Abstract
The Pleiades Promoter Project integrates genomewide bioinformatics with large-scale knockin mouse production and histological examination of expression patterns to develop MiniPromoters and related tools designed to study and treat the brain by directed gene expression. Genes with brain expression patterns of interest are subjected to bioinformatic analysis to delineate candidate regulatory regions, which are then incorporated into a panel of compact human MiniPromoters to drive expression to brain regions and cell types of interest. Using single-copy, homologous-recombination “knockins” in embryonic stem cells, each MiniPromoter reporter is integrated immediately 5′ of the Hprt locus in the mouse genome. MiniPromoter expression profiles are characterized in differentiation assays of the transgenic cells or in mouse brains following transgenic mouse production. Histological examination of adult brains, eyes, and spinal cords for reporter gene activity is coupled to costaining with cell-type–specific markers to define expression. The publicly available Pleiades MiniPromoter Project is a key resource to facilitate research on brain development and therapies.
Keywords: knockin mice, neuroscience, promoter design, selective expression, transcription
The mammalian central nervous system (CNS) is a complex entity comprising diverse neuronal and nonneuronal cell types. The organization of these cell types, as illustrated in brain atlases (e.g., ref. 1) and gene expression maps (e.g., ref. 2), is regional within the brain. The Allen Brain Atlas maps the expression of ≈20,000 genes in the adult mouse brain using in situ hybridization (2). A similar initiative, the GENSAT project, used large-insert, random-insertion transgenic mouse techniques to profile gene expression patterns in the CNS (3). These resources demonstrate the selectivity with which some genes are expressed in the brain. Such specific expression patterns can be driven by modular conserved regulatory regions (RRs) (4). Importantly, patterns of gene expression within the brain tend to be conserved between human and mouse (5).
For decades, researchers have struggled to develop tools that direct specific patterns of gene expression in the brain. There is an increasing availability of genetic applications that are predicted upon targeted gene expression in vivo that require cell-specific promoter constructs to restrain the effect to a region of interest. Despite great demand, the discovery of regulatory sequences to drive gene expression in a region-specific manner in the brain has been slow, primarily arising from low-throughput promoter-deletion studies (e.g., L7/Pcp2, ref. 6 and Camk2a, ref. 7).
The Pleiades Promoter Project resolves this challenge using a higher-throughput, bioinformatically directed parallel design process, producing MiniPromoters (MiniPs) vetted in vivo in the adult mouse brain. The Pleiades Promoter Project adopted a three-step strategy wherein we first identify genes specifically expressed in adult brain regions or cell types of therapeutic interest, then we computationally predict the human RRs responsible for the specific expression and, ultimately, test compact MiniPs in vivo in knockin mice. MiniPs contain human DNA sequences to facilitate studies in human cells; they do not use, for example, any prokaryotic sequences thereby reducing the potential for epigenetic inactivation. The Pleiades resource of brain MiniPs expands dramatically upon the collection of existing promoters for the study of the brain and is publicly available at http://www.pleiades.org. The resource includes a comprehensive dataset of regulatory information on brain-specific genes, the MiniP designs and sequences, the plasmids for Hprt targeting, the targeted embryonic stem cells (ESCs), and the mouse strains to allow researchers comprehensive access to the data and tools.
Results
The Pleiades Promoter Project Approach.
The Pleiades Promoter Project strategy is to computationally identify RRs responsible for the selective expression observed in manually selected brain-region or cell-type enriched genes (8). The goal is not recapitulation of complex endogenous expression patterns; rather we seek to assemble RRs to drive more selective expression patterns. Once candidate RRs are identified, up to seven MiniPs are designed per gene, each including a different combination of one to six regions. To produce physiological levels of expression and define a MiniP entirely derived from human DNA sequence, we use the endogenous promoter of the gene instead of an exogenous promoter. MiniPs are small (≤4 kb) to be easily manipulated and suitable for most space-restricted molecular constructs. The MiniP sequences are cloned 5′ of a reporter gene, such as EGFP, lacZ, or an EGFP/cre fusion protein. The constructs are then introduced by homologous recombination immediately 5′ of the Hprt locus on the X chromosome, as previously described (9, 10), providing a single-copy knockin insertion for reproducible, predictable expression (11).
Validation of the Approach Using a Set of Previously Characterized Promoters and ESC Neural Differentiation.
We selected the CAG ubiquitous promoter as well as five previously characterized multicopy random-insertion promoters that drive expression in a subset of neurons or glial cells, generated vector constructs, and targeted insertion to the Hprt locus (Dataset S1 “Control”). The CAG promoter drives ubiquitous reporter gene activity, demonstrating that widespread, adult expression can be achieved from the locus, consistent with previous studies (10).
Initially, we adopted an ESC-differentiation strategy that generates neural stem cells within 6 d, as confirmed by gene expression patterns, as a test of cell-type specificity from the control promoters inserted 5′ of Hprt (12). Fig. 1 shows the characterization of Ple88 (GFAP RRs) for glial expression and Ple53 (DCX RRs) for neuronal expression (Fig. 1 A and E), selected because both GFAP and DCX are selectively expressed during CNS development and therefore the promoters are likely to drive expression in the differentiation assay. Expression of the MiniPs follows a pattern similar to the corresponding endogenous genes, as shown by RT-PCR (Fig. 1 B and F) and staining (Fig. 1 C and G). The positive ESC prescreening results were then confirmed in the corresponding knockin adult mouse brains (Fig. 1 D and H). This validation test, using these two previously characterized promoters, demonstrates the suitability of the Pleiades Promoter Project approach, using single-copy knockin 5′ of the Hprt locus, for the characterization of brain MiniPs.
Fig. 1.
In vitro neural differentiation for prescreening MiniP designs. (A and E) Ple53 and Ple88 are cloned upstream of EGFP or lacZ, respectively. (B and F) RT-PCR assays across seven time points of ESC neural differentiation for endogenous and reporter genes demonstrate appropriate temporal expression. (C and G) Immunochemistry or X-gal staining demonstrates appropriate spatial expression. [Scale bars, 100 μm (C), 200 μm (G).] (D and H) Germline knockin adult mouse brain sagittal sections analyzed by immunochemistry or X-gal staining confirms expression in the appropriate regions (i.e., olfactory bulb and rostral migratory stream for Ple53 and glia throughout the brain for Ple88).
The five selected control promoters were further characterized in transgenic animals by neurohistological analysis. As detailed in Dataset S1 “Control” and Fig. S1, four of the five controls reproduce the expected expression patterns. The last control MiniP, Ple59 (DDC RRs), does not produce detectable reporter gene expression. As the original study characterizing this construct reports only one of six founder lines expressing the transgene, we conclude that favorable conditions at that site in the genome may permit expression and such insertion-site properties are not present at the other five locations nor at Hprt (13). In conclusion, the analysis of these previously characterized brain-specific promoters validates our knockin strategy 5′ of the Hprt locus as a tool to monitor brain-specific expression.
Brain Region-Specific Gene Selection Using a Regulatory Resolution Score.
To generate new MiniPs expressing with a cell type or regional specificity in the brain, we first identified, using a genomewide approach, 237 candidate genes with region-enriched expression patterns that included 30 target adult brain regions of therapeutic interest (8). To narrow the list of genes, we took into account the potential relevance of each gene to human disease and, importantly, the predicted suitability of the gene for MiniP design. For disease relevance, we reviewed the literature and, when available, examined the phenotypic consequences of mouse gene knockouts. The suitability for MiniP design highlights those genes in which RRs are more easily distinguished. Comparative sequence analysis, or phylogenetic footprinting, has proven useful in delineating RRs with the expectation that sequences under selective pressure are more conserved than those that are not. We thus base our gene prioritization on the following criteria: (i) the existence of known RRs within a gene responsible for a expression pattern in the brain, (ii) transcript evidence supporting the presence of a single transcription initiation site, (iii) the length of sequence to be analyzed, (iv) the number of conserved regions between the analysis boundaries, and (v) how well distinguished conserved regions are relative to the overall conservation level for a gene. Thus, we sought genes containing a small number of well-defined conserved noncoding regions close to the transcription start site. To this end, we developed a “regulatory resolution score” intended to reflect human perception of what constitutes a good candidate gene for MiniP design (SI Materials and Methods and Figs. S2 and S3). The scoring procedure captures aspects of the manual curation process as demonstrated by comparison with scores manually assigned for 100 curated genes (Fig. 2A and SI Materials and Methods). The 57 genes selected for MiniP design are heavily skewed toward higher resolution scores in our set of 237 brain region-selective genes (Fig. 2 B and C).
Fig. 2.
Regulatory resolution score prioritizes genes for MiniP design. (A) Score distribution for 100 manually curated genes. The width of the boxes are proportional to the number of observations in the groups. The increases in scores from 1 to 4 and 5 are significant (P = 1.4e−03 and P = 7e−04, respectively; Wilcoxon test), as well as from 2 to 5 (P = 4.5e−02; Wilcoxon test). (B) Score frequency of the selected 57 genes (black) compared with all other brain region selective genes (white). The dotted gray line shows that the proportion of MiniP genes relative to total increases with the score (linear regression; individual values are marked as gray boxes). (C) Regulatory resolution scores for the genes selected for MiniP design.
MiniPs Are Designed on the Basis of Genomics Analyses.
For some selected genes, published information about their regulation influenced designs. The regulatory literature pertaining to the 57 selected genes was curated and stored in the PAZAR database (14) within the “Pleiades Genes” project. This data collection represents a unique public dataset dedicated to transcriptional regulation in the brain, uniting information about both cis-regulatory sequences and mediating transcription factors (TFs), accompanied by details of the experimental evidence.
A main MiniP design intention is the identification of sequences with the capacity to drive expression in both human and mouse cells. Although gene selection was primarily informed by mouse expression data, MiniPs were designed with human DNA sequences and tested in mice. This dichotomy motivated the use of phylogenetic footprinting between the human and mouse genomes to delineate candidate RRs. Within these conserved regions we generated TF binding site predictions when TFs were known to be important mediators of expression in the specific cells or regions of interest. The TF REST (also known as NRSF) was included in all analyses as it has been described to bind to many neuronal genes to prevent their expression in nonneuronal tissues (15). The TF binding models used in this study were compiled from the JASPAR database (16) supplemented with brain-specific TFs annotated in the PAZAR “Pleiades Genes” project introduced above. As detailed in SI Materials and Methods, the MiniP design pipeline takes into account information from the literature, genome annotations, and computational analyses.
Validation of the Design Strategy Using the Previously Characterized Promoters.
As a proof of principle for our bioinformatics approach, we applied our MiniP design pipeline to the four genes for which we have validated a previously characterized promoter (Dataset S1 “Refined Control,” Fig. 3, and Fig. S4).
Fig. 3.
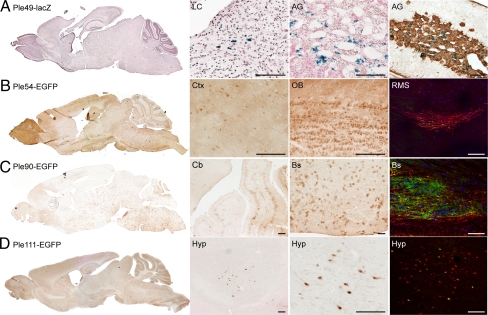
Refined designs of control genes validate the bioinformatics approach. We analyzed adult brain sagittal sections of four positive strains carrying MiniPs designed using our bioinformatics pipeline on control genes. For Ple49, sections of the adrenal gland were also analyzed. EGFP is detected using anti-GFP immunochemistry (brown) and lacZ is detected using X-gal histochemistry (counterstained with neutral red). AG, adrenal gland; Bs, brainstem; Cb, cerebellum; Ctx, cortex; Hyp, hypothalamus; LC, locus coeruleus; OB, olfactory bulb; RMS, rostral migratory stream. (A) Ple49-lacZ (DBH RRs) expression is enriched in the LC (but in cells that do not co-stain with anti-TH) and the AG. The last image shows costaining of X-gal (blue) with tyrosine hydroxylase (brown) in the AG. (B) Ple54-EGFP (DCX RRs) expression is observed in different regions of the brain with enrichment in the OB as seen on the whole brain image. The last image shows costaining of EGFP (green) with the endogenous Dcx protein (red) in the RMS. (C) Ple90-EGFP (GFAP RRs) is expressed in astrocytes throughout the brain. The last image shows costaining of EGFP (green) with the endogenous GFAP protein (red). (D) Ple111-EGFP (HCRT RRs) is specifically expressed in a few cells of the lateral hypothalamus. The last image shows costaining of EGFP (green) with the endogenous HCRT protein (red). (Scale bars, 100 μm.)
For both the DBH and GFAP genes, we identified a minimal promoter and a well-conserved upstream sequence, both included in the previously characterized promoters, and we then fused them to generate Ple49 and Ple90 for DBH and GFAP, respectively. In the case of DCX, two additional MiniPs were designed, Ple54 and Ple55, which include additional putative RRs located within DCX introns. Finally, for the HCRT gene, the previously characterized promoter contains an upstream RR followed by a minimal promoter but our bioinformatics analysis suggests that the minimal promoter alone could be sufficient to induce expression and was designed as Ple111. All our refined control MiniPs express similarly to the original control MiniPs, demonstrating the suitability of the bioinformatics design approach (Fig. 3 and Fig. S1).
Novel MiniPs Based Mostly on Bioinformatics Analyses.
Applying the validated bioinformatics approach to 57 brain region-specific genes, a panel of new MiniPs was designed. This results in the important contribution of 27 novel-expressing MiniPs for which detailed sequence and design information can be found at the project website (http://www.pleiades.org) and in Dataset S1 “novel MiniP.” MiniPs were tested with different reporter genes as the 4-y project progressed: EGFP, EGFP/cre, and LacZ. Each MiniP was characterized in adult brain of germline mouse strains, with selected strains studied for expression in spinal cords and eyes. MiniPs driving expression of the EGFP/cre reporter fusion protein were analyzed for lacZ staining after recombination of the Gt(ROSA)26Sortm1Sor allele (17). Here, lacZ is acting as a historical marker and is visualized in cells descended from progenitors expressing Cre during development. The most recent subset of Pleiades MiniPs was constructed with the lacZ reporter gene to increase detection sensitivity and enable an early-rapid–analysis method in which brains are grossly sectioned in 1-mm slices, stained, and observed using a dissecting microscope. These MiniP-driven lacZ samples are largely taken from chimeras. Where both chimeras and germline animals have been studied, the chimera proves to be a good predictor of the germline expression pattern.
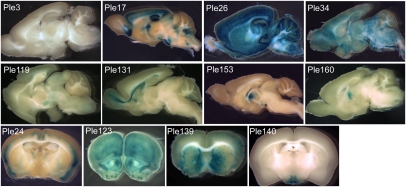
As summarized in Figs. 4 and 5 and Fig. S5 and Dataset S1, the novel MiniPs present unique expression patterns, as exemplified by Ple178, specific to neurons (Fig. 4F), Ple151, specific to glia (Fig. 4C), Ple162, restricted to a discreet region of the brain (Fig. 4D), and Ple34, broadly distributed to cells adjacent to blood vessels (Fig. 5 and Fig. S5D). MiniPs can drive an expression similar to the endogenous source gene (e.g., Ple24; Fig. 5 and Fig. S5I) or an unrelated expression pattern (e.g., Ple140; Fig. 5 and Fig. S5L), but in both cases, the MiniPs provide immediate value in that they direct expression to a subset of brain cells.
Fig. 4.
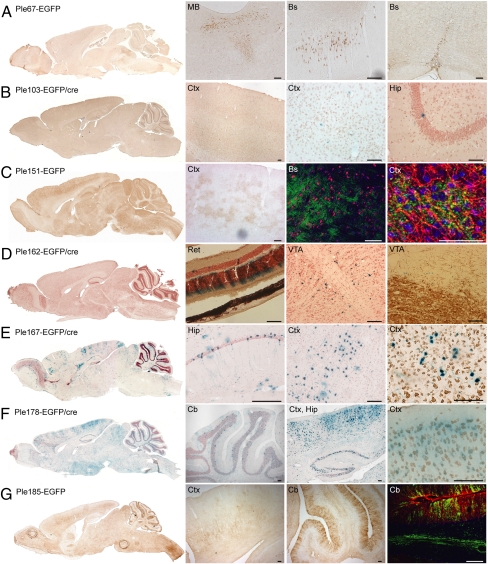
Novel MiniP expression patterns in the adult brain and retina. EGFP is detected using anti-GFP immunochemistry (brown) or EGFP/cre, detected using X-gal histochemistry (counterstained with neutral red). All sections are sagittal unless otherwise stated. Hip, hippocampus; MB, mid brain; Ret, retina; VTA, ventral tegmental area. (A) Ple67-EGFP (FEV RRs) expression is enriched in all raphe nuclei: from left to right, dorsal (coronal section), magnus (sagittal section), pallidus/obscurus (coronal section). (B) Ple103-EGFP/cre (HAP1 RRs) shows sporadic staining in various regions of the brain. (C) Ple151-EGFP (OLIG1 RRs) expresses throughout the brain in a puffy-like manner. The second detail image shows no costaining of EGFP (green) with neuronal marker NeuN (red). The last image shows costaining of EGFP (green) with the myelinating oligodendrocyte marker RIP (red). (D) Ple162-EGFP/cre (PITX3 RRs) is very specifically expressed in cells just dorsal to the VTA as well as the retina. The last image shows no costaining of X-gal (blue) with tyrosine hydroxylase (brown). (E) Ple167-EGFP/cre (POGZ RRs) is expressed in patches of cells across the brain. The last image shows costaining of X-gal (blue) with NeuN (brown). (F) Ple178-EGFP/cre (RGS16 RRs) is expressed in various regions of the brain. The last image shows a costaining of X-gal (blue) with NeuN (brown). (G) Ple185-EGFP (S100B RRs) expresses in Bergmann glia of the cerebellum and myelinated fibers in the cortex. The last image is a costaining of EGFP (green) with the endogenous S100B protein (red). (Scale bars, 100 μm, except C rightmost image, 50 μm.)
Fig. 5.
A view of selected novel MiniP driving lacZ assessed in 1-mm brain slices. At least two chimeric and/or germline brains were analyzed for each strain using X-gal staining of 1-mm brain slices and presented similar phenotypes. More detailed images can be found in Fig. S5 and at http://www.pleiades.org.
To demonstrate the value of the EGFP/cre strains carrying lacZ as a historical marker, we performed a developmental analysis to assess the history of the lacZ-positive neurons located in the mid brain of the Ple162-EGFP/cre (PITX3 RRs) mice (Fig. S6). The expression of the lacZ reporter is observed at E11.5 in the ventricular region at the mesencephalic flexure, but not in neural tissue at E10.5, delineating the onset of Ple162 MiniP expression. In a recent study, several mouse Pitx3 promoter constructs, one overlapping in sequence with Ple162, were analyzed in E12.5 mouse transgenic embryos but the authors do not report a similar brain expression pattern (18).
A Unique Dataset for in Silico Studies.
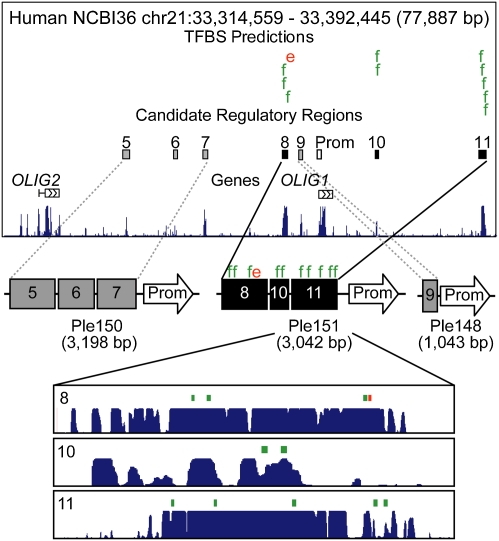
The expression profiles of active MiniPs can provide insights into the mechanisms governing transcriptional regulation in specific cells and regions of the brain. For the MiniPs associated with OLIG1, one MiniP drives expression in oligodendrocytes in the adult brain (Ple151), recapitulating the endogenous OLIG1 expression pattern (19), whereas two other MiniPs did not direct detectable expression in adult brain (Ple148 and Ple150). For oligodendrocyte-enriched TFs, we compared predicted TF binding sites between the positive and negative MiniP sequences (SI Materials and Methods and Tables S1 and S2). Our results highlight the potential involvement of EGR1 (KROX-24) and FOS (AP-1) in the Ple151 expression pattern (Fig. 6 and Table S2). These findings are in agreement with published data suggesting a role for OLIG1, KROX-24, and the AP-1 family of TFs in oligodendrocyte differentiation (20–22) and demonstrate the utility of validated MiniP sequences for predicting gene regulatory mechanisms.
Fig. 6.
Analysis of gene regulation based on MiniP activity. (Top) Human genomic sequence around OLIG1 and OLIG2 together with the candidate RRs included in Ple148, Ple150, and Ple151. Comparisons of TFBS predictions between Ple151 sequences (8, 10, 11) and all others (5–7, 9) identify EGR1 (e; red) and FOS (f; green) binding sites putatively responsible for Ple151-specific expression. The conservation plots were captured from the University of California Santa Cruz Genome Browser.
Discussion
The Pleiades Promoter Project uses a high-throughput, bioinformatically driven approach to produce brain-specific MiniPs. To date, 27 novel MiniPs have demonstrated positive brain expression (32% of constructs tested), greatly increasing the availability of brain promoters for new research initiatives. New promoters for the blood brain barrier (e.g., Ple34), proliferating neurons (e.g., Ple131), and glia (e.g., Ple185) may be particularly impacting.
The bioinformatically driven design of MiniPs for adult-brain gene expression is an important development compared with traditional tedious promoter-deletion studies. It also allows the generation of small constructs by removing nonessential sequences, making these tools potentially more portable and usable in many applications. The GENSAT project generates reporter gene expression using BACs (100–200 kb) driving EGFP regionally in the brain (3), but the results do not specify define RRs. The achieved success of the Pleiades design process was facilitated by the availability of large-scale gene expression studies (2, 23, 24), comparative genomics tools (25), and bioinformatics software for regulatory sequence prediction (26). By introducing the regulatory resolution scoring procedure to target the design efforts on the most tractable genes, the probability of design success was increased, a necessity given the expense of transgenic studies in the developed brain. The bioinformatics approach is not brain specific and therefore could be applied for compact promoter design in other tissues. Moreover, we report on our website both positive and negative constructs to facilitate future designs.
The MiniPs defined in this study are anticipated to be of wide utility. They can be used for brain-, spinal cord-, and eye-directed delivery of molecules such as siRNA, cre recombinase, fluorescent reporters, and research proteins. Driving specific reporters, they can be used in flow-sorting experiment to enrich or exclude cells of specific neural types. We have already demonstrated their function in mouse ESCs and a future critical step will be assaying their function in human stem cells. This will serve as a unique cross-species test of the Pleiades human MiniPs, and, if they function similarly, will deepen our understanding of regulatory program conservation.
The Pleiades mouse strains can also be used as marker strains that can be crossed with mutant strains to reveal specific cellular effect of the mutation. Further characterization of the germline mouse strains will delineate specific cellular subtypes targeted by the MiniPs, as well as expression outside the CNS and over development. As exemplified by the analysis of the OLIG1 data, the MiniPs hold basic utility in ongoing efforts to understand the transcriptional regulatory networks governing cellular phenotypes. Ultimately, the greatest impact of the Pleiades MiniPs is anticipated to be the added specificity for therapeutic gene delivery into the human brain. Although this may be accomplished using viruses, site-specific delivery to the human genome directly or in cell therapy is an area of active research (27, 28) and the suitability of the Pleiades promoters for such therapeutic delivery will require further study. The availability of a large collection of new MiniPs will play an important role in research and treatment for incurable brain diseases.
Materials and Methods
The Pleiades Promoter Project is characterizing 128 MiniPs over a 4-y time frame. The pipeline, involving five specialized laboratories, is summarized below. The general methods used have been described previously (10). Modifications or additions specific to this work are available in SI Materials and Methods.
The MiniP sequences were computationally designed and assembled into DNA molecules at a rate of four per week. The construct DNA was electroporated, typically into B6129F1 ESCs (mEMS1202 or mEMS1204, ref.10), at a rate of seven per week (including controls). A total of 10–15 clones per construct were picked, expanded, and PCR verified, to obtain approximately four correctly targeted ESC lines per construct. ESCs were typically microinjected into E3.5 blastocysts from BAN2, [N2 backcross of B6(Cg)-Tyrc-2J/J (JAX stock 000058) onto CD-1 (Charles River strain 022)], a strain combination selected for high blastocyst yield. Germline females were backcrossed to C56BL/6J (JAX stock 00664). The brains of N2–N3 germline males (8–12 wk) were analyzed using histochemical procedures. A minimum of three brains were processed for each MiniP. Every brain was cryosectioned (20-μm sections at 640-μm intervals) from medial to lateral in the sagittal plane and prepared for brightfield immunodetection of EGFP or lacZ histochemistry. When reporter expression was absent in at least three adult brains, a MiniP strain was classified as negative. Positive MiniP strains underwent further histological analyses to define the cellular pattern of gene expression. Both positive and negative strains were prepared for presentation on the Internet at http://www.pleiades.org.
Dataset S1 provides the list of the mouse strains described in this paper. ESC lines and mouse strains are available at the Mutant Mouse Regional Resource Centers (http://www.mmrrc.org/). All procedures involving animals were in accordance with the Canadian Council on Animal Care (CCAC) and UBC Animal Care Committee (ACC) (protocol no. A05-1258 and A05-1748).
Supplementary Material
Acknowledgments
We thank the Scientific Advisory Board members for their support and advice throughout the project: Steve Brown, Terry Magnuson, Sarah Bronson, Clifford Christians, Mark Ellisman, Yoshihide Hayashizaki, Matthew Kalnik, Gary Stormo, Claes Wahlestedt; Sue Kingsley for work on socioeconomic benefits and her Commercialization Advisory Group Sharon Finch and Martin Peet; Jill Richardson for monthly discussions and input; Tammy Philippo for administrative and management support; Tracey Weir for contributions on pipeline design; Miroslav Hatas for IT management support; Stefanie Butland, Warren Cheung, Christopher Dickman, Andrew Kwon, Anthony McCallum, and Dimas Yusuf for their contribution in the software development; Andrew Jervis and Lisa Findlay-Shirras for work on embryonic tissue; Nickolas Theodoric and Christopher Roach for their contribution in image processing. This work was funded by Genome Canada, Genome British Columbia, GlaxoSmithKline R&D Ltd., BC Mental Health and Addiction Services, Child and Family Research Institute, University of British Columbia (UBC) Institute of Mental Health, and UBC Office of the Vice President Research. We also acknowledge support from Canadian Research Chairs (to D.G. and E.M.S.), Canadian Institutes of Health Research New Investigator Award (to W.W.W.), Canadian Institutes of Health Research Canada Graduate Scholarships (to C.N.d.L. and D.L.F.), Michael Smith Foundation for Health Research Awards (to C.N.d.L., S.J.H.S., D.L.F., S.J.M.J., R.A.H., and W.W.W.), and Natural Sciences and Engineering Research Council of Canada (to S.J.H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009158107/-/DCSupplemental.
References
- 1.Sidman RL, Angevine JBJ, Taber Pierce E. Atlas of the Mouse Brain and Spinal Cord. Cambridge, MA: Harvard Univ Press; 1971. [Google Scholar]
- 2.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 3.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 4.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strand AD, et al. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007;3:e59. doi: 10.1371/journal.pgen.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson GW, et al. Purkinje cell protein-2 cis-elements mediate repression of T3-dependent transcriptional activation. Mol Cell Endocrinol. 1997;131:79–87. doi: 10.1016/s0303-7207(97)00095-6. [DOI] [PubMed] [Google Scholar]
- 7.Mima K, Deguchi S, Yamauchi T. Characterization of 5′ flanking region of alpha isoform of rat Ca2????dependent protein kinase II gene and neuronal cell type specific promoter activity. Neurosci Lett. 2001;307:117–121. doi: 10.1016/s0304-3940(01)01941-3. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza CA, et al. Identification of a set of genes showing regionally enriched expression in the mouse brain. BMC Neurosci. 2008;9:66. doi: 10.1186/1471-2202-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronson SK, et al. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang GS, et al. Next generation tools for high-throughput promoter and expression analysis employing single-copy knock-ins at the Hprt1 locus. Genomics. 2009;93:196–204. doi: 10.1016/j.ygeno.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Farhadi HF, et al. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J Neurosci. 2003;23:10214–10223. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barberi T, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 13.Chatelin S, et al. Neuronal promoter of human aromatic L-amino acid decarboxylase gene directs transgene expression to the adult floor plate and aminergic nuclei induced by the isthmus. Brain Res Mol Brain Res. 2001;97:149–160. doi: 10.1016/s0169-328x(01)00318-7. [DOI] [PubMed] [Google Scholar]
- 14.Portales-Casamar E, et al. PAZAR: A framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenherr CJ, Anderson DJ. Silencing is golden: Negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 16.Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Coulon V, et al. A muscle-specific promoter directs Pitx3 gene expression in skeletal muscle cells. J Biol Chem. 2007;282:33192–33200. doi: 10.1074/jbc.M706119200. [DOI] [PubMed] [Google Scholar]
- 19.Arnett HA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 21.Sock E, et al. Expression of Krox proteins during differentiation of the O-2A progenitor cell line CG-4. J Neurochem. 1997;68:1911–1919. doi: 10.1046/j.1471-4159.1997.68051911.x. [DOI] [PubMed] [Google Scholar]
- 22.Barnett SC, et al. Differential regulation of AP-1 and novel TRE-specific DNA-binding complexes during differentiation of oligodendrocyte-type-2-astrocyte (O-2A) progenitor cells. Development. 1995;121:3969–3977. doi: 10.1242/dev.121.12.3969. [DOI] [PubMed] [Google Scholar]
- 23.Magdaleno S, et al. BGEM: An in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visel A, Thaller C, Eichele G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32(Database issue):D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 27.Thyagarajan B, et al. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells. 2008;26:119–126. doi: 10.1634/stemcells.2007-0283. [DOI] [PubMed] [Google Scholar]
- 28.Kuduvalli PN, Mitra R, Craig NL. Site-specific Tn7 transposition into the human genome. Nucleic Acids Res. 2005;33:857–863. doi: 10.1093/nar/gki227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.