Abstract
Intake of toxic cadmium (Cd) from rice caused Itai-itai disease in the past and it is still a threat for human health. Therefore, control of the accumulation of Cd from soil is an important food-safety issue, but the molecular mechanism for the control is unknown. Herein, we report a gene (OsHMA3) responsible for low Cd accumulation in rice that was isolated from a mapping population derived from a cross between a high and low Cd-accumulating cultivar. The gene encodes a transporter belonging to the P1B-type ATPase family, but shares low similarity with other members. Heterologous expression in yeast showed that the transporter from the low-Cd cultivar is functional, but the transporter from the high-Cd cultivar had lost its function, probably because of the single amino acid mutation. The transporter is mainly expressed in the tonoplast of root cells at a similar level in both the low and high Cd-accumulating cultivars. Overexpression of the functional gene from the low Cd-accumulating cultivar selectively decreased accumulation of Cd, but not other micronutrients in the grain. Our results indicated that OsHMA3 from the low Cd-accumulating cultivar limits translocation of Cd from the roots to the above-ground tissues by selectively sequestrating Cd into the root vacuoles.
Cadmium (Cd) is a highly toxic heavy metal for all organisms. In humans, Cd exposure has been associated with cancers of the prostate, lungs, and testes, kidney tubule damages, rhinitis, emphysema, osteomalacia, and bone fractures (1, 2). Cd also inhibits plant growth and development by binding to free sulfhydryl residues and interfering with homeostasis of essential elements, such as zinc, calcium, and iron, or causing their displacement from proteins (3). There is a difference in the level of Cd toxicity between plants and humans; accumulation of Cd to levels endangering human health could occur in crops without showing phytotoxicity (4). Therefore, it is necessary to limit Cd into the food chain from soil to reduce potential health risks to humans.
Rice, an important staple food for nearly a half of the world's population, is a major source of Cd intake (5, 6). The Codex Alimentarius Commission of Food and Agriculture Organization/World Health Organization, which has a responsibility for the safety of food and human health, has set 0.4 mg Cd kg−1 in the polished rice grain as the limit for human intake (7). However, rice grain with Cd exceeding this value is produced on soils contaminated with Cd because of industrial and agro-chemical usages and anthropogenic activities. Furthermore, straw is fed to livestock. Therefore, reducing Cd transport to the above-ground tissues of rice is an important health issue for minimizing the intake of Cd.
There is a large variation in the Cd accumulation of the shoots between rice cultivars (8). Several quantitative trait loci (QTLs) for Cd accumulation have been identified using populations derived from low- and high-Cd cultivars. For example, three putative QTLs on chromosomes 3, 6, and 8 for Cd concentration in brown rice were detected by using chromosome-segment substitution lines derived from Koshihikari and Kasalath (9). By using a similar approach with Kasalath/Nipponbare backcross inbred lines, three different QTLs for Cd concentration in upper plant parts of rice, two on chromosome 4 and one on chromosome 11, were reported (10). A major QTL controlling shoot Cd concentration was also detected on chromosome 11 in a population derived from two contrasting rice cultivars: Badari Dhan and Shwe War (8). More recently, a major QTL for Cd accumulation was detected on the short arm of chromosome 7 in a population derived from a low Cd-accumulating cultivar, Nipponbare, and a high Cd-accumulating cultivar, Anjana Dhan (11). This QTL explains 85.6% of the phenotypic variance in the shoot Cd concentration of the F2 population (11). However, the gene responsible for this QTL has not been cloned so far. In the present study, we isolated the QTL gene on chromosome 7 for Cd accumulation with a map-based cloning technique and characterized this gene in terms of transport activity, expression pattern, and cellular localization. We found that the gene from the low-accumulating cultivar encodes a transporter, which functions as a “firewall” to limit translocation of Cd from the roots to the shoots by sequestrating Cd into root vacuoles. In contrast, the allelic gene from the high Cd-accumulating cultivar lost the function to act as a firewall because of a single amino acid mutation.
Results and Discussion
Map-Based Cloning of the QTL Gene.
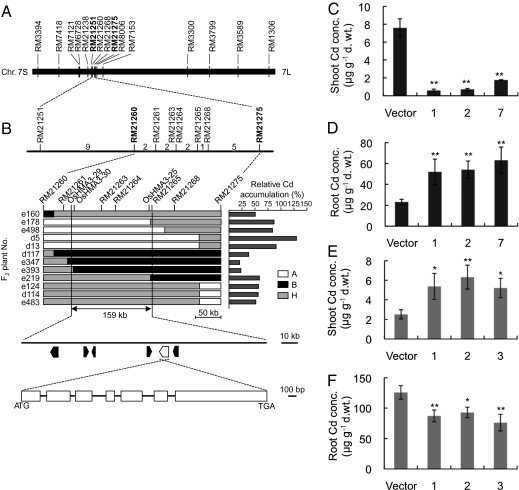
To isolate the gene responsible for Cd accumulation, we used a population derived from an indica cultivar, Anjana Dhan and a japonica cultivar, Nipponbare, which was used for QTL analysis previously (11). Anjana Dhan showed severalfold higher accumulation of Cd than Nipponbare in the shoot (leaf blades and sheaths) and brown rice (de-husked grain) when grown in the same field (11). A physiological study revealed that this difference in the Cd accumulation results from root-to-shoot translocation. Nipponbare, a low Cd-accumulating cultivar, takes up higher Cd into the roots, but retains most Cd in the roots compared with a high Cd-accumulating cultivar, Anjana Dhan (11). To delimit and eventually clone the gene at this QTL, we first analyzed genetically 965 F2 plants. Because there is a good correlation between Cd concentration in the shoots and grains (11), we used Cd concentration in the shoots for mapping the gene. QTL analysis using 67 plants with recombinant between SSR markers RM21238 and RM7153 suggested that the QTL was located near SSR markers RM21260 or RM21268 in the interval defined by RM21251 and RM21275 (SI Appendix, Tables S1 and S2). Based on the association between the genotype of closely linked markers and the level of relative Cd accumulation (SI Appendix, Table S1), plants heterozygous at the candidate region of the QTL showed a slightly increased level of relative Cd accumulation, suggesting that relative Cd accumulation is inherited as a semidominant manner. Because the phenotypic effect of the Anjana Dhan allele at the QTL was very large, we could perform linkage mapping as a single Mendelian factor in further analysis. Plants exhibiting greater than 70% relative Cd accumulation could be classified into a homozygous class of the Anjana Dhan allele at the QTL. The second screening of recombinant from an additional 808 plants could define the candidate region to 159 kb between the markers OsHMA3-29 and RM21265 (Fig. 1 A and B; primer sequences for markers are listed in SI Appendix and Table S4). According to the database of rice cultivar Nipponbare (Rice Annotation Project Database, http://rapdb.dna.affrc.go.jp/), there are six annotated genes (Os07g0231900, Os07g0232200, Os07g0232300, Os07g0232800, Os07g0232900, and Os07g0233300) in this region (SI Appendix, Table S3). Among them, Os07g0232800 and Os07g0232900 encode putative heavy-metal transporters, OsZIP8 and OsHMA3, respectively. OsZIP8 is reported to encode an influx transporter of zinc (12). At first, we isolated the full-length cDNA of OsZIP8 from Anjana Dhan (OsZIP8a) and Nipponbare (OsZIP8n). The deduced polypeptides of OsZIP8a and OsZIP8n were 387 and 390 amino acids, respectively (SI Appendix, Fig. S1) and showed high identity (96.4%). There was no difference in the expression level of OsZIP8 in the roots between two cultivars (SI Appendix, Fig. S2A). Furthermore, when expressed in yeast, both proteins did not show transport activity for Cd (SI Appendix, Fig. S2 B and C), in contrast to AtNramp4 as a positive control, which showed Cd transport activity. All these data indicate that OsZIP8 is not responsible for differential Cd accumulation between Anjana Dhan and Nipponbare.
Fig. 1.
Delimitation of QTL for Cd accumulation. (A) A QTL gene for Cd accumulation was mapped on short arm of chromosome 7 between markers RM21251and RM21275. (B) Candidate genomic region for the QTL and gene structure. A linkage map with the number of recombinants between the molecular markers is indicated (Top Right). Graphical genotypes of plants having recombination in the target region and their relative Cd accumulation are shown. Cd accumulation in the shoots was phenotyped for QTL analysis. “A,” “B,” and “H” indicate homozygous Anjana Dhan, homozygous Nipponbare, and heterozygous allele, respectively. The candidate genomic region was defined 159 kb between OsHMA3-29 and RM21265. The candidate gene consists of seven exons (white box) and six introns (black horizontal lines). (C and D) Complementation test. Cd concentration in the shoots (C) and roots (D) of three independent transgenic lines carrying OsHMA3n and empty vector in Anjana Dhan background. (E and F) Effect of decreased expression of OsHMA3n on Cd accumulation. Cd concentration in the shoots (E) and roots (F) of three independent RNAi lines and vector control plants in Nipponbare background. The plants were exposed to 50 nM Cd for 10 d. Data for C and D are mean ± SD of four independent biological replicates, and for E and F are mean ± SD of three independent biological replicates. The values followed by asterisks are statistically different from the vector control according to a Dunnett's test (*P < 0.05; **P < 0.01).
We then considered Os07g0232900 as a candidate gene. We isolated the full-length cDNA of Os07g0232900 from Anjana Dhan and Nipponbare, and designated as them OsHMA3a (from Anjana Dhan) and OsHMA3n (from Nipponbare), respectively. OsHMA3a and OsHMA3n share 91.4% identity at amino acid level (SI Appendix, Fig. S3). The gene from Nipponbare is predicated to encode a membrane protein, belonging to P1B-ATPase (13). The amino acid sequence similarity of OsHMA3 to HMA3 proteins from other plants is very low, with 39.4% identity to the closest homolog in Arabidopsis (AtHMA3 from ecotype Wassilewskija) and AhHMA3 from Arabidopsis halleri, a Zn-hyperaccumulating plant (SI Appendix, Figs. S4 and S5). Comparison of sequence between OsHMA3n and OsHMA3a showed that OsHMA3a has a 53-amino acid deletion in the C-terminal region in addition to several amino acid changes (SI Appendix, Fig. S3), but this deletion is not responsible for the loss of function of this gene in Anjana Dhan, as shown later in this article.
To demonstrate that Os07g0232900 is a gene responsible for the differential Cd accumulation observed in two cultivars, we introduced this gene from Nipponbare to Anjana Dhan. Analysis with three independent transgenic lines showed that introduction of OsHMA3n into Anjana Dhan resulted in decreased Cd accumulation in the shoots (P < 0.01) (Fig. 1C), but increased Cd accumulation in the roots (P < 0.01) (Fig. 1D). However, there was no difference in the concentrations of other micronutrients and macronutrients in both the roots and shoots between vector control and transgenic lines (SI Appendix, Figs. S6 and S7). This complementation test demonstrated that Os07g0232900 is the gene responsible for differential Cd accumulation between cultivars.
Furthermore, when OsHMA3 expression was knocked down in Nipponbare by RNAi, the concentration of Cd in the shoot was increased by 2.1- to 2.5-times in the RNAi lines compared with the empty-vector control plants (P < 0.05) (Fig. 1E) and the root Cd concentration was decreased by 74 to 60% in the RNAi line (P < 0.05) (Fig. 1F). There was also no difference in the concentration of other micronutrients, including Zn, Cu, Mn, and Fe in both the roots and the shoot between RNAi and vector control lines (SI Appendix, Fig. S8). These results further confirm that OsHMA3 is a gene responsible for differential Cd accumulation observed in two cultivars.
Expression Pattern Analysis.
We determined the expression level of two allelic genes in different tissues of both cultivars by using quantitative real time RT-PCR. OsHMA3 is mainly expressed in the roots at a similar level in two cultivars contrasting in Cd accumulation (SI Appendix, Fig. S9A). Spatial analysis shows that there is no difference in the expression of OsHMA3 among different root segments: 0 to 1 cm, 1 to 2 cm, and 2 to 3 cm from the apex of both cultivars (SI Appendix, Fig. S9B). Furthermore, the expression is not affected by Cd exposure (SI Appendix, Fig. S9B), indicating constitutive expressions of OsHMA3 in two cultivars.
Cellular and Subcellular Localization of OsHMA3.
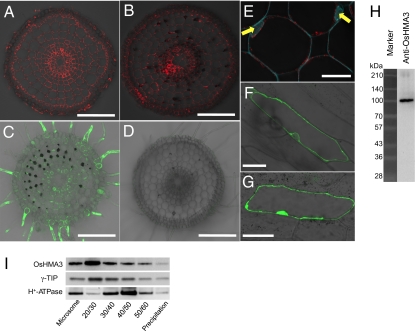
We investigated the localization of the OsHMA3 protein by immunostaining and by using promoter-GFP transgenic rice. Immunostaining with anti-OsHMA3 polyclonal antibodies common for two cultivars showed that OsHMA3 is localized in all root cells and there was no difference in the localization between Anjana Dhan and Nipponbare (Fig. 2 A and B). In the overexpressing line, the signal was greatly enhanced (SI Appendix, Fig. S10A), but the signal was very weak in the RNAi line (SI Appendix, Fig. S10B), indicating the specificity of the antibody. Observation with transgenic plants carrying GFP under the control of the OsHMA3n promoter also showed that OsHMA3n is expressed in all root cells (Fig. 2C). No GFP signal was observed in the wild-type rice (Fig. 2D). Further examination showed that OsHMA3 is localized to the tonoplast (the signal is observed outside of the nucleus) (Fig. 2E). Transient expression of OsHMA3-GFP fusion in onion epidermal cells also showed that OsHMA3 from either cultivar is localized to the tonoplast (Fig. 2F and SI Appendix, Fig. S11A). Coexpression of GFP-OsHMA3 from either cultivar with DsRed-HDEL, an endoplasmic reticulum (ER) marker, further demonstrated the subcellular localization of OsHMA3 at the tonoplast, but not the ER (SI Appendix, Fig. S12). Western blot analysis with antibody against OsHMA3 showed a single band at the predicated size, further indicating the specificity of this antibody (Fig. 2H). Sucrose-density gradient analysis showed that OsHMA3 was present in the same fraction as γ-TIP, a tonoplast marker (Fig. 2I). Taken together, all these results indicate that OsHMA3 is localized to the tonoplast of rice root cells.
Fig. 2.
Localization of Cd transporter OsHMA3 in rice roots. (A and B) OsHMA3 immunolocalization stained with anti-OsHMA3 polyclonal antibody in rice roots of Anjana Dhan (A) and Nipponbare (B). Red color indicates the OsHMA3-specific signal. (C and D) Fluorescence of GFP protein in the roots of pOsHMA3n-GFP transgenic (C) and wild-type Nipponbare (D). (E–I) Subcellular localization of OsHMA3n. (E) Immunostaining of OsHMA3 (red) in the roots of Nipponbare. Cyan color indicates cell wall autofluorescence and nucleus (yellow arrow) stained by DAPI. (F and G) Fluorescence of GFP in onion epidermal cells expressing GFP-OsHMA3n (F) or GFP alone (G). (Scale bars: 100 μm in A–D, F, and G; 10 μm in E). (H and I) Western blot analysis. Microsome extracted from whole roots of OsHMA3n overexpressing lines generated from Nipponbare (137 d old) were used for Western blot analysis with anti-OsHMA3n antibody. (H) Specificity of anti-OsHMA3 antibody. (I) Sucrose-density gradient analysis. The microsome fraction was fractionated by sucrose-density gradient. Polyclonal antibodies of anti-OsHMA3, anti-γ-TIP (tonoplast marker), and anti-H+-ATPase (plasma membrane marker) were used.
Heterologous Assay in Yeast.
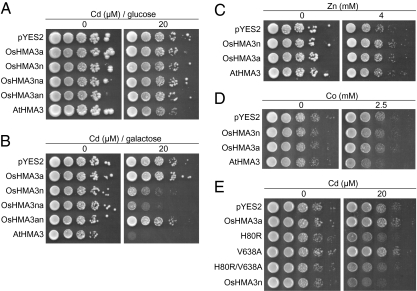
To understand the large difference in Cd accumulation between Anjana Dhan and Nipponbare carrying allelic genes, we expressed each gene in yeast for functional assay. Gene expression from the GAL1 promoter in pYES2 vector is induced in the presence of galactose, but repressed in the presence of glucose (14, 15). There was no difference in the Cd sensitivity between yeast (BY4741 strain) carrying plasmids containing OsHMA3a and OsHMA3n in the presence of glucose (Fig. 3A). However, in the presence of galactose, yeast expressing OsHMA3n showed increased sensitivity to Cd (Fig. 3B), but that expressing OsHMA3a showed similar growth as vector control. Expression of OsHMA3a or OsHMA3n in ycf1, a Cd-sensitive mutant, also yielded the same results (SI Appendix, Fig. S13). These results are consistent with OsHMA3n acting as a Cd transporter in yeast, whereas OsHMA3a appears not to act as a Cd transporter. Expression of either OsHMA3n or OsHMA3a did not alter the sensitivity to Zn in Δzrc1, a Zn-sensitive mutant strain (Fig. 3C) and to Co in the Δcot1 strain (Fig. 3D). In contrast, expression of AtHMA3 as a positive control resulted in enhanced sensitivity to Co (Fig. 3D).
Fig. 3.
Yeast functional assay for Cd and other metals. (A and B) Growth of wild-type yeast cells transformed with empty vector pYES2, OsHMA3n, OsHMA3a, or two chimeric genes of OsHMA3a and OsHMA3n in the presence of glucose (A) to suppress the expression of transformed gene, or galactose (B) to induce the expression of transformed gene. Chimera proteins including N-OsHMA3n-C-OsHMA3a (OsHMA3na) and N-OsHMA3a-C-OsHMA3n (OsHMA3an) were fused at the position of 501. The yeast was grown in the presence of 0 or 20 μM CdSO4. (C) Growth of Δzrc1 strain transformed with empty vector pYES2, OsHMA3n, OsHMA3a, or AtHMA3 in the presence of 0 or 4 mM ZnSO4 and galactose. (D) Growth of Δcot1 strain transformed with empty vector pYES2, OsHMA3n, OsHMA3a, or AtHMA3 as a positive control in the presence of 0 or 2.5 mM CoCl2 and galactose. (E) Growth of wild-type yeast cells transformed with empty vector pYES2, OsHMA3a, site-directed mutagenesized genes at a position of 80 and 638 (OsHMA3aH80R, OsHMA3aV638A, OsHMA3aH80R/V638A), or OsHMA3n in the presence of 0 or 20 μM CdSO4 and galactose. All strains were grown at 30 °C for 3 d.
OsHMA3 is localized at the tonoplast in rice roots (Fig. 2 E–I). If the localization is similar in yeast, expression of functional OsHMA3 should increase the tolerance to Cd. However, yeast expressing OsHMA3n showed increased Cd sensitivity (Fig. 3B). This discrepancy could be attributed to the mislocalization of OsHMA3 in the yeast. OsHMA3 is likely localized to the ER in yeast (SI Appendix, Fig. S14). Therefore, functional OsHMA3 transports Cd into the ER, resulting in increased Cd sensitivity (Fig. 3B).
To dissect the mechanism underlying the loss of function of OsHMA3a, we prepared two chimera proteins between OsHMA3a and OsHMA3n. Because the most different part between OsHMA3a and OsHMA3n is that in the C terminus, missing 53 amino acid residues in OsHMA3a within the putative metal-binding domain repeat (nine repeats in OsHMA3n and six repeats in OsHMA3a) (SI Appendix, Fig. S3), we examined the role of this part in transport activity by fusing the N terminus from OsHMA3a with the C terminus of OsHMA3n or fusing the N terminus from OsHMA3n with the C terminus of OsHMA3a at the position of 501 (SI Appendix, Fig. S15). In N-OsHMA3n-C-OsHMA3a chimera, the Cd sensitivity was increased as observed in OsHMA3n (Fig. 3B). However, the N-OsHMA3a-C-OsHMA3n chimera did not change the Cd sensitivity (Fig. 3B). These results indicate that not missing 53 residues in the C terminus of OsHMA3a, but in the N-terminal region in OsHMA3n, is important for the function.
We then further compared the N-terminal part of two allelic genes. With the help of the transmembrane domain prediction program (SOSUI; http://bp.nuap.nagoya-u.ac.jp/sosui/), we found that only mutations of amino acids at the position of 80 and 638 in OsHMA3a result in change of predicted transmembrane domain numbers (SI Appendix, Fig. S16). To determine whether these mutations are responsible for the loss-of-function of OsHMA3a, we performed a site-directed mutagenesis analysis using yeast expression system. When His at the position of 80 in OsHMA3a was substituted to Arg (H80R), the enhanced sensitivity to Cd was observed (Fig. 3E). However, substitution of Val at the position of 638 to Ala (V638A) did not alter the Cd sensitivity (Fig. 3E). This result is consistent with that of the chimera experiment (Fig. 3B). Substitution of amino acids at both 80 and 638 (H80R/V638A) also gave increased sensitivity to Cd (Fig. 3E). These results indicate that the amino acid at the position of 80 (H80R) might be critical for the function of OsHMA3n. This amino acid may cause topology change as predicated by the SOSUI program; OsHMA3n has eight predicted transmembrane domains, whereas mutation of amino acid at position 80 from Arg to His results in an additional transmembrane domain between TM1 and 2 (SI Appendix, Fig. S16). This amino acid is well conserved in all Zn/Cd transporting subgroup of HMAs (SI Appendix, Fig. S4). However, further work is needed to demonstrate this predication.
Based on these results, we conclude that OsHMA3n from a low Cd-accumulating cultivar (Nipponbare) functions as a firewall by sequestrating Cd into the vacuoles in the roots, keeping the Cd away from the above-ground tissues. In contrast, probably because of the mutation of amino acid at the position of 80 in OsHMA3a from high Cd-accumulating cultivar (Anjana Dhan), this transporter has lost its function to sequester Cd into the vacuoles, resulting in high translocation of Cd from the roots to the shoots. Cd is also supposed to be transported into the vacuoles by Cd2+/H+ antiporters, such as CAX2, CAX4, and MHX (16, 17), but the importance of these transporter activities are not clear, although overexpression of these genes resulted in enhanced accumulation of Cd in tobacco roots. Cd may be formed as a complex with phytochelatins or glutathione and then transported to the vacuoles through an unidentified ABC transporter (18). The form of Cd transport by OsHMA3 remains to be determined.
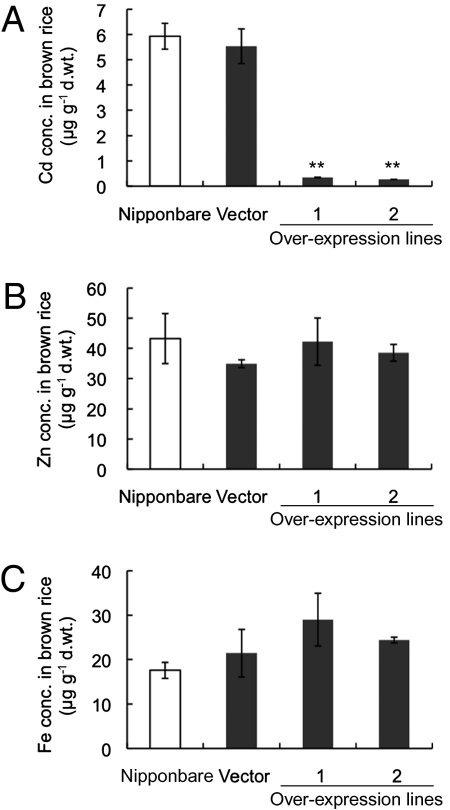
Our results raised a possibility that the Cd accumulation in the rice grains could be reduced by manipulation of the OsHMA3 expression level. To test this possibility, we overexpressed OsHMA3n in a low Cd-accumulating cultivar, Nipponbare, under the control of maize ubiquitin promoter (SI Appendix, Fig. S17). The Cd concentration in the de-husked grains (brown rice) was greatly reduced in the overexpressing lines compared with vector control and the nontransgenic line, Nipponbare, when grown in a Cd-contaminated soil (P < 0.01) (Fig. 4A). The concentration of Zn and Fe in the grains did not differ between the vector control and overexpressing lines (Fig. 4 B and C). A similar trend was observed in the shoots (SI Appendix, Fig. S18).
Fig. 4.
Effect of overexpression of OsHMA3n on Cd accumulation and other metals. Two independent lines overexpressing OsHMA3n in Nippobare, vector control, and a nontransgenic line (cv. Nipponbare) were cultured in a Cd-contaminated soil (1.5 mg Cd kg−1) for 5 mo. Concentration of Cd (A), Zn (B), and Fe (C) in the brown rice is shown. Error bars represent ± SD of three independent biological replicates. The values followed by asterisks are statistically different from the vector control according to a Dunnett's test (**P < 0.01).
Because Cd is a nonessential metal ion, plants are not expected to have specific uptake systems for this metal (19). The uptake of Cd from the soil seems to be mediated by transporters for essential cations, such as Ca, Fe, Mn, and Zn (20). For example, IRT1, a ferrous iron uptake transporter, is also able to transport Cd in Arabidopsis and rice (21, 22). The translocation of Cd from the roots to the shoots is mediated by AtHMA4 and AtHMA2 in Arabidopsis (23). However, these transporters also lack high specificity because they transport Zn in addition to Cd (23, 24). Therefore, it is difficult to control the influx of Cd from soil into the root cells and the release of Cd into the xylem without preventing transport of other cations. However, our results reveal that some cultivars of rice have been evolved to selectively limit Cd taken up by the roots to the above-ground tissues through sequestration of Cd into vacuoles.
The identification of this transporter as being highly specific for Cd is unique. In the group of P1B-ATPase (SI Appendix, Fig. S5), other members show a broad substrate-specificity. For example, AtHMA3, one of the closest homologs of OsHMA3n in Arabidopsis, transports Co, Pb, Cd, and Zn (25). Overexpression of AtHMA3 resulted in increased accumulation of Cd in Arabidopsis shoot (25), which is different from our results on OsHMA3 (Fig. 4A and SI Appendix, Fig. S18). Furthermore, the expression pattern of AtHMA3 is also different from that of OsHMA3. AtHMA3 is expressed in guard cells, hydathodes, vascular tissues, and the root apex (25), whereas OsHMA3 is mainly expressed in all cells of whole roots (Fig. 2 and SI Appendix, Fig. S9). AhHMA3 from a Zn-hyperaccumulating plant, A. halleri, shows transport activity for Zn, but not Cd (26). These differences in the transport substrate specificity may result from the low similarity between OsHMA3 and other P1B-type ATPases (SI Appendix, Figs. S4 and S5). Different from other P-type ATPases, P1B-ATPase is characterized by possessing eight transmembrane domains, a CPx/SPC motif in the transmembrane domain six, and putative transition metal-binding domains at the N and C termini (27). It is proposed that the metal specificity of P1B-ATPases is associated with transmembrane domain six at the CPx/SPC motif and those in transmembrane domains seven and eight (28). However, OsHMA3n has all these features as other members (SI Appendix, Fig. S3), suggesting that the high specificity of OsHMA3 for Cd is regulated by other factors. P1B-ATPases also exhibit about 70-aa HMA domain (referred as PS01047 by PROSITE), including a metal binding domain at the N-terminal region. AtHMA2, AtHMA3, and AtHMA4 contain an HMA domain with a cysteine pair in the sequence GICC(T/S)SE instead of the conserved CxxC pattern and mutations in this domain impair function of AtHMA4 (28, 29). OsHMA3a and -n also have a similar motif but substituted by GVCCSAE (SI Appendix, Fig. S4). These differences possibly affect metal specificity of OsHMA3, although further work is required to explore this theory.
Cd is an important industrial metal in the production of batteries, pigments, coatings, and plating, and as stabilizers for plastics. However, its production and usage cause contamination in the environment, especially in soils as a result of mining, industrial waste disposal, use and disposal of batteries and sludge, and application of pesticides and phosphate fertilizers (30). Because of regulation by law, most soils are not heavily contaminated by Cd, but slightly and moderately contaminated. These levels may not affect the growth of plants, but accumulation of Cd in the plants affect human health through the food chain. Phytoremediation by using plants to remove toxic metals is a promising approach, but it is a time-consuming process. Therefore, the best way to lower Cd accumulation may be to prevent toxic Cd from entering the above-ground tissues, even when crops are grown on Cd-contaminated soils. Identification of the selective transporter for sequestration of Cd in the roots in this study provides an efficient way to breed rice and other crops with low Cd accumulation.
Materials and Methods
An F2 population, derived from a cross between a high Cd-accumulating cultivar (Anjana Dhan) as a female parent and a low Cd-accumulating cultivar (Nipponbare) as a male parent of rice (Oryza sativa L.), was used for fine mapping. The ORF of OsHMA3n was amplified by RT-PCR. The full-length OsHMA3a cDNA was generated by the RACE method from total RNA of Anjana Dhan seedlings (SMART RACE cDNA amplification kit; Clontech) using gene-specific primers, 5′-TGCCAATGTCCTTCTGTTCCCA-3′ for 5′-RACE and 5′-TCCATCCAACCAAACCCGGAAA-3′ for 3′-RACE. Sequence alignment was analyzed by ClustalW (http://clustalw.ddbj.nig.ac.jp/). To generate the hairpin RNAi construct, we cloned a 511-bp fragment (893–1,407 bp from transcriptional start) of OsHMA3n cDNA as inverted repeats into the pANDA vector under control of the maize ubiquitin1 promoter. To generate a construct carrying a ubiquitin promoter, we amplified OsHMA3n cDNA and NOS terminator by PCR. To construct a translational OsHMA3-GFP fusion, we amplified 2 kb of upstream region (34 to 2,094 bp from the translational start codon) of the OsHMA3 gene by PCR from Nipponbare genomic DNA. All constructs were introduced into rice calluses derived from Nipponbare by means of Agrobacterium-mediated transformation. To investigate the expression pattern of OsHMA3 genes, we extracted RNA from the shoots and roots of both cultivars. Spatial expression of OsHMA3 was examined by excising the roots at different segments (0–1 cm, 1–2 cm, and 2–3 cm) of rice exposed to 0 or 1 μM CdSO4 for 24 h. The roots of both cultivars were used for immunostaining of OsHMA3 protein. Fluorescence of secondary antibody (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) was observed with a confocal laser scanning microscopy (LSM700; Carl Zeiss). Saccharomyces cerevisiae reference strain BY4741 (Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and mutant strains Δzrc1 (Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YMR243c::kanMX4) and Δcot1 (Mat a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YOR316c::kanMX4) were used for transport assay. For further details, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kazuko Ono for technical assistance in generating transgenic rice. This research was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan, Genomics for Agricultural Innovation Grant QTL-4005 (to J.F.M), and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan 21248009 (to J.F.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005396107/-/DCSupplemental.
References
- 1.Bertin G, Averbeck D. Cadmium: Cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88:1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Nawrot T, et al. Environmental exposure to cadmium and risk of cancer: A prospective population-based study. Lancet Oncol. 2006;7:119–126. doi: 10.1016/S1470-2045(06)70545-9. [DOI] [PubMed] [Google Scholar]
- 3.DalCorso G, Farinati S, Maistri S, Furini A. How plants cope with cadmium: Staking all on metabolism and gene expression. J Integr Plant Biol. 2008;50:1268–1280. doi: 10.1111/j.1744-7909.2008.00737.x. [DOI] [PubMed] [Google Scholar]
- 4.Chaney RL. Health risks associated with toxic metals in municipal sludge. In: Bitton G, Damro BL, Davidson GT, Davidson JM, editors. Sludge—Health Risks of Land Application. Ann Arbor, MI: Ann Arbor Science; 1980. pp. 59–83. [Google Scholar]
- 5.Cheng F, et al. Cadmium and lead contamination in japonica rice grains and its variation among the different locations in southeast China. Sci Total Environ. 2006;359:156–166. doi: 10.1016/j.scitotenv.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, et al. Gender-related difference, geographical variation and time trend in dietary cadmium intake in Japan. Sci Total Environ. 2004;329:17–27. doi: 10.1016/j.scitotenv.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Codex Alimentarius Commission of Food and Agriculture Organization . Report of the 29th Session of the Codex Alimentarius Commission. Rome: Codex Alimentarius Commission; 2006. [Google Scholar]
- 8.Ueno D, et al. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa) New Phytol. 2009;182:644–653. doi: 10.1111/j.1469-8137.2009.02784.x. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa S, Ae N, Yano M. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa) New Phytol. 2005;168:345–350. doi: 10.1111/j.1469-8137.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 10.Kashiwagi T, Shindoh K, Hirotsu N, Ishimaru K. Evidence for separate translocation pathways in determining cadmium accumulation in grain and aerial plant parts in rice. BMC Plant Biol. 2009;9:8. doi: 10.1186/1471-2229-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno D, et al. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant Cell Physiol. 2009;50:2223–2233. doi: 10.1093/pcp/pcp160. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Huang J, Jiang Y, Zhang HS. Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.) Mol Biol Rep. 2009;36:281–287. doi: 10.1007/s11033-007-9177-0. [DOI] [PubMed] [Google Scholar]
- 13.Baxter I, et al. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003;132:618–628. doi: 10.1104/pp.103.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West RW, Jr, Yocum RR, Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984;4:2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 16.Korenkov V, Hirschi K, Crutchfield JD, Wagner GJ. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta. 2007;226:1379–1387. doi: 10.1007/s00425-007-0577-0. [DOI] [PubMed] [Google Scholar]
- 17.Berezin I, et al. Overexpression of AtMHX in tobacco causes increased sensitivity to Mg2+, Zn2+, and Cd2+ ions, induction of V-ATPase expression, and a reduction in plant size. Plant Cell Rep. 2008;27:939–949. doi: 10.1007/s00299-007-0502-9. [DOI] [PubMed] [Google Scholar]
- 18.Song WY, et al. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol. 2003;21:914–919. doi: 10.1038/nbt850. [DOI] [PubMed] [Google Scholar]
- 19.Palmgren MG, et al. Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 2008;13:464–473. doi: 10.1016/j.tplants.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi H, et al. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Science and Plant Nutrition. 2006;52:464–469. [Google Scholar]
- 23.Hussain D, et al. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verret F, et al. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Morel M, et al. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becher M, Talke IN, Krall L, Krämer U. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 2004;37:251–268. doi: 10.1046/j.1365-313x.2003.01959.x. [DOI] [PubMed] [Google Scholar]
- 27.Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta. 2000;1465:104–126. doi: 10.1016/s0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]
- 28.Williams LE, Mills RF. P(1B)-ATPases—An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Verret F, et al. Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett. 2005;579:1515–1522. doi: 10.1016/j.febslet.2005.01.065. [DOI] [PubMed] [Google Scholar]
- 30.Dahmani-Muller H, van Oort F, Gélie B, Balabane M. Strategies of heavy metal uptake by three plant species growing near a metal smelter. Environ Pollut. 2000;109:231–238. doi: 10.1016/s0269-7491(99)00262-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.