peptide bond (25–27). This process is known as intein-mediated protein splicing, which is an autocatalytic event (25–27). Mutation of inteins at their N or C terminus allows controllable cleavage of peptide bonds at either the C or N terminus of the inteins. These mutant inteins have been used to isolate recombinant proteins possessing either a C-terminal thioester moiety or an N-terminal cysteine for a variety of applications in biotechnology and chemical biology (28–30). Herein, we used an Asn 198 to Ala mutant of the miniintein, encoded by the Mycobacterium xenopi GyrA gene (Mxe GyrA) that leaves a C-terminal thioester in the target protein upon cleavage of the peptide bond between the C-terminal residue of the target protein and the N terminus of the intein (31, 32).
We exploited the unique thioester group to install an ATRP initiator solely at the C terminus of a protein, followed by in situ ATRP of a polymer to yield a stoichiometric, C-terminal protein-polymer conjugate. We note that although intein-mediated protein ligation has been used for C-terminal protein modification (28–30), it has not been exploited for the in situ growth of a polymer from the C terminus of the protein. We show that a protein-polymer conjugate can be synthesized with high yield, good purity, good retention of protein activity using this methodology, and that the specific polymer chosen for this study -poly(OEGMA)- also has the desirable attribute of significantly improving the plasma pharmacokinetics and in vivo tissue distribution of its conjugate relative to the protein.
Results and Discussion
GFP was selected as a model protein for proof-of-principle of this methodology for the following reasons: (i) GFP is a fluorescent protein, so that it can be easily tracked by absorbance or fluorescence measurements; (ii) the fluorescence of GFP requires proper folding of the protein (33), so that any detrimental changes to its tertiary structure upon attachment of the initiator or the growth of a polymer would be reflected in changes in its absorbance and fluorescence spectrum; and (iii) GFP can be easily expressed with high yield as a recombinant protein in Escherichia coli (E. coli), which made it possible to devise an intein-mediated strategy to install a C-terminal ATRP initiator at the C terminus of GFP.
The C terminus of GFP was fused with the N terminus of the Mxe GyrA miniintein mutant. A purification tag, an elastin-like polypeptide (ELP), was fused to the C terminus of the intein at the gene level (34). We appended an ELP at the C terminus of the intein to enable easy purification of the ternary fusion protein by a nonchromatographic purification method we have developed, inverse transition cycling (ITC) (35). We chose poly(oligo(ethylene glycol) methyl ether methacrylate) (poly(OEGMA)) as the polymer for in situ ATRP because we had previously shown that a myoglobin-poly(OEGMA) conjugate has significantly better pharmacokinetics than the native protein (21).
Intein-Mediated GFP Ligation with an ATRP Initiator at the C terminus.
A two-step procedure was used to attach the ATRP initiator, 2-Amino-N-[2-(2-bromo-2-methyl-propionylamino)-ethyl]-3-mercapto-propionamide (ABMP) to the C terminus of GFP (Fig. 1). First, the ternary GFP-Mxe-ELP fusion was obtained by recombinant overexpression in E. coli, and was purified by ITC. SDS-PAGE analysis of the purification process showed that one round of ITC was sufficient to purify the soluble fusion protein from cell lysate, though we typically used 3–4 rounds to ensure high purity and lack of endotoxins (36) (Fig. S1). After purification by ITC, two bands were observed (lanes 4 and 5 in Fig. S1): a band with a molecular weight (MW) of 88,000 daltons that corresponds to GFP-Mxe-ELP, and a lower MW band with a MW of 60,000 daltons. This lower MW band corresponds to Mxe-ELP, and is a consequence of premature intracellular cleavage of ∼30% of the ternary fusion to yield GFP and Mxe-ELP. The free GFP liberated as a consequence of the premature cleavage of the protein from the intein was not observed in SDS-PAGE because only proteins that have an appended ELP tag are recovered by ITC.
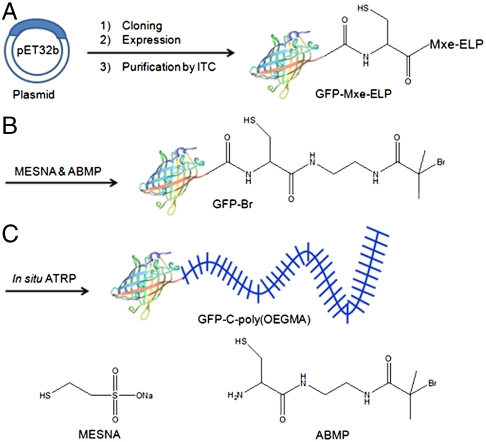
Fig. 1.
Schematic illustration of the synthetic route for GFP-C-poly(OEGMA). (A) Recombinant overxpression of a GFP-Mxe-ELP fusion in E. coli and its purification by ITC. (B) Cleavage of GFP from intein-ELP with a mixture of MESNA and ABMP to create the GFP-Br macroinitiator. (C) In situ growth of poly(OEGMA) from the C terminus of GFP by ATRP to form GFP-C-poly(OEGMA).
Attachment of the ATRP initiator ABMP to the C terminus of GFP was then carried out by addition of ABMP to the GFP-Mxe-ELP fusion protein in the presence of 2-mercaptoethane sulfonate sodium (MESNA) that was added to cleave the intein from the GFP. SDS-PAGE clearly showed that a new band corresponding to the ATRP initiator-modified GFP (named GFP-Br for brevity) appeared with a MW of 28,000 daltons, while the band for GFP-Mxe-ELP almost disappeared and the band corresponding to Mxe-ELP showed an increase in intensity (Fig. 2A). These results indicate that the cleavage efficiency of GFP from the intein was almost quantitative. The crude product was further purified by ITC to remove Mxe-ELP and unreacted GFP-Mxe-ELP, followed by dialysis against PBS to remove low MW impurities. One round of ITC was sufficient to remove residual GFP-Mxe-ELP and Mxe-ELP from GFP-Br (Fig. 2A). In a control experiment, GFP-Mxe-ELP was cleaved solely with MESNA to yield the thioester-terminated GFP (abbreviated to GFP-SO3H). This protein was subsequently utilized as a negative control for the ATRP of OEGMA.
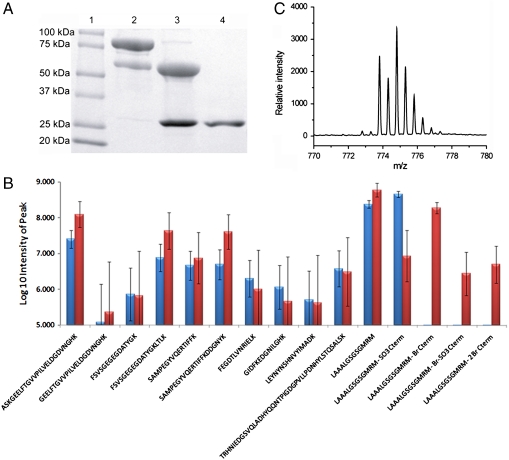
Fig. 2.
Attachment of the ATRP initiator (ABMP) to the C terminus of GFP. (A) SDS-PAGE analysis of cleavage of GFP from Mxe-ELP with a mixture of MESNA and ABMP. Lane 1: protein marker; Lane 2: GFP-Mxe-ELP fusion; Lane 3: GFP-Mxe-ELP fusion after cleavage; Lane 4: GFP-Br after purification by ITC. (B) Log10 intensity of fragment peptides after cleavage by Endo-C protease Blue column: GFP-SO3H; red column: GFP-Br. Note: Log10 = 5 is the approximate S/N limit, (C) Experimental isotope distribution of the C-terminal peptide K.LAAALGSGSGMRM-Br of GFP-Br, wherein M was oxidized.
The purified GFP-Br conjugate was next characterized by electrospray-ionization (ESI)-MS. A major peak was observed at 28,631 daltons, which is close to the theoretical MW of 28,650 daltons of GFP-Br (Fig. S2A). The ESI-MS determined mass of the control, GFP-SO3H was 28,460 daltons (Fig. S2B), which is close to its theoretical MW of 28,476 daltons. Next, the site-specificity of the modification at the C terminus was confirmed by subjecting GFP-Br and GFP-SO3H to proteolytic digestion with endoproteinase Lys-C. Only the C-terminal fragment was observed as a brominated cation (Fig. 2B) by analysis of the resulting peptide fragments of GFP-Br by liquid chromatography/mass spectrometry/mass spectrometry (LC-MS/MS), which indicated that only the C terminus of GFP was modified with ABMP. The experimental isotopic distribution of the C-terminal fragment (Fig. 2C) was comparable to the theoretical bromine isotopic distribution (Fig. S2C), which further confirmed the incorporation of bromine into this C-terminal fragment. In addition, the cysteine residue at the C terminus was partially oxidized with either MESNA or ABMP to form two other possible derivatives (Fig. S2D), but their intensity in LC/MS-MS was almost two orders of magnitude lower than that of the primary brominated cation. The peptide fragments from the control sample GFP-SO3H were experimentally indistinguishable from those observed for GFP-Br except that the C-terminal group was sulfonate, indicating the cleavage occurred only at the C terminus. These data confirm that the ATRP initiator was solely attached to the C terminus of the protein to form a C-terminal brominated macroinitiator (GFP-Br), although there were trace amounts of byproducts that are likely to constitute a few percent of the primary product.
In situ ATRP at the C Terminus of GFP.
Poly(OEGMA) was directly grown by in situ ATRP from the GFP-Br macroinitiator in buffer. The progress of ATRP was monitored as a function of polymerization time by size exclusion chromatography (SEC) (Fig. 3A and Fig. S3A–C) and SDS-PAGE (Fig. S3D). The SEC traces for detection of UV-visible absorbance at 280 nm (Fig. 3A) clearly showed that a new peak was observed with a lower overall elution time than the peak corresponding to GFP-Br, and that this peak shifted to lower elution times with increasing ATRP time. Furthermore, integration of the peak areas showed that the GFP-poly(OEGMA) conjugate constituted ∼80% of the product, with only residual, ∼20% fraction of GFP and GFP-Br, which indicated the high efficiency of ATRP from the C terminus of GFP. The lower overall elution time of this peak with increasing polymerization time also indicated that the MW of the polymer in the conjugate increased with increasing ATRP time. These results were confirmed by detection at 475 nm corresponding to the extinction peak of the GFP chromophore (Fig. S3A), refractive index (RI) (Fig. S3B), and fluorescence emission at 507 nm (Fig. S3C).
Fig. 3.
In situ ATRP at the C terminus of GFP and physical characterization of conjugate. (A) In situ ATRP of OEGMA from the ATRP-initiator-modified C terminus of GFP as a function of polymerization time. The polymerization mixtures were directly analyzed by SEC by UV absorbance at 280 nm. Black trace: GFP-Br; Red trace: 5 min ATRP; Green trace: 15 min ATRP; Blue trace: 120 min ATRP. (B) SEC traces with UV detection at 280 nm, of GFP-C-poly(OEGMA) before and after purification. Black trace: before purification; Red trace: after purification. (C) DLS analysis of GFP and purified conjugate. Black: GFP; Red: conjugate. (D) Fluorescence of GFP and GFP-C-poly(OEGMA) conjugate at a protein concentration of 1 μM at an excitation wavelength of 460 nm and an emission wavelength of 507 nm.
SDS-PAGE analysis also clearly showed that the bands for GFP-C-poly(OEGMA) conjugates had a larger size than GFP-Br (Fig. S3D) and their size increased with increasing ATRP time. Together, the SEC and SDS-PAGE results confirmed the successful ATRP of OEGMA from GFP-Br and the increase in the MW of the polymer with increasing polymerization time. In a control experiment, where GFP-SO3H was used for ATRP under the same polymerization conditions as were used for GFP-Br, the SEC peak traces of the protein did not shift to lower elution times even after 1.5 h (Fig. S4), clearly indicating that ATRP did not occur without an ATRP initiation site on the protein. The absence of a peak corresponding to free polymer also indicated that removal of residual ABMP from the macroinitiator mixture was successful, so that there was no free polymer generated in these ATRP experiments. 1H NMR analysis of the conjugate confirmed the synthesis of poly(OEGMA) from the protein.
In order to further characterize the MW, size, polydispersity, and protein activity of the conjugate, a GFP-C-poly(OEGMA) conjugate that was synthesized by in situ ATRP for 1 h was separated from the residual protein by SEC. The SEC traces for UV absorbance detection at 280 nm of the purified conjugate (red trace, Fig. 3B) showed only one peak with an elution time of 8 min and the absence of a peak corresponding to the unreacted GFP (black trace, Fig. 3B), indicating that the unreacted protein had been completely removed from GFP-C-poly(OEGMA). These results were confirmed by absorbance detection at 475 nm (Fig. S5A), RI (Fig. S5B) and fluorescent detection as well (Fig. S5C). Dynamic light scattering (DLS) analysis also confirmed the efficient purification of the conjugate from the protein. Only one distribution of particles with a hydrodynamic radius (Rh) of 21 nm with a polydispersity of 21% was detected by DLS after purification, with the absence of a species corresponding to GFP, which has a Rh of 3.0 nm as shown in Fig. 3C. Elemental analysis of the GFP-C-poly(OEGMA) conjugate indicated the absence of copper in the conjugate, consistent with our previous work (21). We confirmed the retention of the functional activity of the conjugate by quantifying the fluorescence of GFP and the conjugate at the same concentration of protein (1 μM) (Fig. 3D). The lack of change in the fluorescence of the GFP chromophore clearly indicate that the fluorescence of GFP was retained after in situ growth of a stoichiometric (1∶1) poly(OEGMA) conjugate at the C terminus of the protein.
In vivo Pharmacokinetics.
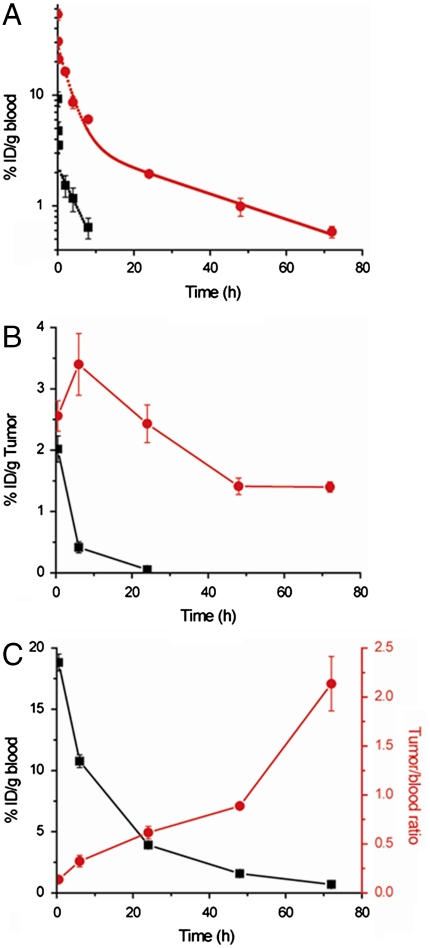
Most proteins and peptide drugs -with the notable exception of antibodies, albumin (37), and transferrin (38)- are rapidly cleared from circulation after their i.v. or s.c. injection. One of the critical issues in improving the therapeutic efficacy of this class of therapeutic molecules is to enhance their systemic circulation or “plasma half-life.” We hence investigated the blood circulation of one of the GFP-C-poly(OEGMA) conjugates synthesized herein by analyzing blood samples at various time points after i.v. injection of 125I labeled GFP and its poly(OEGMA) conjugate ((Rh = 21 nm) into female BALB/c mice. The GFP or conjugate present in the blood, calculated as the percentage of injected dose per gram of blood (% ID/g blood), was plotted against time (Fig. 4A). Unmodified GFP was rapidly cleared from blood with a high clearance rate of 7 mL/h, and exhibited a short distribution phase (t1/2α = 0.1 h) and a rapid terminal elimination phase (t1/2β = 4 h) (Table 1). In contrast, the clearance rate of the conjugate decreased to 0.5 mL/h, the distribution phase of the conjugate increased by 20 times (t1/2α = 2 h) relative to GFP, and the terminal elimination phase was prolonged to 26 h. These pharmacokinetic differences resulted in a 15-fold increase in the area under the curve (AUC) of the conjugate (214%h/mL) compared to unmodified GFP (14%h/mL), which indicates that C-terminal site-specific poly(OEGMA) modification significantly improved the blood exposure of GFP. It is interesting to compare these results with a recent report on an unstructured polypeptide -XTEN- that was developed by high throughput screening for high-level overexpression and a long plasma half-life; a GFP-XTEN fusion exhibited a terminal half-life in rats of 29 h upon i.v. injection (39). Although cross-species comparison of pharmacokinetics is somewhat problematic, evaluation of the pharmacokinetic data for GFP-C-poly(OEGMA) in mice with that of XTEN fused to GLP-1 and GFP across different species suggests that a once-a-week dosing schedule for many peptide drugs such as GLP-1 and other rapidly cleared biologic drugs should be attainable by a C-terminal conjugate of poly(OEGMA). Another important point to note about the poly(OEGMA) conjugates is that because the ATRP kinetics are linear, it is simple to tune the pharmacokinetics of a poly(OEGMA) conjugate length simply by controlling the polymerization time, and hence MW of the polymer. In addition, similar to polypeptide fusions of proteins, poly(OEGMA) is likely to be biodegradable because the oligo(ethylene glycol) (OEG) side chain is linked to the backbone by enzyme-degradable and hydrolyzable ester bonds, so that it may be cleared from the body by renal filtration, though verification of this hypothesis awaits experiments.
Fig. 4.
In vivo pharmacokinetics and tumor accumulation of GFP and GFP-C-poly(OEGMA). (A) Blood concentration as a function of time postinjection. Black square: GFP (Rh = 1,5 nm); Red circle: GFP-C-poly(OEGMA) (Rh = 21 nm). Black and red curves are fits of the data with a two-compartmental model to quantify the elimination and distribution response of GFP and GFP-C-poly(OEGMA). (B) Tumor accumulation as a function of time postinjection. Black curve: GFP; Red curve: GFP-C-poly(OEGMA) conjugate. (C) Tumor to blood ratio as a function of time postinjection of GFP-C-poly(OEGMA) conjugate (Rh = 21 nm). Black square: blood concentration-time course; Red circle: tumor/blood ratio-time course.
Table 1.
Pharmacokinetic parameters calculated from a two-compartment model
| Protein | T1/2α, h | T1/2β, h | AUC, % h/mL | CL, mL/h |
| GFP | 0.1 | 4 | 14 | 7 |
| Conjugate | 2 | 26 | 214 | 0.5 |
The data in Fig. 4A were fitted with a two-compartment model shown as a solid line, to quantify the distribution and elimination response of GFP and GFP-C-poly(OEGMA).
Tumor Accumulation.
One of the critical issues in the delivery of therapeutic molecules is the low accumulation of the drug at diseased sites. In cancer therapy, the accumulation of macromolecules within tumors can be enhanced relative to normal tissues by virtue of the enhanced permeability and retention (EPR) effect, which is a consequence of the leaky vasculature of tumors, and the lack of a fully functional lymphatic system (40, 41). Despite the EPR effect, the accumulation of macromolecules in tumors is typically suboptimal for effective therapy. We hypothesized that the significantly longer blood exposure of the GFP-C-poly(OEGMA) conjugate, coupled with the EPR effect should translate into significantly greater accumulation of the GFP-C-poly(OEGMA) conjugate in solid tumors relative to the native protein. We hence quantified the tumor accumulation of a GFP-C-poly(OEGMA) conjugate and unmodified GFP in female nude BALB/c mice that were implanted with s.c. C26 murine tumors (Fig. 4B). After bolus injection, the concentration of unmodified GFP in the tumor rapidly decreased from 2% ID/g at 30 min to 0.4% ID/g at 6 h. In contrast, the concentration of the conjugate in the tumor increased from 2.5% ID/g tumor tissue at 30 min to 3.4% ID/g tumor tissue at 6 h, and was 50 times greater than that of unmodified GFP at 24 h (Fig. 4B). Similarly, the tumor/blood ratio of the conjugate increased by 21 times from 0.1 at 30 min to 2.1 at 72 h (Fig. 4C). In contrast, unmodified GFP showed a decrease in its tumor/blood ratio with increasing time and the % ID/tumor of GFP was below the detection limit beyond 24 h (Fig. S6). These data clearly indicate that the increasing concentration of the GFP-C-poly(OEGMA) conjugate in the tumor as compared to GFP over the course of 24 h is consistent with its accumulation via the EPR effect, which is typically marked by a slow, steady increase in intratumoral accumulation of a macromolecule over 24–48 h (41, 42). These results on the enhanced tumor accumulation of the GFP-C-poly(OEGMA) conjugate relative to GFP go well beyond our previous paper (21), by demonstrating that an important consequence of a long plasma half-life is enhanced accumulation of the protein-polymer conjugate in a solid tumor, and hence strongly suggest that this protein conjugation technology may be useful to improve the efficacy of protein and peptide drugs and targeting agents under development for cancer imaging and therapy such as interferon α-2b (22), L-asparaginase (43), ) IL-12 (44), and TNF-related apoptosis-inducing ligand (45).
This paper rounds-off the technology platform on site-specific living polymerization from ubiquitous sites on a protein’s scaffold: while our previous paper used a chemical strategy to attach an initiator and grow a polymer from the N terminus (21), this paper, in contrast, demonstrates the utility of posttranslational protein splicing to install a polymerization initiator at the C terminus, followed by in situ living polymerization in buffer from that site alone. Together, these two approaches provide a complete toolbox to grow site-specific and stoichiometric conjugates of peptide and protein therapeutics with improved pharmacological properties.
This paper also illustrates the utility of an ancillary enabling technology for the nonchromatographic purification of recombinant proteins that we have developed that can greatly facilitate site-specific, in situ polymerization from a protein. We showed a decade ago that ELP fusion proteins show stimulus-responsive solubility behavior, which provides a very simple and scalable batch process methodology -ITC- to purify proteins at the 100 mg-gram scale in the laboratory (35). We used the ELP tag to purify the GFP-intein-ELP fusion by exploiting the phase transition behavior of the ELP, then cleaved the intein-ELP from the GFP, and purified away the thioester-modified GFP from the intein-ELP fusion by another round of ITC. Together, the ELP purification technology with that of inteins provides a simple and scalable recombinant methodology that when combined with in situ living polymerization provides a seamless workflow for the expression, purification, and efficient site-specific modification of a protein with a polymer.
Conclusions
We report a unique and general route to synthesize a site-specific (C-terminal) and stoichiometric (1∶1) conjugate at a defined and ubiquitous site on the protein scaffold -the C terminus- by intein-mediated protein ligation to install a polymerization initiator at the C terminus followed by in situ living polymerization of a pharmacologically useful polymer -poly(OEGMA)- with high yield and good retention of protein activity. The results reported herein are significant because: this paper is the first demonstration of the in situ growth of a polymer solely from the C terminus of a protein to create stoichiometric C-terminal polymer conjugate. Second, pharmacologically useful conjugates are obtained by ATRP with a polymerization time that ranges from minutes to an hour in aqueous buffer, with no waste polymer by-product generated so that this methodology is likely to be useful in a manufacturing setting, as it will prove to be relatively cheap, easy to scale-up, and environmentally benign. Third, the C-terminal poly(OEGMA) conjugate significantly prolongs the plasma half-life of a protein, so that this methodology is likely to be useful to improve the pharmacological profiles of a diverse set of protein and peptide therapeutics; examples include interferon alpha, glucagon-like peptide-1 (GLP-1), exendin-4, and parathyroid hormone. Finally, we demonstrate for the first time that the consequence of a long plasma-half-life translates to the enhanced accumulation of the protein-C-poly(OEGMA) conjugate in a solid tumor in a mouse model, which is likely to be useful for the delivery of recombinant protein and peptide therapeutics for cancer therapy.
Methods
Synthesis of ATRP Initiator.
The ATRP initiator 2-Amino-N-[2-(2-bromo-2-methyl-propionylamino)-ethyl]-3-mercapto-propionamide (ABMP) was synthesized by a three-step procedure as described in the SI Text.
Cloning, Expression, and Purification of GFP-Mxe-ELP.
The synthesis of the GFP-Mxe-ELP gene, its expression and purification were based on a method reported previously (33). Details are contained in SI Text.
Cleavage of GFP from Mxe-ELP and Attachment of ABMP.
1 mL of a 250 μM GFP-Mxe-ELP solution in 50 mM Tris-HCl buffer, pH 7.4 was mixed with 1 mL of 5 mM ABMP and 20 mM MESNA in Tris-HCl buffer and allowed to sit without agitation overnight at room temperature. The cleaved GFP-Br was purified by ITC and ultrafiltration (Amicon Ultra-15 centrifugal filter, 3000 daltons molecular weight cut off (MWCO)).
Proteolytic Digestion of GFP.
Protein samples were analyzed using a miniBradford assay to determine protein concentration (Bio-Rad, Inc.) followed by brief digestion with Lys-C by incubation of the protein with Lys-C at a 25∶1 (w/w) ratio for 1 h at 37 °C in 50 mM ammonium bicarbonate. Samples were then acidified in 0.1% formic acid to stop proteolysis. Approximately 2.5 pmol of GFP equivalent (by Bradford assay) was analyzed by LC-MS/MS using a Waters nanoAcquity and Thermo LTQ-Orbitrap XL mass spectrometer (Thermo Corporation).
In situ ATRP from GFP-Br.
A deoxygenated solution of 1.05 mmol OEGMA (MW = 475, Sigma-Aldrich), 0.02 mmol CuCl, 0.044 mmol CuCl2, and 0.087 mmol 1,1,4,7,10,10-hexamethyltriethylenetetramine in 1 mL of PBS was transferred into 4 mL of 100 μM deoxygenated GFP-Br in PBS. The polymerization was allowed to proceed for a specified time under argon and then exposed to air. The polymerization mixture was further purified with HPLC (AKTA, GE Life Science) by SEC.
Attachment of Radiohalogen to GFP.
Iodination of GFP with 125I was performed with a Pierce IODO-Gen precoated tube and purified by gel-filtration on a dextran column (Pierce). Briefly, 100 μL of 27 μM GFP (or conjugate) was added to 1 mCi Na[125I] loaded into IODO-Gen precoated tubes. The tubes were shaken every 30 s for 15 min. The iodinated protein or conjugate was purified by gel-filtration chromatography on a dextran column.
Pharmacokinetics.
All animals were treated in accordance with the NIH Guide for the Care and Use of Laboratory Animals under protocols approved by the Duke University Institutional Animal Care and Use Committee. Typically, eight BALB/c mice were randomly divided into two groups of four mice. Each animal was weighed before injection. Groups received intravenous bolus injections of either 125I labeled GFP or 125I labeled GFP-C-poly(OEGMA) conjugate. The injected dose was ∼0.3 μg GFP (or GFP molar equivalent) with ∼5 μCi of 125I. The injection volume of the sample solution was calculated by (Body weight/9) × 51. Blood samples (10 μL) were collected from the tail vein of the mice at 40 s, 5 min, 15 min, 2 h, 4 h, 8 h, 24 h, 48 h, and 72 h after injection. The blood samples were counted for 125I activity with an automated gamma counter (LKB-Wallac). The blood concentration time-course was analyzed with a standard two-compartment pharmacokinetic model to approximate both the distribution and elimination phase of the molecules.
Tumor Accumulation.
C26 Cells were cultured at 37 °C with 5% carbon dioxide in RPMI-1640 (R8758; Sigma), supplemented with 10% fetal bovine serum (F0392; Sigma), 4.5 g L-1 D-glucose (G8769; Sigma), 10 mM Hepes (15630-080; Invitrogen), and 1 mM sodium pyruvate (11360-070, Invitrogen). Cells were maintained in T-75 flasks (Corning) and passaged twice a week after washing with 7 mL Dulbecco’s PBS without calcium or magnesium (14190-144; Invitrogen) and 2 mL of 0.05% trypsin +0.5 mM EDTA (25300-054; Invitrogen). After 5 min at 37 °C, detached cells were suspended in media, trypsin was removed by centrifugation at 100 x g for 5 min, and cells were resuspended in media. Prior to in vivo implantation, cells were resuspended twice in Minimum Essential Media (51200-038; Invitrogen) and subcutaneously injected (5 × 105 cells in a 30 μL volume) into the back of female nude BALB/c mice. Tumors were allowed to grow to ∼100 mm3 in diameter before starting treatment, typically about 10 d after inoculation. Tumor volume was determined using the equation: volume = (width)2 × length × 0.52 by measuring the size by caliper. After the tumors grew to a size of ∼100 mm3, the mice were randomly divided into groups of five mice each. Each group received intravenous bolus injections of ∼0.34 μg GFP (or GFP molar equivalent) and ∼5 μCi 125I as either 125I labeled GFP or 125I GFP-C-poly(OEGMA) conjugate. At selected time points after i.v. administration, mice were euthanized by decapitation under anesthesia overdose (pentobarbital 250 mg/kg, intraperitoneal (i.p.)). Tumor and blood were collected for γ-counting.
Supplementary Material
Acknowledgments.
We thank David Gooden at the Duke University Small Molecule Synthesis Facility for synthesis of the ATRP initiator, ABMP; M. Arthur Moseley, J. Will Thompson, Erik J. Soderblom, and Laura G. Dubois at the Proteomics facility in the Duke Institute for Genome Sciences and Policy for protein sequencing. This work was supported by a Grant from the National Institutes of Health (NIH) (R01 GM-61232) (to A.C.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006044107/-/DCSupplemental.
References
- 1.Duncan R. The dawning era of polymer therapeutics. Nat Rev Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 2.Duncan R. Polymer conjugates as anticancer nanomedicines. Nat Rev Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 3.Peer D, et al. Nanocarrriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 4.Uchio T, et al. Site-specific insulin conjugates with enhanced stability and extended action profile. Adv Drug Deliver Rev. 1999;35:289–306. doi: 10.1016/s0169-409x(98)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Chilkoti A, Chen G, Stayton PS, Hoffman AS. Site-specific conjugation of a temperature-sensitive polymer to a genetically-engineered protein. Bioconjugate Chem. 1994;5:504–507. doi: 10.1021/bc00030a004. [DOI] [PubMed] [Google Scholar]
- 6.Stayton PS, et al. Control of protein-ligand recognition using a stimuli-responsive polymer. Nature. 1995;378:472–474. doi: 10.1038/378472a0. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Stewart RJ, Kopecek J. Hybrid hydrogels assembled from synthetic polymers and coiled-coil protein domains. Nature. 1999;397:417–420. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RN, Kopeckova P, Kopecek J. Synthesis and evaluation of multivalent branched HPMA copolymer-Fab2 conjugates targeted to the B-cell antigen CD20. Bioconjugate Chem. 2009;20:129–137. doi: 10.1021/bc800351m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzi SC, Hubbell JA. Recombinant protein-co-PEG networks as cell-adhesive and proteolytically degradable hydrogel matrixes. Part I: Development and physicochemical characteristics. Biomacromolecuels. 2005;6:1226–1238. doi: 10.1021/bm049614c. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, et al. Efficient delivery of bioactive antibodies to the cytoplasm of living cells by charge-conversional polyion complex micelles. Angewandte Chemie International Edition. 2010;49:2552–2555. doi: 10.1002/anie.200905264. [DOI] [PubMed] [Google Scholar]
- 11.Hinds K, et al. Synthesis and characterization of poly(ethylene glycol)-insulin conjugates. Bioconjugate Chem. 2000;11:195–201. doi: 10.1021/bc9901189. [DOI] [PubMed] [Google Scholar]
- 12.Nucci ML, Shorr R, Abuchowski A. The therapeutic value of poly(ethylene glycol)- modified proteins. Adv Drug Deliver Rev. 1991;6:133–151. [Google Scholar]
- 13.Harris JM, Martin NE, Modi M. PEGylation: a novel process for modifying pharmacokinetics. Clin Pharmacokinet. 2001;40:539–551. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- 14.Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)-protein conjugates. Adv Drug Deliver Rev. 2003;55:1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- 15.Lele BS, Murata H, Matyjaszewski K, Russell AJ. Synthesis of uniform protein-polymer conjugates. Biomacromolecules. 2005;6:3380–3387. doi: 10.1021/bm050428w. [DOI] [PubMed] [Google Scholar]
- 16.Bontempo D, Maynard HD. Streptavidin as a macroinitiator for polymerization: in-situ protein-polymer conjugate formation. J Am Chem Soc. 2005;127:6508–6509. doi: 10.1021/ja042230+. [DOI] [PubMed] [Google Scholar]
- 17.Heredia KL, et al. In situ preparation of protein-“smart” polymer conjugates with retention of bioactivity. J Am Chem Soc. 2005;127:16955–16960. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, et al. In situ formation of protein-polymer conjugates through reversible fragmentation chain transfer polymerization. Angewandte Chemie International Edition. 2007;46:3099–3103. doi: 10.1002/anie.200604922. [DOI] [PubMed] [Google Scholar]
- 19.Boyer C, et al. Well-defined protein-polymer conjugates via in situ RAFT polymerization. J Am Chem Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- 20.De P, et al. Temperature-regulated activity of responsive polymer-protein conjugates prepared by grafting-from via RAFT polymerization. J Am Chem Soc. 2008;130:11288–11289. doi: 10.1021/ja804495v. [DOI] [PubMed] [Google Scholar]
- 21.Gao W, et al. In situ growth of a stoichiometric PEG-like conjugate at a protein’s N terminus with significantly improved pharmacokinetics. Proc Natl Acad Sci USA. 2009;106:15231–15236. doi: 10.1073/pnas.0904378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YS, et al. Structural and biological characterization of PEGylated recombinant interferon alpha-2b and its therapeutic implications. Adv Drug Deliver Rev. 2002;54:547–570. doi: 10.1016/s0169-409x(02)00027-3. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe Y, et al. Structure-activity relationships of glucagon-like peptide-1(7-36)amide: insulinotropic activities in perfused rat pancreases, and receptor binding and cyclic AMP production in RINm5F cells. Journal of Endocrinology. 1994;140:45–52. doi: 10.1677/joe.0.1400045. [DOI] [PubMed] [Google Scholar]
- 24.Marx UC, et al. Structure-activity relation of NH2-terminal human parathyroid hormone fragments. J Biochem. 1998;273:4308–4316. doi: 10.1074/jbc.273.8.4308. [DOI] [PubMed] [Google Scholar]
- 25.Kane PM, et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 26.Hirata R, et al. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+- translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 27.Perler FB, et al. Protein splicing elements: inteins and exteins—a definition of terms and recommended nomenclature. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 30.Esser-Kahn AP, Francis MB. Protein-cross-linked polymeric materials through site-selective bioconjugation. Angewandte Chemie International Edition. 2008;47:3751–3754. doi: 10.1002/anie.200705564. [DOI] [PubMed] [Google Scholar]
- 31.Evans JTC, Benner J, Xu MQ. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans JTC, Benner J, Xu MQ. The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. J Biol Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 34.Ge X, et al. Self-cleavable stimulus responsive tags for protein purification without chromatography. J Am Chem Soc. 2005;127:11228–11229. doi: 10.1021/ja0531125. [DOI] [PubMed] [Google Scholar]
- 35.Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptide. Nat Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 36.Mchale MK, Setton LA, Chilkoti A. Synthesis and in vitro evaluation of enzymatically cross-linked elastin-like polypeptide gels for cartilaginous tissue repair. Tissue Eng. 2005;11:1768–1779. doi: 10.1089/ten.2005.11.1768. [DOI] [PubMed] [Google Scholar]
- 37.Peters T., Jr Serum albumin. Adv Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 38.Awai M, Brown EB. Studies of the metabolism of I-131-labeled human transferrin. J Lab Clin Med. 1963;61:363–396. [PubMed] [Google Scholar]
- 39.Schellenberger V, et al. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat Biotechnol. 2009;27:1186–1192. doi: 10.1038/nbt.1588. [DOI] [PubMed] [Google Scholar]
- 40.Maeda H, et al. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 41.Dreher MR, et al. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J Natl Cancer Inst. 2006;98:335–344. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 42.Mackay JA, et al. Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish tumours after a single injection. Nat Mater. 2009;8:993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasut G, Sergi M, Veronese FM. Anti-cancer PEG-enzymes: 30 years old, but still a current approach. Adv Drug Deliver Rev. 2008;60:69–78. doi: 10.1016/j.addr.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 44.DelVecchio M, et al. Interleukin-12: biological properties and clinical application. Clin Cancer Res. 2007;13:4677–4685. doi: 10.1158/1078-0432.CCR-07-0776. [DOI] [PubMed] [Google Scholar]
- 45.Walczak H, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nature Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.