Abstract
The covalent attachment of SUMO (small ubiquitin-like modifier) to other intracellular proteins affects a broad range of nuclear processes in yeast and animals, including chromatin maintenance, transcription, and transport across the nuclear envelope, as well as protects proteins from ubiquitin addition. Substantial increases in SUMOylated proteins upon various stresses have also implicated this modification in the general stress response. To help understand the role(s) of SUMOylation in plants, we developed a stringent method to isolate SUMO-protein conjugates from Arabidopsis thaliana that exploits a tagged SUMO1 variant that faithfully replaces the wild-type protein. Following purification under denaturing conditions, SUMOylated proteins were identified by tandem mass spectrometry from both nonstressed plants and those exposed to heat and oxidative stress. The list of targets is enriched for factors that direct SUMOylation and for nuclear proteins involved in chromatin remodeling/repair, transcription, RNA metabolism, and protein trafficking. Targets of particular interest include histone H2B, components in the LEUNIG/TOPLESS corepressor complexes, and proteins that control histone acetylation and DNA methylation, which affect genome-wide transcription. SUMO attachment site(s) were identified in a subset of targets, including SUMO1 itself to confirm the assembly of poly-SUMO chains. SUMO1 also becomes conjugated with ubiquitin during heat stress, thus connecting these two posttranslational modifications in plants. Taken together, we propose that SUMOylation represents a rapid and global mechanism for reversibly manipulating plant chromosomal functions, especially during environmental stress.
Keywords: mass spectrometry, SUMO, TOPLESS, chromatin remodeling, stress
Posttranslational modification of proteins has emerged as a central regulatory mechanism that underpins a wide range of cellular processes. One essential modification in eukaryotes involves the reversible attachment of the ∼100-amino-acid protein small ubiquitin-like modifier (SUMO) to other intracellular proteins (1, 2). SUMO becomes covalently linked by an isopeptide bond between its C-terminal glycine and the ε-amino group of lysines within the target via an ATP-dependent reaction cascade involving the sequential action of single E1-activating and E2-conjugating enzymes, and a diverse collection of E3-ligase enzymes. SUMO addition can then be reversed by a family of de-SUMOylating enzymes which cleave the isopeptide linkage. Most often a single SUMO moiety is attached to the target but in some cases polymeric SUMO chains are assembled (1, 2). Many attachment sites conform to a consensus ΨKXE sequence, where Ψ is a large hydrophobic amino acid and K represents the lysine that binds SUMO (3).
Over a decade of work, primarily in yeast and mammalian cell cultures, shows that SUMOylation controls a broad spectrum of cellular activities through the modification of predominantly, but not exclusively, nuclear proteins. These include roles in gene expression, maintenance of chromatin integrity, signal transmission, nuclear trafficking, and in stabilizing proteins by protecting them from ubiquitylation (1, 2). A substantial increase in SUMO conjugates can also be observed during environmental stress, suggesting a link between SUMOylation and the general stress response (4). Particularly informative have been proteomic studies which have identified hundreds of mammalian and yeast proteins that become SUMOylated (e.g., refs. 5–8). In some cases, the added SUMO modifies the activity of the target, whereas in other cases, the SUMO moiety promotes interactions with proteins bearing SUMO-interacting motifs (SIMs) (2).
Studies with Arabidopsis thaliana have shown that a similar SUMOylation pathway exists in plants. The core pathway is comprised of four expressed isoforms of SUMO (SUMO1-3, and 5), the E1 heterodimer (SAE1a/b and SAE2), a single E2 (SCE1), at least two E3s (SIZ1 and MMS21/HPY2), and a collection of de-SUMOylating enzymes including ESD4 and OTS1/2 (9–14). Genetic studies have confirmed that SUMO conjugation is essential in plants (12), and uncovered roles for specific components in flowering, the cell cycle, abscisic acid signaling, and stress responses induced by heat, cold, drought, salinity, ethanol, phosphate starvation, and invasion by pathogens (10, 13–16). Particularly intriguing is the rapid and robust increase in SUMOylated proteins when plants are stressed. For example, within minutes of heat stress, a dramatic rise in SUMO1/2 conjugates can be observed in Arabidopsis seedlings, which is subsequently reversed upon return to nonstress temperatures (11, 12). Together with the observation that most, if not all, soluble conjugates are nuclear localized (12), stress-induced SUMOylation may represent an early response that globally affects stress-regulated gene expression. Because so few of the myriad of SUMO targets have been identified to date [e.g., PHR1, ICE1, FLD, and ABI5 (15, 16)], the effect(s) of this SUMOylation is unclear.
Clearly, a comprehensive catalog of SUMOylated proteins, especially during stress, is needed to more fully appreciate the functions of SUMO in plants. Here, we describe an efficient method to enrich for SUMO conjugates in Arabidopsis that exploits a tagged variant of SUMO1 designed to faithfully replace wild-type SUMO1 and 2 yet affords both stringent purification and the ability to map SUMO attachment sites. Combined with sensitive tandem MS techniques, we examined the profile of SUMO conjugates in both nonstressed plants and plants subjected to heat or oxidative stress. The list of 357 SUMO targets, some of which are condition specific, was substantially enriched for nuclear components that participate in a wide range of processes. Similar to studies with nonplants (17), we found that Arabidopsis SUMO1 becomes ubiquitylated and provide evidence linking this process to heat stress. Based on the array of substrates, we propose that SUMOylation plays a pervasive stress-protective role during plant gene expression and in chromatin maintenance.
Results and Discussion
Development of a SUMO Variant for Affinity Purification.
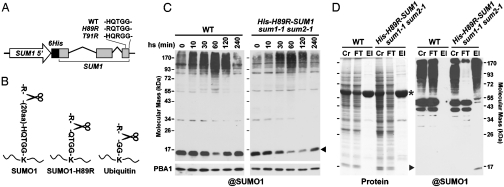
Key to our proteomic analysis in Arabidopsis was the development of a fully functional SUMO variant that can be exploited for affinity purification and subsequent MS analyses. In preliminary studies, we tested variants of SUMO1 expressed via its native promoter that contained an assortment of N-terminal affinity tags, including 6His, Flag, and TAP (Fig. 1A and Fig. S1A). We also engineered amino acid substitutions near the C-terminus that would simplify MS/MS detection of SUMO attachment sites on lysines following trypsinization (SUMO footprints) (7). Whereas the footprint generated with wild-type SUMO1 (K + 25 amino acids) would be too large for standard MS identification, the H89R variant used here would leave only a four-residue footprint (K + QTGG). Unlike another variant we tested (T91R), the footprint generated from the H89R protein would be distinct from that generated with Ub (K + GG), thus allowing us to map both modifications simultaneously (Fig. 1B).
Fig. 1.
Use of the His-H89R-SUMO1 variant to enrich for SUMO1 conjugates in Arabidopsis. (A) A schematic of the His-H89R-SUM1 transgene. Lines indicate introns. Black and gray boxes represent 6His and SUMO1 coding regions, respectively. The C-terminal sequences of the T91R and H89R variants are shown. (B) Diagrams of the trypsin footprints generated from proteins modified with SUMO1, H89R-SUMO1, and Ub. (C) Accumulation of SUMO conjugates during and after a 30 min heat stress (37 °C) of wild-type (WT) and His-H89R-SUM1 sum1-1 sum2-1 seedlings. Crude extracts were subjected to immunoblot analysis with anti-SUMO1 or anti-PBA1 antibodies (loading control). (D) Enrichment of SUMO1 conjugates from His-H89R-SUM1 sum1-1 sum2-1 plants by Ni-NTA affinity chromatography. The crude extracts (Cr), column flow through (FT), and eluates (El) from WT and rescued lines were separated by SDS/PAGE and stained for total protein (Left) or subjected to immunoblot analysis with anti-SUMO1 antibodies (Right). ▸, free His-H89R-SUMO1. *, large subunit of RUBISCO.
SUMO1 is the most actively expressed isoform, which along with its closest paralog SUMO2 becomes quickly incorporated into conjugates during heat and presumably other stresses in a SIZ1 E3-dependent reaction (11, 12). Arabidopsis requires either SUMO1 or 2 (12), thus allowing us to use the viability of sum1-1 sum2-1 plants to easily screen for functional rescue. When transgenes expressing 6His-, Flag-, or TAP-tagged SUMO1 with or without the H98R or T91R substitutions were introduced, we found that only combinations containing the 6His and H89R alterations (H89R-SUM1 and the His-H89R-SUM1) provided complete replacement in a double homozygous sum1-1 sum2-1 background (Fig. 1C and Figs. S1 and S2). The rescued plants had completely normal growth, were fully fertile, and accumulated SUMO1 conjugates during heat stress indistinguishable to wild type, indicating that the His-H89R SUMO1 protein is fully functional. In contrast, the Flag-SUM1- and T91R-SUM1-rescued plants were stunted and accumulated less SUMO1 conjugates, whereas the TAP-SUM1 plants were phenotypically normal but accumulated less conjugates. In addition to the full-length TAP-SUMO1 protein, the TAP-SUM1 plants contained a smaller SUMO1 fragment presumably missing the TAP tag (Figs. S1B), which might be responsible for the phenotypic rescue. Importantly, the His-H89R-SUMO1 protein retained a functional 6His sequence. When crude extracts from His-H89R-SUM1 sum1-1 sum2-1 plants were subjected to nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography, SUMO1 conjugates were strongly enriched as compared to extracts from wild-type plants (Fig. 1D).
Stringent Affinity Purification of SUMO Conjugates.
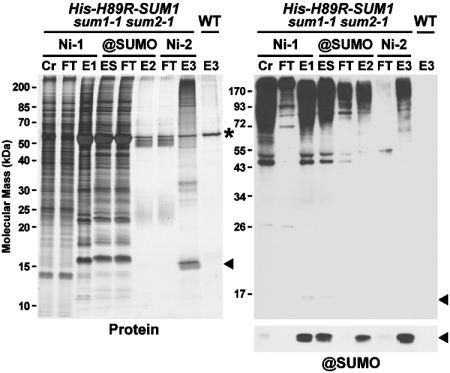
Using homozygous His-H89R-SUM1 sum1-1 sum2-1 seedlings, we developed a stringent three-step purification procedure to enrich for SUMO conjugates. To avoid isolating proteins that bound nonspecifically to the columns or would copurify via their association with SUMOylated proteins, and to minimize conjugate breakdown by endogenous de-SUMOylating enzymes, strong denaturants (e.g., hot 7 M guanidine or 8 M urea) and 10 mM iodoacetamide (IAA) were included in the initial extraction and most subsequent manipulations. The protocol included two Ni-NTA affinity chromatography steps sandwiching anti-SUMO1 antibody affinity chromatography. Most purification was achieved by the first two steps with the second Ni-NTA step primarily helping remove the large subunit of RUBISCO, which adheres nonspecifically to the beads, and the crosslinked immunoglobulins which bleed from anti-SUMO1 antibody columns in SDS and urea. As shown in Fig. 2, this protocol robustly enriches for both free SUMO and SUMOylated proteins from His-H89R-SUM1 sum1-1 sum2-1 but not from wild-type seedlings as detected by both protein staining and immunoblot analysis with anti-SUMO1 antibodies.
Fig. 2.
Affinity purification of SUMOylated proteins from Arabidopsis. Crude extracts (Cr) from wild-type (WT) and the His-H89R-SUM1 sum1-1 sum2-1 seedlings were subjected to sequential Ni-NTA (Ni-1), anti-SUMO1 (@SUMO), and Ni-NTA affinity chromatography (Ni-2). The flow through (FT), eluate (E1-3), and solubilized E1 (ES) fractions were separated by SDS-PAGE and either stained for total protein (Left) or subjected to immunoblot analysis with anti-SUMO1 antibodies (Right). Immunoblot loads were proportionally adjusted to allow direct comparisons among samples. A longer exposure of the immunoblot is included to show the levels of His-H89R-SUMO1. ▸, free His-H89R-SUMO1. *, large subunit of RUBISCO.
MS Analysis of SUMOylated Arabidopsis Proteins.
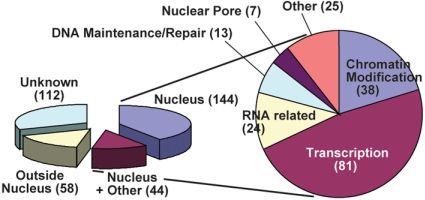
Using the three step procedure, we enriched for SUMOylated proteins from His-H89R-SUM1 sum1-1 sum2-1 seedlings grown under normal conditions, or exposed to heat stress (37 °C) or oxidative stress (50 mM H2O2). After trypsinization, the resulting peptides were eseparated by reversed-phase liquid chromatography followed by MS/MS sequencing, with the peptide sequences searched against the Arabidopsis proteome database. We also generated a background dataset from a duplicate purification using heat-stressed, wild-type seedlings (Table S1). After subtracting this background, a final list of 357 putative SUMOylated proteins was generated that included all treatments (Table S2). Confidence that the list included bona fide SUMO substrates was provided by significant enrichment of proteins bearing the SUMO-binding ΨKXE motif and by predictions that most are nuclear localized. As compared to the entire Arabidopsis proteome, 80% of the SUMOylated proteins contained at least one ΨKXE sequence (average of 2.24 sites per protein) versus 47% for the whole proteome (0.84 sites per protein) (Table 1), an enrichment similar to that found for SUMOylated proteins in cultured human cells (5). Of the 245 SUMOylated proteins with known or predicted intracellular locations, 187 (76%) associated with the nucleus (Table 2), which agreed with the observations that most Arabidopsis SUMOylated proteins copurify with this compartment (12).
Table 1.
Enrichment of proteins with consensus SUMO attachment sites
Table 2.
| Functional group | SUMO substrate |
| SUMO pathway | SUMO1, SAE2, SCE1, SIZ1, ESD4 |
| Transcription factors | |
| WRKY family | WRKY3, WRKY4, WRKY6, WRKY33, WRKY72 |
| HSF family | HSFA1D, HSFA2, HSFB2B |
| Global TF family | GT2, GTB1, GTE1, GTE4, GTE7, GTL1 |
| ANACONDA family | ANACO50, ANACO51, ANACO80/ATAF2 |
| Homeodomain | BEL1, BEL10, KNAT3, HB6, ANL2 |
| Others of note | EIN3, ARR1, PHR1, ERF6, BIM1, HUA2 |
| Transciptional coregulators | |
| TOPLESS family | TPL, TPR1, TPR2, TPR4 |
| Others of note | LEUNIG, LUH, SEUSS |
| Chromatin modifiers | |
| Chromatin methylation-related | SUVR1, SUVR2, SUVH2, SUVH9, KTF1, IBM1 |
| SWI/SNF complex | SWI3A, SWI3C, SWI3D, PICKLE, PKR1, CHR11 |
| Histone acetylation-related | SNL2, SNL4, SNL5, HAC1, GCN5, HDA19(HD1), ADA2A, ADA2B |
| Histone 2b-related | Histone2B, NRP1, UBP26, SPT16 |
| DNA main/repair | LIG1, DRT111, KU80, POLD3, TOP1, TRB1 |
| RNA-related | SERRATE, LACHESIS, XRN3, DRH1, SDN3 PRP40B, LA1, PARL1, STA1 |
| Nuclear pore | IMP-6, IMPA1, WIP1 |
| Cell cycle regulators | SYN4, ILP1, ILP2, RHL1, DPB, CDC5, CDC48, RPA1 |
*See Table S2 or the Scaffold file (Dataset S1) available at http://www.genetics.wisc.edu/node/558 for complete list.
†Number of unique proteins are in parentheses.
Consistent with their nuclear localization, many SUMOylated proteins could be assigned functions within the nucleus, including roles in transcription, chromatin modification, RNA-related processes, DNA maintenance/repair, and nuclear pore assembly (Table 2 and Table S2). For a number of targets in the dataset which are encoded by gene families, multiple members were identified as SUMOylated, including representatives of the WRKY, heat-shock factor (HSF), homeodomain and ANACONDA transcription factor families, and the SWI/SNF ATP-dependent chromatin remodeling and the SUVR chromatin methylase families to name a few (Table 2). An unbiased motif analysis of the candidates also found a statistical enrichment for nuclear-related domains, including those present in: (i) transcription factors (SANT, MADF, BZIP_1, and WRKY), (ii) components of RNA metabolism (RRM_1, Helicase_C, ResIII, and DEAD), (iii) chromatin binding/remolding proteins (Bromodomain, WD40, PHD, PWWP, SET, SNF2_N, and JmjC), and (iv) cell cycle regulators (LisH) (Table S3 and Fig. S3). Many of these domains are similarly enriched in human SUMOylated proteins (5), suggesting that a conserved set of SUMOylation targets exists among eukaryotes (Fig. S3). One intriguing example is the SANT domain, which physically interacts with the SUMO E2 and a SIZ1-type E3 (18).
Inspection of the SUMOylated protein list identified a number of critical cell regulators (Table 2 and Table S2). Like observations with yeast and mammalian cells (5, 6), some of the most frequently identified substrates are components that direct SUMO conjugation, including the SAE2 subunit of the E1 heterodimer, SCE1, and SIZ1. Although less abundant, we also detected the de-SUMOylating enzyme ESD4. This enrichment could reflect the inadvertent modification of SUMOylating enzymes during conjugation, or more intriguingly could reflect an autoregulatory loop that modulates SUMOylation. Furthermore, given the prevalence of SUMO pathway enzymes and their interactors (SANT domain-containing) in our dataset, it is possible that other heretofore unknown pathway components are present, including additional SUMO E3s.
A variety of factors involved in chromatin modification and DNA maintenance/repair are in our Arabidopsis SUMOylation dataset, implying that SUMO addition can profoundly alter the accessibility and integrity of plant genomes. Particularly relevant are histone 2b, the GCN5 histone acetyltransferase, the HDA19(HD1) histone deacetylase, and the deubiquitylating enzyme UBP26, which removes Ub bound to histone 2b. The modification of these proteins strongly suggests that SUMOylation is a key part of the plant histone code which controls chromatin accessibility. We also detected enzymes involved in RNA-directed DNA methylation (SUVH2, SUVH9, and KTF1) and DNA repair (KU80, LIG1, DRT111, and RPA1), thus implicating SUMO in various DNA modification events. The SUMOylation dataset is most enriched for transcription factors (58% of the known nuclear proteins) (Table 2 and Table S2). The diverse array of process that these factors individually control, ranging from cytokinin (ARR1) and ethylene signaling (EIN3) to ovule development (BEL1), phosphate starvation (PHR1), and heat-shock responses (HSFs), implies a far-reaching role for SUMOylation in regulating the Arabidopsis transcriptome. SUMO was first identified as a posttranslational modifier by its attachment to the mammalian nuclear pore protein RanGAP1 with the E3 responsible, RanBP2, also found associated with the pore complex (19). Although Arabidopsis RanGAPs do not appear to be SUMOylated in planta and an ortholog of RanBP2 is not obvious, it is noteworthy that other components of the plant nuclear pore appear to be SUMOylated [importin (IMP)-6, IMPα1, WIP1, ESD4, and two proteins related to yeast RanBP1].
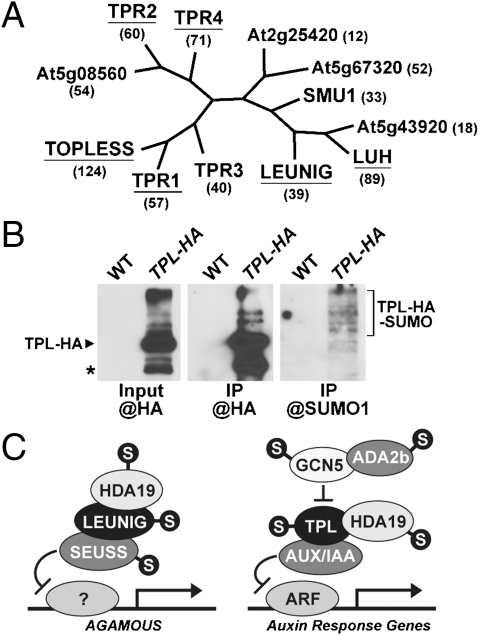
Some of the more abundant proteins in our SUMOylation dataset are members of the Groucho/Tup1 family of transcriptional corepressors, which include TOPLESS (TPL) and LEUNIG (20). Of the 12 Arabidopsis representatives, 6 were found to be SUMOylated with TPL being one of the most frequently detected targets (Table 2, Table S2, and Fig. 3A). We confirmed the SUMOylation of TPL by coimmunoprecipitation of a functional hemagglutinin (HA)-tagged TPL variant expressed in the tpl-2 background (21). HA antibodies enriched for the HA-TPL protein and for a ladder of high molecular mass SUMO1 conjugates from HA-TPL tpl-2 plants but not from wild-type plants (Fig. 3B).
Fig. 3.
Members of the TOPLESS (TPL)/LEUNIG corepressor family are targets of SUMOylation. (A) Phylogenetic tree of the Arabidopsis TPL/LEUNIG family (20) with SUMOylated members underlined. The number of transcripts for each locus in the Arabidopsis EST database is in parentheses. TPR, TPL-related. (B) Detection of TPL-SUMO1 conjugates. TPL was immunoprecipitated from heat-stressed wild-type (WT) and TPL-HA expressing seedlings with anti-HA antibodies and subjected to immunoblot analysis with anti-HA or anti-SUMO1 antibodies. Input, crude extracts before immunoprecipitation. Location of the free TPL-HA and TPL-HA-SUMO1 conjugates are indicated. * identifies a breakdown production of TPL (21). (C) The proposed genetic and/or biochemical interactions of LEUNIG and TPL with their respective partners and target genes highlighting the proteins modified with SUMO1 (S).
TPL, LEUNIG, and presumably others in the Groucho/Tup1 family work as corepressors by binding to gene-specific repressors and then recruiting components involved in histone deacetylation such as HDA19 (20) (Fig. 3C). For TPL, its targets include members of the AUX/INDOLE-3-acetic acid (IAA) family that repress auxin-regulated gene expression by binding to and inactivating the family of ARF transcription factors (21). At least genetically, TPL is positively regulated by HDA19 and negatively regulated by the GCN5 histone acetyltransferase and its coregulator ADA2a/b (20, 22). In the case of LEUNIG, it forms a repression complex with SEUSS and HDA19 which limits AGAMOUS expression during flower development by inhibiting a yet unknown transcription factor (20). Intriguingly, almost all of the additional components of these two transcriptional regulatory circuits are present in our SUMOylation dataset (Table 2 and Fig. 3C), including SEUSS, HDA19, GCN5, and ADA2a and b, suggesting that the entire process is under SUMO control. Effects on LEUNIG may even extend to the positive regulator of AGAMOUS—HUA2 (23), which is also in our SUMOylation dataset.
Changes in SUMOylation Patterns During Stress.
A general feature of SUMOylation is the substantial increase in conjugates during stress (5, 11, 12). Strikingly, many proteins present in our dataset from nonstressed plants were also present in the datasets generated from heat- and oxidative-stressed plants [138 of 357 (Fig. S4A)], implying that stress mainly increases the levels of the SUMOylated forms. However, normalization of specific peptide counts relative to total peptides in each dataset revealed that some targets likely become selectively SUMOylated under specific conditions. DNA repair/maintenance and nuclear pore components in particular were more prevalent in the stressed versus nonstressed datasets (Fig. S4B). For specific examples, peptides for histone 2b were significantly enriched in nonstressed and oxidative-stressed datasets but low in the heat-stressed dataset, implying that heat induces the de-SUMOylation of this histone (Fig. S4 C and D). Conversely, peptides for the RNA-binding protein AtLA1 were abundant in the heat-stressed but absent in nonstressed or oxidative-stressed datasets, whereas peptides from the ESD4 de-SUMOylating enzyme were only detected in the oxidative-stressed dataset.
Use of His-H89R SUMO1 to Detect SUMO-Ligation Sites.
By reducing the SUMO1 tryptic footprint from a 25 amino acid fragment to only the QTGG sequence, we hoped to simplify identification of SUMO attachment sites. MS/MS analysis of a synthetic peptide of SUMO1 (NQEEDKK*PGDGG) bearing QTGG isopeptide-linked to Lys10 (*) confirmed this possibility along with the discovery that the N-terminal glutamine cyclizes to generate a pyro-Glu derivative (Fig. S5B). Subsequent searches of our MS/MS spectra for the pyro-QTGG footprint (K + 326 m/z) detected 17 potential SUMOylation sites on 14 targets (Fig. 4A and Fig. S5C–E). Intriguingly, some of the SUMO-footprint peptides were found only under certain conditions. Ten of the 17 sites are within the consensus SUMO-binding sequence ΨKXE (3), further confirming that this motif is also preferred by the plant SUMOylation machinery (Fig. 4A).
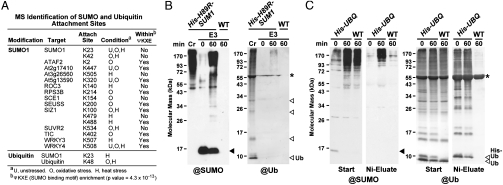
Fig. 4.
MS identification of SUMO1 attachment sites and detection of SUMO1-Ub conjugates. (A) List of proteins identified to have SUMO1 or Ub attachment sites based on MS/MS detection of the respective footprints. The target name, site of SUMO or Ub attachment, seedling treatment conditions where the footprint was detected, and whether or not the SUMO footprint is within the canonical ΨKXE attachment site (3) are listed. (B) Immunoblot detection of SUMO1 conjugates modified with Ub. WT and His-H89R-SUM1 sum1-1 sum2-1 and seedlings were collected before (0) and 30 min after a 30 min heat stress and enriched for SUMO conjugates by the 3-step affinity method shown in Fig. 2. Cr, crude seedling extracts from heat-stressed plants before purification. (C) Immunoblot detection of Ub conjugates modified with SUMO1. WT and transgenic seedlings expressing His-Ub were collected before (0) and 30 min after a 30 min heat stress and enriched for Ub conjugates by a single Ni-NTA affinity step. Start represents the crude extracts before Ni-NTA purification. Samples in (B) and (C) were subjected to SDS-PAGE and immunoblot analysis with anti-SUMO1 (Left) or anti-Ub antibodies (Right). ▸, free SUMO1. ⊳, free Ub, His-Ub and poly-Ub chains. *, large subunit of RUBISCO.
Of interest are the footprints identified in SUMO1 itself at K23 and K42 (Fig. S5 C and D), thus providing in vivo support for the proposal that plants assemble polymeric SUMO chains (12, 24). SUMOylation of K10 in Arabidopsis SUMO1 has also been proposed (24), but the 6His tag interfered with MS analysis of this region. We also failed to detect peptides specific to SUMO3 and 5, suggesting that mixed SUMO chains are not assembled in planta. ΨKXE motifs in the N-terminal tails of yeast Smt3 and human SUMO2/3 have been shown to provide concatenation sites for SUMO chain formation (5, 25). In contrast, none of the Arabidopsis SUMO isoforms contain the ΨKXE motif (Fig. S5A), implying that the assembly mechanism(s) and architecture of SUMO polymers differ in plants.
Detection of Proteins Both SUMOylated and Ubiquitylated after Heat Stress.
Recent nonplant studies have revealed a cross talk between the Ub and SUMO pathways, and identified specific Ub ligases that ubiquitylate SUMOylated proteins (17, 26). That we consistently detected Ub tryptic peptides in our SUMOylation datasets from heat- and oxidative-stressed plants, indicated that Arabidopsis also simultaneously modifies some proteins with both SUMO and Ub, especially during stress. Some SUMOylated proteins likely become modified with K48-linked Ub polymers based on our detection of Ub peptides bearing a Ub footprint (K + 114 m/z) at K48 (Fig. 4A).
To confirm the possibility that SUMOylated proteins are ubiquitylated under stress, we subjected proteins purified by our three-step affinity protocol from nonstressed and heat-stressed plants to immunoblot analysis with anti-Ub antibodies. As shown in Fig. 4B, a smear of high molecular mass Ub conjugates was detected after heat stress of His-H89R-SUM1 sum1-1 sum2-1 seedlings but not in the transgenic seedlings before heat stress nor in wild-type seedlings subjected to heat stress. For the reciprocal experiment, we affinity-purified Ub conjugates from His-UBQ plants (27) both before and after heat stress and subjected the samples to immunoblot analysis with anti-SUMO1 antibodies (Fig. 4C). Even though heat stress did not increase the level of total Ub conjugates, a substantial increase in ubiquitylated proteins containing SUMO was observed after heat stress, which was not evident before heat stress nor in samples from heat-stressed wild-type plants.
Subsequent analyses of our SUMOylation dataset detected a Ub footprint on SUMO1 itself, indicating that Arabidopsis SUMO1 is a direct target of ubiquitylation (Fig. 4A and Fig. S5F). The ubiquitylated lysine (K23) is also used for SUMOylation, indicating that the two conjugation machineries may compete for this residue. Interestingly, ubiquitylation of SUMO1 may be specific to heat stress because the SUMO1 peptide bearing the K23 Ub footprint was detected only in these samples. Taken together with the evidence that a subset of SUMOylated proteins are polyubiquitylated during heat stress, it is plausible that the Ub polymers are attached directly to SUMO1 previously conjugated to other proteins. Yeast and animals contain SUMO-dependent Ub ligases Slx5/8 and Rnf4, respectively, that can direct this ligation (26), but whether orthologous activities exist in plants is not yet clear.
Future Perspective.
Our development of a robust purification protocol based on the complete replacement of SUMO1/2 with a novel variant now allows for the in-depth analysis of SUMO conjugates in plants. Particularly noteworthy are our abilities to conduct the purification under stringent denaturing conditions, thus mitigating the indirect isolation of nonmodified proteins simply by their association with SUMOylated proteins, and to identify SUMO attachment sites through an engineered SUMO footprint that is distinct from that generated by Ub. The poor MS coverage of SUMO conjugates by a previous study (28) may be explained by the use of functionally compromised tagged SUMO that competed poorly with native SUMO coupled with insufficient enrichment. We note that SUMO1 attachment sites were detected on only a limited number of targets (14 of 357) despite the use of the H89R variant, a similarly low number to those found by MS for Ub attachment sites (27) and SUMO attachment sites in other organisms (5–8). The reason(s) behind this difficulty could include novel MS fragmentation patterns for such bifurcated peptides, and/or the inability of the MS algorithms to correctly identify them.
From our in-depth MS/MS analyses, we identified a large collection of SUMOylated proteins, with some modified only under specific conditions. Of future interest will be similar analyses of specific tissues and plants compromised in various steps of the SUMO pathway (e.g., siz1, mms21, ots1/2, and esd4 backgrounds) or expressing other SUMO isoforms. Our discovery that SUMO1 itself is a target for both SUMOylation and ubiquitylation further connects these two posttranslational modifications. The ubiquitylation of SUMO targets during heat stress implies that the Ub/26S proteasome pathway may eventually degrade some SUMOylated proteins, with the SUMO moiety potentially representing a secondary degron.
Important questions yet to be answered are: Why do plants SUMOylate such a wide array of proteins? And why does this SUMOylation dramatically increase during stress in a reversible manner? Given the role of many targets in nucleic acid metabolism, one possibility is that SUMOylation provides a rapid and reversible mechanism to globally affect gene expression and chromatin stability both before and during various environmental stresses. Most of the stress-induced SUMOylation is driven by the SIZ1 E3 (12), which may explain the pleiotropic nature of siz1 plants (12, 15, 16). SUMO modification of DNA repair enzymes and various DNA and histone modification complexes could enhance their activities/stabilities or promote their binding to chromatin via association with SIM-containing proteins. The SUMOylation of transcription factors and coactivators/corepressors could down regulate the expression of housekeeping and developmental genes (e.g., BEL1, LEUNIG, PICKLE, and TPL), while simultaneously upregulating expression of genes responsible for stress protection (e.g., EIN3, PHR1 and HSFs). Thus, through the use of a small set of SUMO-binding motifs such as ΨKXE combined with a signaling system that activates/represses the central SUMO ligation/de-SUMOylation machineries, a thorough protective response could be rapidly elicited and reversed.
Materials and Methods
Construction of Tagged SUMO1 Variants.
The SUM1 transgene from A. thaliana ecotype Col-0 was from (12) and altered by PCR-based mutagenesis. Tags were added in-frame either by PCR-based extension (6His and TAP) with appropriate oligonucleotides or by LR recombination into the pEarleyGate 202 vector (Flag) (29). The constructions were used to rescue a sum1-1-/- sum2-1-/- line as described (12). TPL-HA and 6His-Ub expressing lines were from (27, 21). Plants were grown under continuous white light for 7 d at 25 °C in shaking liquid Gamborg’s B5 medium (GM) (Invitrogen) (11). For heat stress, the cultures were heated to 37 °C for 30 min followed by a 30 min recovery at 25 °C. For oxidative stress, the plants were exposed to 50 mM H2O2 for 4 hrs. Immunoblot analyses with antibodies against Ub, PBA1, SUMO1, and HA were as described (11, 12). Monoclonal anti-HA antibody (MMS-101P) was purchased from Covance.
Purification of SUMOylated Proteins.
Pulverized frozen tissue (50 g) was extracted for 1 hr at 55 °C with 100 mL of Extraction Buffer (EB) (100 mM Na2HPO4, 10 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM IAA) containing 7 M guanidine-HCl, 10 mM sodium metabisulfite, and 2 mM PMSF. The crude extract was clarified by centrifugation at 15,000 × g, filtered, made 10 mM in imidazole, and incubated with rotation overnight at 4 °C with 5 mL of Ni-NTA beads (Qiagen). The beads were washed with EB containing 6 M guanidine-HCl and 0.25% Triton X-100, followed by EB containing 0.25% Triton X-100 and 8 M urea. Proteins were eluted with 6 M urea, 100 mM Na2HPO4, 10 mM Tris-HCl (pH 8.0), 300 mM imidazole and 10 mM IAA, and concentrated to ∼500 μL by ultrafiltration (Amicon Ultracel-10 K, Millipore). The concentrate was diluted 25 fold in RIPA buffer (1% NP40, 0.5% deoxycholate, 0.1% SDS, 10 mM Tris-HCl (pH 7.2), 50 mM Na2HPO4, 100 mM NaCl, 10 mM IAA, and 2 mM PMSF) and incubated overnight with rotation at 4 °C with anti-SUMO1 IgG beads [1 mg IgG/mL of Affigel-10 resin (BioRad)] (11). The beads were washed with RIPA buffer followed by PBS (50 mM Na2HPO4 (pH 7.4), 100 mM NaCl, and 10 mM IAA). Protein was sequentially eluted at 65 °C with 1% SDS and 10 mM IAA followed by 8 M urea, 100 mM Na2HPO4, 10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 10 mM IAA. Pooled eluates were made 10 mM in imidazole and sodium metabisulfite and incubated with 0.3 mL of Ni-NTA beads at 4 °C. Beads were washed with EB containing 8 M urea. SUMO conjugates were eluted with 6 M urea, 100 mM Na2HPO4, 10 mM Tris-HCl (pH 8.0), 300 mM imidazole, and 10 mM IAA, and concentrated to 250 μL by ultrafiltration. HA-TPL was enriched as described (21) using Red Anti-HA affinity resin E6779 (Sigma).
Tandem Mass Spectrometry.
SUMOylated proteins were reduced, carboxylmethylated and digested in solution with trypsin as described (12). Tryptic peptides were acidified with 6 μL trifluoroacetic acid (TFA), and desalted and concentrated by C18 solid-phase extraction (Varian Omix tips, 70 μg capacity) using 100 μL of 75% acetonitrile and 0.1% TFA for elution. Vacuum dried peptides were resuspended in 10 μL of 0.1% TFA and subjected to LC/MS using an LTQ-Orbitrap (ThermoFisher Scientific) hybrid mass spectrometer coupled to an Agilent 1100 Nanoflow HPLC equipped with a 75 μm × 15 cm Magic C18 microcapillary column (Michrom Bioresources). Peptides were eluted over 4 hrs using 0–95% acetonitrile gradient in 0.1 M acetic acid. MS and MS/MS spectra were acquired in data-dependent mode with MS spectra acquired in the Orbitrap at 100,000 resolving power and 5 MS/MS spectra acquired per MS in the LTQ linear ion trap. MS/MS precursor selection required that the charge state be known and ≥2. Dynamic exclusion was enabled to prevent repeated MS/MS on the same precursor, with exclusion duration set at 20 sec. Peptide sequences were assigned using the MASCOT software (version 2.2, Matrix Science) against the Arabidopsis protein database (www.NCBI.nlm.NIH.gov/RefSeq/) and common contaminants (e.g., human keratin and trypsin). Search parameters included a precursor mass tolerance of 2.5 Da, fragment ion mass tolerance of 0.5 Da, up to 3 missed trypsin cleavages, carbamidomethylation of Cys, oxidation of Met, and variable modification of Lys residues by either ubiquitylation (Gly-Gly, +114 m/z) or SUMOylation (pyroGlu-Thr-Gly-Gly, +326 m/z). Data were curated using Peptide Prophet in Scaffold 2 (Proteome Software) with putative SUMOylated peptides verified by manual inspection. Proteins isolated from wild type required at least one peptide of ≥50% confidence. Proteins isolated from the His-H89R-SUM1 line required a 99% protein probability and at least one peptide with ≥95% confidence. SUMO-binding sites were predicted by SUMOsp2.0 (30).
Supplementary Material
Acknowledgments.
We thank Dr. Jeffrey Long for the HA-TPL line, Joseph Walker, Dr. Scott Saracco, and Dr. Mark Scalf for advice, and Gregory Blythe and Daniel Kassera for technical help. This work was supported by a National Science Foundation (NSF) Arabidopsis 2010 Program Grant (MCB-0929100) to R.D.V, a National Institutes of Health sponsored predoctoral training fellowship to the University of Wisconsin Genetics Training Program (M.J.M.), and a NSF Grant (MCB 0929395) to Dr. Michael Sussman for the ESI-LTQ-Orbitrap MS facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004181107/-/DCSupplemental and http://www.genetics.wisc.edu/node/558.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Bio. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 4.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 5.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 6.Denison C, et al. A proteomic strategy for gaining insights into protein SUMOylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR. Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J Proteome Res. 2006;5:761–770. doi: 10.1021/pr050451o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Murtas G, et al. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conti L, et al. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurepa J, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 12.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishida T, et al. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Miura K, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 18.Tiefenbach J, et al. SUMOylation of the corepressor N-CoR modulates its capacity to repress transcription. Mol Biol Cell. 2006;17:1643–1651. doi: 10.1091/mbc.E05-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Karmarkar V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008;13:137–144. doi: 10.1016/j.tplants.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 22.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Meyerowitz EM. HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell. 1999;3:349–360. doi: 10.1016/s1097-2765(00)80462-1. [DOI] [PubMed] [Google Scholar]
- 24.Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 26.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Saracco SA, et al. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 2009;59:344–358. doi: 10.1111/j.1365-313X.2009.03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budhiraja R, et al. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on deSUMOylation. Plant Physiol. 2009;149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 30.Ren J, et al. Systematic study of protein SUMOylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.