Abstract
Mutations in ASPM (abnormal spindle-like microcephaly associated) cause primary microcephaly in humans, a disorder characterized by a major reduction in brain size in the apparent absence of nonneurological anomalies. The function of the Aspm protein in neural progenitor cell expansion, as well as its localization to the mitotic spindle and midbody, suggest that it regulates brain development by a cell division-related mechanism. Furthermore, evidence that positive selection affected ASPM during primate evolution has led to suggestions that such a function changed during primate evolution. Here, we report that in Aspm mutant mice, truncated Aspm proteins similar to those causing microcephaly in humans fail to localize to the midbody during M-phase and cause mild microcephaly. A human ASPM transgene rescues this phenotype but, interestingly, does not cause a gain of function. Strikingly, truncated Aspm proteins also cause a massive loss of germ cells, resulting in a severe reduction in testis and ovary size accompanied by reduced fertility. These germline effects, too, are fully rescued by the human ASPM transgene, indicating that ASPM is functionally similar in mice and humans. Our findings broaden the spectrum of phenotypic effects of ASPM mutations and raise the possibility that positive selection of ASPM during primate evolution reflects its function in the germline.
Keywords: evolution, cerebral cortex, fertility, neural stem cells, germ cells
To understand the expansion of the cerebral cortex during mammalian evolution, it is necessary to identify the genes that determine cerebral cortical size during development (1). Genes that influence neural progenitor proliferation vs. differentiation are of particular interest, because the shifts in the balance between proliferation and differentiation may explain differences in brain size among mammals (2, 3). Genes in which mutations cause autosomal recessive primary microcephaly in humans, in which patients exhibit a major reduction in brain size in the apparent absence of nonneurological anomalies (4), can provide insight into mechanisms of progenitor cell division that operate in brain development (3).
ASPM (abnormal spindle-like microcephaly associated) (5) is an intriguing candidate gene for the regulation and evolution of brain size in the primate lineage (6) because ASPM mutations cause a substantial reduction in brain size, and the ASPM gene has been the target of positive selection during primate evolution (7–9). Of importance with regard to the underlying mechanism, the reduction in brain size in patients with ASPM mutations concerns all regions of the cerebrum and results in a reduced cortical surface area and a simplified gyral pattern (4, 10). This points to a defect in progenitor proliferation. Consistent with this, knockdown of Aspm in embryonic neural progenitors leads to an increase in asymmetric cell division and premature differentiation (3, 11), implicating the Aspm protein in the regulation of symmetric vs. asymmetric cell division, a crucial process in the balancing of progenitor proliferation vs. differentiation (3). In line with a role in cell division, Aspm localizes to mitotic spindle poles and the midbody (11–13).
ASPM mutations identified in microcephaly patients typically lead to protein truncation, with no correlation between the severity of the disorder and the length of the truncated protein (14, 15). This is consistent with the notion that the lack of the C-terminal domain of ASPM, which mediates midbody localization (13), may be sufficient to cause microcephaly in humans. However, although some nonneurological effects in microcephaly patients with ASPM mutations have been described (10, 16), it is unclear why other tissues are apparently much less affected than the brain although Aspm is expressed in many proliferating tissues (12, 17). Moreover, Aspm expression levels correlate with tumor progression (18, 19), and its knockdown leads to reduced proliferation of glioblastomas (20). In this context, mammalian Aspm may functionally differ from the Drosophila ortholog asp (abnormal spindle), which when mutated causes metaphase arrest in larval neuroblasts (21). In contrast, mouse neuroepithelial cells do not exhibit metaphase arrest upon knockdown (11).
To address these questions, we generated Aspm mutant mouse lines that mimic mutations found in human microcephaly patients. Furthermore, we introduced a human ASPM transgene into these mice to explore the function of human ASPM in the mouse.
Results and Discussion
Mutations in Aspm Cause Microcephaly in Mice.
To study the function of Aspm in the development of the cerebral cortex and elsewhere, we generated two mutant mouse lines from gene trap ES cells (22), in which the endogenous protein is truncated and fused to a β-galactosidase and neomycin phosphotransferase fusion protein (β-geo). Basic characterization of these mouse lines with regard to Aspm mRNA expression and the Aspm–β-geo fusion proteins is described in SI Results (Fig. S1). The insertion site of the gene trap vector in ES cell line AJ0069 [AspmGt(AJ0069)Wtsi] was found to be between exons 25 and 26, and for AA0137 [AspmGt(AA0137)Wtsi] between exons 7 and 8 (Fig. S1A). The mutant mice generated from AJ0069 and AA0137 will be referred to as Aspm1-25 and Aspm1-7 (1-25 and 1-7 in figures), respectively. The truncated Aspm1-7 protein will contain only the microtubule-binding domain (Fig. S1A). In contrast, the predicted mutant protein in Aspm1-25 mice only lacks the C-terminal amino acids encoded by the three 3′ exons but will retain the N-terminal microtubule-binding domain, the calponin homology domains, and the calmodulin-binding isoleucine-glutamine (IQ) repeats (12) (Fig. S1A). In this context, it is worth noting that all ASPM mutations identified in human microcephaly patients (with the exception of a missense mutation) truncate the protein in, or before, the region encoded by exon 26 (15, 23).
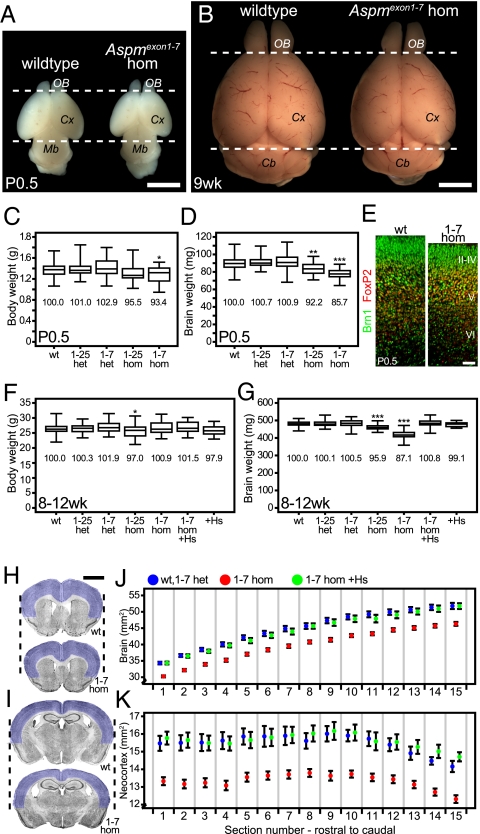
We first addressed whether mutations in Aspm cause microcephaly in mice. Because the clinical definition of human primary microcephaly is a reduction in brain size at birth, which does not progress in severity with age (4), we examined whether microcephaly is observed in newborn mice, on postnatal day (P) 0.5, and in adult mice (8–12 wk). In newborn mice, brains of Aspm1-25- and Aspm1-7-hom (homozygotes) were significantly reduced in weight compared with WT littermates (Fig. 1 A and D), and interestingly, body weight was also reduced (Fig. 1C). Immunostaining for FoxP2, a deep-layer marker, and Brn1, an upper-layer marker, revealed normal cortical layering in Aspm1-7-hom, in an apparently thinner cortex (Fig. 1E), indicating that the reduced brain weight was not due to defects in cortical layering or neuronal migration. Adult brains of homozygous Aspm1-7 mutants showed a significant reduction in brain weight compared with WT (Fig. 1G), similar in proportion as in newborn mice (Fig. 1D); however, body weight was largely unchanged (Fig. 1F). As in humans, the effects of the mutant alleles were recessive because heterozygous (het) mice were indistinguishable from WT mice.
Fig. 1.
Brain size analysis of Aspm mutant and human ASPM transgenic mice. (A and B) Brains from newborn (P0.5) and adult (9 wk) WT and Aspm1-7-hom. Dashed lines delimit the rostrocaudal extent of the WT cerebral cortex (Cx). OB, olfactory bulb; Mb, midbrain; Cb, cerebellum. (Scale bar, 3 mm.) (C and D) Boxplot of whole body (C) and brain (D) weight of P0.5 WT and Aspm1-25 (1-25) and Aspm1-7 (1-7) heterozygous (het) and homozygous (hom) mice. Data from (left to right) 51, 34, 58, 18, and 22 mice. Mean weights compared with WT are indicated as percentages. (E) Double immunofluorescence for Brn1 (green) and FoxP2 (red) of vibratome sections (6-μm optical sections) of P0.5 neocortex. Cortical layers are indicated by II-VI. (Scale bar, 50 μm.) (F and G) Boxplot of whole-body (F) and brain (G) weight of adult (8–12 wk) WT, 1-25, and 1-7 het and hom, Aspm1-7-hom with human ASPM BAC (1-7 hom +Hs), and WT mice with human ASPM BAC (+Hs). Data from (left to right) 65, 37, 27, 35, 59, 42, and 24 mice. Mean weights compared with WT are indicated as percentages. (H and I) Nissl staining of coronal 50-μm vibratome sections of adult brains from WT and 1-7 hom. Dashed lines delimit the lateral extent of the WT sections. Neocortex is highlighted in blue. Note the smaller size of the 1-7 hom brain. (Scale bar, 2 mm.) (J and K) Quantification of whole-brain (J) and neocortex (K) area across 15 sections along the rostrocaudal axis of control (WT and 1-7 het, n = 9, blue), 1-7 hom (n = 20, red), and 1-7 hom +Hs (n = 12, green). Sections in H and I are representative of sections 1 and 15 in the area quantifications, respectively. Data points indicate mean area (square millimeters); error bars indicate SEM. All data points of the 1-7 hom are significantly reduced (P < 0.001) compared with control. In boxplots (C, D, F, and G), the line within the box indicates the median value, the box spans the interquartile range, and whiskers extend to data extremes. *P < 0.05; **P < 0.01; ***P < 0.001.
To analyze whether certain brain regions were affected more than others, we performed histological measurements on adult (8–9 wk) male brains. Serial coronal brain sections were Nissl stained, and a total of 15 sections, anatomically matched between brains, covering much of the rostrocaudal extent of the cerebral cortex, were analyzed (examples of rostral section in Fig. 1H, caudal section in Fig. 1I). Whole-section area (Fig. 1J) and the area of the six-layered neocortex (Fig. 1K, indicated in blue in Fig. 1 H and I) were measured. Data from WT and Aspm1-7 heterozygotes (Aspm1-7-het) were pooled and treated as control and were compared with littermate Aspm1-7-hom. Results showed a significant reduction in whole-section (Fig. 1J) and neocortex (Fig. 1K) area in Aspm1-7-hom compared with control. The mean area of the whole section and of the neocortex of Aspm1-7-hom was reduced by a similar proportion (88.4% and 86.2% of control, respectively).
The same experiment was performed for Aspm1-25-hom (n = 10) compared with littermate controls (WT and Aspm1-25-het, n = 8). Whole-section area of the homozygote was 96.1% (not statistically significant) and of neocortex was 95.0% (P < 0.05) of control.
Thus, mutations in Aspm reduce brain size in mice, similar in nature to, albeit with less severity than, human primary microcephaly. Possible origins of this difference in severity, assuming that the Aspm1-25 and Aspm1-7 alleles are loss-of-function alleles equivalent to ASPM alleles causing microcephaly in humans, may lie in the difference in brain size itself. With a 1,000-fold larger cortical surface area, a longer neurogenic interval, and more progenitor divisions in humans compared with mice (1), potential effects caused by lack of Aspm function may accumulate and be more detrimental the more the progenitor cells divide. Alternatively, differences in the structure and/or the regulation of Aspm between humans and mice could be responsible for the different effects on brain size.
Human ASPM Transgene Rescues the Microcephaly Phenotype but Does Not Produce a Gain of Function in Mice.
To be able to directly test the functional significance of the amino acid substitutions that have affected ASPM in the primate lineage (7–9) as well as to test unknown differences in Aspm regulation, we generated transgenic mice with a bacterial artificial chromosome (BAC) that contains the human ASPM locus. Qualitatively, embryos transgenic for human ASPM expressed ASPM mRNA (Fig. S1B) with the same spatial pattern as endogenous mouse Aspm (Fig. S2).
Mice transgenic for the human ASPM BAC exhibited normal brain (Fig. 1G) and body (Fig. 1F) weights, both in the WT background (+Hs in figures) and Aspm1-7 homozygous background (1-7 hom +Hs in figures). Therefore, human ASPM can rescue the effects of mouse Aspm mutations. Results were further substantiated by the histological brain size measurements described above, which showed that the human ASPM BAC can fully rescue the microcephaly phenotype but does not produce a gain of function (Fig. 1 J and K). The full rescue, but lack of an increase in brain size beyond the WT level, has interesting implications as to the role of ASPM evolution with regard to brain development. In light of the evidence for positive selection of ASPM, our data do not exclude that the human protein may exert functions in the development of the human gyrencephalic brain that are not exerted by the mouse protein and to which the rodent model is not receptive. Be this as it may, our results imply that human ASPM can exert the function of mouse Aspm in the development of the rodent lissencephalic brain.
Midbody Localization Defects of Mutant Aspm Proteins.
Immunofluorescence analysis of the subcellular localization of the Aspm1-25 and Aspm1-7 mutant proteins in mitotic neuroepithelial cells of embryonic day (E) 11.5 dorsal telencephalon showed that although the mutant proteins could localize to spindle poles in metaphase (Fig. S3A), they failed to localize to the midbody in telophase (Fig. S3B) (for details, see SI Results). These findings indicate that the N-terminal portion of Aspm encoded by the first seven exons is sufficient to direct metaphase spindle pole localization, whereas the lack of the C-terminal domain encoded by the last three exons is sufficient to cause a midbody localization defect (13). Furthermore, we found that the conserved C-terminal domain contains Armadillo repeats (Fig. S3C), which are lacking in the Aspm1-7 mutant protein and disrupted in the Aspm1-25 mutant protein, and disrupted in the most C-terminal mutation identified to date in microcephaly patients (10059C→A) (15, 23). Other proteins with Armadillo repeats have been shown to localize to the midbody and are crucial for cytokinesis (24). Because in the case of the Aspm1-25 mutant, and in human patients, disruption of this domain alone causes microcephaly, this suggests that the Armadillo repeat may be crucial for proper Aspm function.
Analysis of Neural Progenitor Cell Function.
Dysregulation of vertical cleavage planes of neuroepithelial and radial glial cells (collectively referred to as apical progenitors) to a more oblique and horizontal orientation, and hence their switch from symmetric to asymmetric division, can lead to premature differentiation and have consequences for brain size (3). However, quantitation of cleavage plane orientation of neuroepithelial cells of E11.5 dorsal telencephalon of Aspm1-25 and Aspm1-7 mutant mice revealed no major alterations compared with WT (Fig. S4 A and B). Likewise, no significant change in the ratio of symmetric vs. asymmetric cell divisions of Aspm1-7-hom compared with WT was observed (Fig. S4 C–E) (for details, see SI Results).
Precocious differentiation of apical progenitors into neurons or downstream basal (intermediate) progenitors may lead to depletion of the apical progenitor pool and result in brain size reduction (2, 3). To address whether the microcephaly observed in Aspm mutants is due to premature differentiation, we quantified the proportion of Tis21-GFP+, neurogenic progenitors, and Tbr2+ basal progenitor cells in the embryonic dorsal telencephalon (Fig. S5 A–C and D, Upper). No significant change was observed in Aspm1-7-hom compared with WT. The mitotic index of apical and basal progenitors also did not show a significant change (Fig. S5D, Lower) (for details, see SI Results).
In Drosophila asp mutants, neuroblasts become arrested in metaphase (21). By contrast, analysis of E11.5 dorsal telencephalon neuroepithelial cells of Aspm1-25 and Aspm1-7 mutant mice did not reveal a detectable metaphase block or defect in mitotic progression (Fig. S5 E and F) (for details, see SI Results).
In the cell counting at E11.5 and E13.5 described above (Figs. S4 and S5), no obvious increase in pyknotic nuclei, revealed by DAPI staining, was noticed in the two homozygotes compared with WT, indicating that an increase in apoptosis is not a likely explanation for the origin of the microcephaly.
The present results are in contrast to RNAi knockdown of Aspm, which led to increased asymmetric division (11). One possibility for this difference is that the present mutants generate truncated Aspm proteins that still localize to the mitotic spindle (Fig. S3A) and thus may partially fulfill the function of the full-length Aspm protein. Furthermore, alterations in mitotic cleavage plane orientation may only be achieved by acute ablation, such as by RNAi. In this context, it remains to be determined whether symmetric vs. asymmetric division is altered in human fetuses with ASPM mutations.
Mutations in Aspm Reduce Fertility in Males and Females.
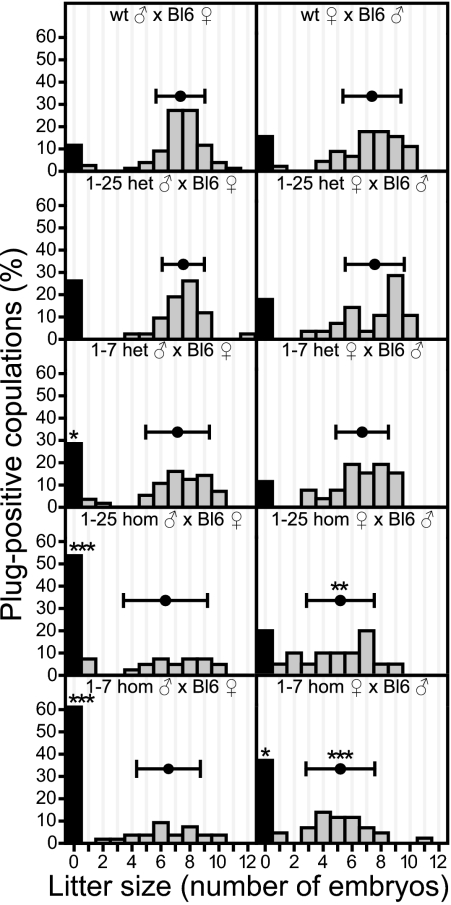
During the course of this study, we noticed that Aspm homozygous mutant mice exhibited a reduction in breeding efficiency. To systematically investigate the effects of Aspm mutations on fertility, mutant males and females were paired with C57BL/6 (BL6) females and males, respectively (Fig. 2). Plugs were observed in females of all genotypes at similar frequency, indicating that copulation frequency was not affected. Aspm1-25 and Aspm1-7 homozygous males mated with BL6 females exhibited a much lower pregnancy rate (number of pregnancies per plug) and also subtle changes in litter size (Fig. 2, Left). Interestingly, Aspm1-25 and Aspm1-7 heterozygous males exhibited an intermediate phenotype in pregnancy rate when mated with BL6 females. Aspm1-25 and Aspm1-7 homozygous females mated with BL6 males had significantly fewer embryos, and in the case of Aspm1-7-hom females, also pregnancies (Fig. 2, Right). Taken together, results show that Aspm mutant males and females exhibit a reduction in pregnancy rate and offspring number.
Fig. 2.
Mutations in mouse Aspm reduce fertility in males and females. Frequency of occurrence of various litter sizes, expressed as percentage of all vaginal plug-positive copulations, for WT and 1-25 and 1-7 het and hom males (Left) and females (Right) paired with C57BL/6 females and males, respectively. Black columns indicate the percentage of females with no embryos (litter size = 0), gray columns the percentage of litters with embryos. Mean number of embryos in pregnant females is illustrated above the gray columns; horizontal error bars indicate SD. Data from (left, top to bottom) 74, 40, 55, 35, and 54 matings and (right, top to bottom) 45, 28, 26, 20, and 39 matings. *P < 0.05; **P < 0.01; ***P < 0.001.
Reduction in Sperm Count and Motility in Aspm Mutant Mice.
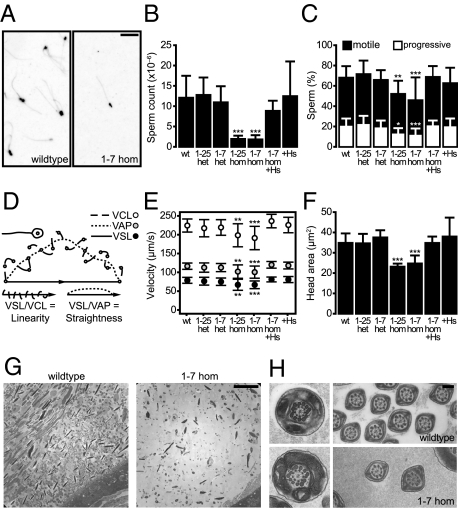
Given the reduced fertility in Aspm homozygous males, we proceeded with an analysis of sperm. In live epididymal sperm observed by dark-field microscopy, we observed a much lower number of sperm in Aspm1-7-hom compared with WT (Fig. 3A). To quantify epididymal sperm count, motility parameters, and morphometric features, freshly isolated epididymal sperm were analyzed in a sperm analyzer. The sperm count of Aspm1-25- and Aspm1-7-hom was ≈5- to 10-fold lower than in WT (Fig. 3B).
Fig. 3.
Sperm analysis in Aspm mutant mice. (A) Dark-field image (inverted) of live epididymal sperm from WT and 1-7 hom mice. (Scale bar, 50 μm.) (B, C, E, and F) Epididymal sperm parameters of adult (8–12 wk) WT, 1-25 and 1-7 het and hom, 1-7 hom +Hs, and +Hs mice, obtained using the IVOS sperm analyzer. Data are the mean of (left to right) 21, 10, 11, 8, 13, 7, and 8 mice; error bars indicate SD; *P < 0.05; **P < 0.01; ***P < 0.001. (B) Sperm count. (C) Percentage of sperm categorized as motile (black columns), of which a subset is categorized as progressive (white column segments). (D) Schematic illustration of sperm velocity parameters defined by the sperm analyzer: the track speed of motile sperm defined as the VCL (dashed lines), VAP (dotted lines), and VSL (solid line with arrowhead) calculated from the distance between the start and the end of the track. Furthermore, sperm movement can be characterized by the ratios VSL/VCL (linearity) and by VSL/VAP (straightness). (E) Sperm velocity parameters as defined in D: VCL (white circles), VAP (gray circles), and VSL (black circles). (F) Sperm head area. (G and H) Transmission electron microscopy of adult epididymis from adult WT (G, Left; H, Upper) and 1-7 hom (G, Right; H, Lower) mice. Note the reduction in cell density within the epididymis (G) and the normal axoneme ultrastructure (H) of the 1-7 hom compared with WT. (Scale bar, 10 μm in G; 200 nm in H.)
Aspm1-25- and Aspm1-7-hom showed a significant reduction in the proportion of total sperm that were motile or progressive (Fig. 3C). In addition, the following velocity parameters were measured: the track speed of motile sperm defined as the curvilinear velocity (VCL; Fig. 3D, dashed line), a smoothed average path velocity (VAP; Fig. 3D, dotted line), and the straight line velocity (VSL; Fig. 3D, solid line with arrowhead), which is calculated from the distance between the start and the end of the track. The two homozygotes showed a significant reduction in all three classes of sperm velocity parameters (Fig. 3E). Furthermore, sperm movement can be characterized by the ratios VSL/VCL, defined as linearity, which represents the linearity of the movement itself, and by VSL/VAP, defined as straightness, which represents the straightness of the track from beginning to end (Fig. 3D). No significant differences were observed in these two ratios (Fig. 3E), indicating that despite the reduction in velocity, the nature of movement did not differ. Interestingly, sperm from the two homozygotes showed a significant reduction in head area (Fig. 3F). However, the head shape, as defined by elongation (ratio of the minor to major axis of sperm head) did not differ, indicating that, despite the reduction in size, the sperm heads were morphologically normal. Importantly, all of the sperm parameter changes observed in Aspm1-7-hom described above were rescued back to WT levels in Aspm1-7-hom transgenic for the human ASPM BAC (Fig. 3 B, C, E, and F).
We next addressed, by transmission electron microscopy, whether the reduction in sperm motility and velocity was due to defects in the axoneme structure of the tail. Within the intact epididymis of Aspm1-7-hom, the density of cells appeared drastically reduced compared with WT (Fig. 3G). Despite a reduction in sperm cells, the homozygote exhibited normal axoneme structure (Fig. 3H).
Aspm Mutations Cause Major Defects in the Male and Female Germlines.
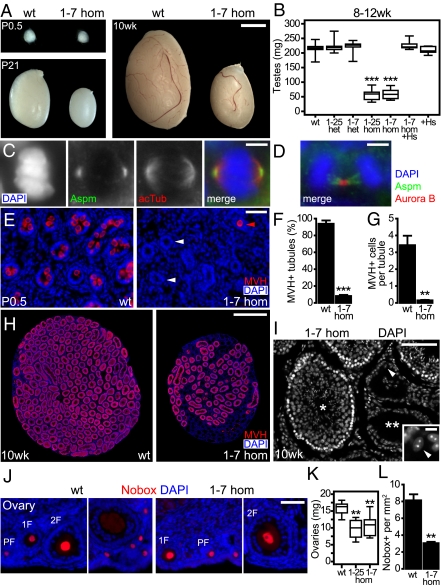
We next investigated testes from newborn (P0.5), juvenile (P21), and adult (8–12 wk) homozygous mutant mice (Fig. 4A). At P0.5, testes from Aspm1-7-hom appeared slightly reduced in size, and at P21 and in adults a dramatic reduction was observed. Testis weight was drastically reduced in Aspm1-25- and Aspm1-7-hom and rescued back to WT levels in Aspm1-7-hom transgenic for the human ASPM BAC (Fig. 4B). In the seminiferous tubules of adult testes, Aspm was found to localize to the spindle poles of spermatogenic cells in metaphase (Fig. 4C) and at the midbody in telophase (Fig. 4D); these are subcellular localizations identical to those in neuroepithelial cells (Fig. S3 A and B).
Fig. 4.
Aspm mutations cause major defects in the male and female germlines. (A) Testes from newborn (P0.5), juvenile (P21), and adult (10 wk) WT and 1-7 hom mice. (Scale bar, 2 mm.) (B) Boxplot of testis weight (sum of left and right testis) of adult (8–12 wk) WT, 1-25 and 1-7 het and hom, 1-7 hom +Hs, and +Hs mice. Data from (left to right) 34, 13, 13, 12, 16, 7, and 10 mice. (C and D) Double immunofluorescence for Aspm (green) with acetylated α-tubulin (acTub, red) in C, and with Aurora B (red) in D, combined with DAPI staining (blue), of 10-μm cryosections (epifluorescence) of an adult testis from a WT mouse. A spermatogenic cell in metaphase is shown in C, and in telophase in D. (Scale bar, 5 μm.) (E) Immunofluorescence for MVH (red) with DAPI staining (blue) of 10-μm cryosections (epifluorescence) of testes from P0.5 WT (Left) and 1-7 hom (Right) mice. In WT, numerous MVH+ cells are found in the seminiferous tubules, whereas in 1-7 hom, most tubules lack germ cells (white arrowheads), and only a few tubules contain MVH+ gonocytes (red arrowhead). (Scale bar, 50 μm.) (F and G) Quantification of seminiferous tubules containing MVH+ cells, expressed as a percentage of the total number of tubules contained in testis sections (F), and the average number of MVH+ cells per seminiferous tubule in testis sections (G), from P0.5 WT and 1-7 hom mice. Data are the mean of testes from three mice (sum of five sections per testis). (H) Immunofluorescence for MVH (red) combined with DAPI staining (blue) of 10-μm cryosections (epifluorescence) of testes, cut orthogonally to the longitudinal axis, from 10-wk-old WT (Left) and 1-7 hom (Right) mice. In WT, essentially every seminiferous tubule contains MVH+ spermatogenic cells, whereas in 1-7 hom, only approximately half of the tubules contain MVH+ cells. (Scale bar, 1 mm.) (I) DAPI staining of a 10-μm cryosection (epifluoresence) of a testis from a 10-wk-old 1-7 hom mouse, showing a tubule containing spermatogenic cells (single asterisk), and empty tubules (double asterisk) containing only few Sertoli cells, identified by the characteristic centromeric heterochromatin condensed in two chromocenters revealed by DAPI staining (Inset). (Scale bar, 100 μm, 10 μm in Inset.) (J) Immunofluorescence for Nobox (red) combined with DAPI staining (blue) of 10-μm cryosections (epifluorescence) of ovaries from adult WT (Left) and 1-7 hom (Right) mice. All follicular stages—primordial (PF), primary (1F), secondary (2F), and antral follicles—were observed in WT and 1-7 hom ovaries. (Scale bar, 50 μm.) (K) Boxplot of ovary weight (sum of left and right ovary) of adult (10–12 wk) WT and 1-25 and 1-7 hom mice. Data from (left to right) 7, 4, and 9 mice. (L) Quantification of Nobox+ cells per total ovary section area from adult (10–12 wk) WT and 1-7 hom mice. Data are the mean of ovaries from three mice (sum of five sections per ovary). In boxplots (B and K), the line within the box indicates the median value, the box spans the interquartile range, and whiskers extend to data extremes. **P < 0.01; ***P < 0.001.
A reduction in testicular size may be a reflection of the reduction in the number of germ cells within the seminiferous tubules (25). To investigate this, P0.5 testis sections were immunostained for the mouse vasa homolog (MVH; Fig. 4E), at this stage a marker of gonocytes or prespermatogonia (26). In WT P0.5 testes, essentially every tubule contained MVH+ cells (Fig. 4E, Left), whereas most tubules of Aspm1-7-hom were lacking MVH+ cells (Fig. 4E, Right, and F). The number of MVH+ cells per tubule also showed a strong reduction (Fig. 4G). Thus, there was massive reduction of germ cells in Aspm1-7-hom at birth.
Considering the reduction in adult testis size of the homozygotes, we investigated whether germ cells were still present in the seminiferous tubules of the adult testis by MVH immunostaining, at this stage a marker of spermatogenic cells from the spermatogonia to the round spermatid stage (26). In Aspm1-7-hom, although spermatogenic cells were still present, many tubules did not contain MVH+ cells (Fig. 4H, Right). By DAPI staining (Fig. 4I), these empty tubules contained only Sertoli cells, which were identified by their characteristic centromeric heterochromatin condensed in two chromocenters (27) (Fig. 4I, Inset).
To investigate the origin of reduced fertility in Aspm homozygous mutant females, ovaries from adult (10–12 wk) mice were analyzed. Ovary weight was significantly reduced in Aspm1-25- and Aspm1-7-hom (Fig. 4K). We also analyzed whether there is a reduction in oocyte number, by immunostaining ovary sections for Nobox (Fig. 4J), a marker of oocytes in primordial, primary, secondary, and tertiary (antral) follicles (28). Aspm1-7-hom ovaries appeared histologically normal, with follicles at the various stages readily identifiable (Fig. 4J). However, quantification of the number of Nobox+ oocytes (pooling all of the follicle stages) per ovary section area (including the corpus luteum) revealed an approximately 3-fold reduction in oocyte number in Aspm1-7-hom compared with WT (Fig. 4L).
Because a large reduction in germ cells was observed already at birth in the male germline, that is, before the onset of germ cell differentiation and meiosis, this indicates that the origin of the defect is in either one, or a combination of (i) initial specification of primordial germ cells at E6.25–7.25, (ii) the subsequent proliferation of these cells, (iii) the migration of these cells to the genital ridge, and/or (iv) increased apoptosis during any of the above processes (29, 30). The additional germline defects observed, such as the reduction in sperm motility and head size, indicate further phenotypes seemingly unrelated to the initial loss of gonocytes.
Drosophila asp mutants also exhibit major germline defects, specifically in spermatogenesis and oocyte differentiation due to abnormal meiotic spindles, and also in germ cell proliferation (31, 32). Furthermore, studies in Caenorhabditis elegans show a role for Aspm in meiotic spindle organization (33). These phenotypes raise the possibility of pleiotropic effects of Aspm mutations in several aspects of the mammalian germline. Aspm mutant mice also showed a reduction in body weight at birth, which has been described for some human patients (10, 16), pointing to the possibility of additional defects in other organs.
Conclusion
The magnitude of the brain and germline phenotypes in the mice are in strong contrast, with the brain exhibiting only mild microcephaly, whereas the gonads exhibit a massive loss of germ cells. Primordial germ cells and undifferentiated primitive gonocytes are believed to be closely related to multipotent cells, because they express key markers of pluripotent cell lineages and give rise to teratomas and embryonic carcinomas in vivo and multipotent stem cells in vitro (34). Because the number of gonocytes was strongly reduced, Aspm mutations may be more detrimental to undifferentiated multipotent cells than to committed progenitor cells, such as those predominating in the developing brain during neurogenesis, which did not show any observable abnormality in the Aspm mutant mice. Accordingly, the mild microcephaly observed in the Aspm mutant mice may reflect a reduction in neural stem cells that took place already before the onset of neurogenesis; this would also explain the proportionality of the reduction in brain size. Furthermore, the finding that the proliferation of primordial germ cells seems to be affected upon Aspm mutation, together with the facts that ASPM is highly expressed in many cancers (12) and its expression levels correlate with tumor progression (18, 19), suggests a role for ASPM in the proliferative expansion of many different cell types.
The observation that Aspm mutations dramatically affect germline function raises a number of issues that should be addressed. The question arises whether germline defects similar to those seen in Aspm mutant mice are also present in patients with primary microcephaly caused by mutations in ASPM. Furthermore, our results raise the possibility that the positive selection of ASPM in the primate lineage may be related to the reproductive system rather than brain size (35). In fact, the detection of positive selection in ASPM relies on the observation of a high rate of nucleotide substitutions that affect the amino acid sequence of ASPM normalized to the rate of nucleotide substitutions that do not affect the protein. This effect is seen on the human evolutionary lineage, where it correlates with an increase in brain size, but also in chimpanzees and gorillas (8). Because evolutionary changes frequently affect spermatogenesis (36), these points taken together raise the possibility that selection on fertility, which is frequent among primates (37), may have influenced the evolution of ASPM in apes and humans.
Materials and Methods
Mouse Lines.
Mouse lines were maintained in pathogen-free conditions in the animal facility of the Max Planck Institute of Molecular Cell Biology and Genetics (Dresden, Germany). Experiments were performed in accordance with German animal welfare legislation. Gene trap ES cells were obtained from the Sanger Institute Gene Trap Resource (Hinxton, Cambridge, United Kingdom). Details on mouse lines and genotyping are described in SI Materials and Methods.
Molecular, Morphological, and Functional Analyses.
Tissue dissection, fixation, cryosectioning, vibratome sectioning, immunofluorescence, and Nissl staining were performed as previously described (SI Materials and Methods). Other methods of molecular, morphological, and functional analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Sanger Institute Gene Trap Resource (Hinxton, Cambridge, United Kingdom) for the gene trap ES cells; D. H. Douglas-Hamilton (Hamilton Thorne Inc.) and Drs. YoonJeung Chang and Jérôme Gilleron for discussions. J.N.P. and J.L.F. were members of the International Max Planck Research School for Molecular Cell Biology and Bioengineering. W.B.H. was supported by Deutsche Forschungsgemeinschaft Grant SFB 655, A2, the Deutsche Forschungsgemeinschaft-funded Center for Regenerative Therapies Dresden, and the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010494107/-/DCSupplemental.
References
- 1.Rakic P. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 3.Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains—the evolution of neural-progenitor-cell division. J Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- 4.Woods CG. Human microcephaly. Curr Opin Neurobiol. 2004;14:112–117. doi: 10.1016/j.conb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 6.Ponting C, Jackson AP. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr Opin Genet Dev. 2005;15:241–248. doi: 10.1016/j.gde.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouprina N, et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mekel-Bobrov N, et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 10.Saadi A, et al. Compound heterozygous ASPM mutations associated with microcephaly and simplified cortical gyration in a consanguineous Algerian family. Eur J Med Genet. 2009;52:180–184. doi: 10.1016/j.ejmg.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouprina N, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet. 2005;14:2155–2165. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- 13.Paramasivam M, Chang YJ, LoTurco JJ. ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle. 2007;6:1605–1612. doi: 10.4161/cc.6.13.4356. [DOI] [PubMed] [Google Scholar]
- 14.Bond J, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas AK, et al. The molecular landscape of ASPM mutations in primary microcephaly. J Med Genet. 2009;46:249–253. doi: 10.1136/jmg.2008.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passemard S, et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology. 2009;73:962–969. doi: 10.1212/WNL.0b013e3181b8799a. [DOI] [PubMed] [Google Scholar]
- 17.Lüers GH, Michels M, Schwaab U, Franz T. Murine calmodulin binding protein 1 (Calmbp1): Tissue-specific expression during development and in adult tissues. Mech Dev. 2002;118:229–232. doi: 10.1016/s0925-4773(02)00253-8. [DOI] [PubMed] [Google Scholar]
- 18.Hagemann C, et al. Expression analysis of the autosomal recessive primary microcephaly genes MCPH1 (microcephalin) and MCPH5 (ASPM, abnormal spindle-like, microcephaly associated) in human malignant gliomas. Oncol Rep. 2008;20:301–308. [PubMed] [Google Scholar]
- 19.Lin S-Y, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14:4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 20.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.do Carmo Avides M, Glover DM. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- 22.Skarnes WC, et al. International Gene Trap Consortium. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gul A, et al. Novel protein-truncating mutations in the ASPM gene in families with autosomal recessive primary microcephaly. J Neurogenet. 2007;21:153–163. doi: 10.1080/01677060701508594. [DOI] [PubMed] [Google Scholar]
- 24.Wolf A, et al. The armadillo protein p0071 regulates Rho signalling during cytokinesis. Nat Cell Biol. 2006;8:1432–1440. doi: 10.1038/ncb1504. [DOI] [PubMed] [Google Scholar]
- 25.Chubb C. Genes regulating testis size. Biol Reprod. 1992;47:29–36. doi: 10.1095/biolreprod47.1.29. [DOI] [PubMed] [Google Scholar]
- 26.Toyooka Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 27.Hsu TC, Cooper JE, Mace ML, Jr, Brinkley BR. Arrangement of centromeres in mouse cells. Chromosoma. 1971;34:73–87. doi: 10.1007/BF00285517. [DOI] [PubMed] [Google Scholar]
- 28.Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111:137–141. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 29.McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 30.Saitou M. Germ cell specification in mice. Curr Opin Genet Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Casal J, Gonzalez C, Wandosell F, Avila J, Ripoll P. Abnormal meiotic spindles cause a cascade of defects during spermatogenesis in asp males of Drosophila. Development. 1990;108:251–260. doi: 10.1242/dev.108.2.251. [DOI] [PubMed] [Google Scholar]
- 32.Riparbelli MG, Massarelli C, Robbins LG, Callaini G. The abnormal spindle protein is required for germ cell mitosis and oocyte differentiation during Drosophila oogenesis. Exp Cell Res. 2004;298:96–106. doi: 10.1016/j.yexcr.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 33.van der Voet M, et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- 34.Kanatsu-Shinohara M, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Ponting CP. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 2006;22:1031–1035. doi: 10.1093/bioinformatics/btl022. [DOI] [PubMed] [Google Scholar]
- 36.Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays. 2010;32:26–36. doi: 10.1002/bies.200900127. [DOI] [PubMed] [Google Scholar]
- 37.Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.