Abstract
The photoreceptor and PAS/LOV protein VIVID (VVD) modulates blue-light signaling and influences light and temperature responses of the circadian clock in Neurospora crassa. One of the main actions of VVD on the circadian clock is to influence circadian clock phase by regulating levels of the transcripts encoded by the central clock gene frequency (frq). How this regulation is achieved is unknown. Here we show that VVD interacts with complexes central for circadian clock and blue-light signaling, namely the WHITE-COLLAR complex (WCC) and FREQUENCY-interacting RNA helicase (FRH), a component that complexes with FRQ to mediate negative feedback control in Neurospora. VVD interacts with FRH in the absence of WCC and FRQ but does not seem to control the exosome-mediated negative feedback loop. Instead, VVD acts to modulate the transcriptional activity of the WCC.
Keywords: blue light, entrainment, photoreceptor, phase, circadian
Light, in addition to providing an energy source for many life forms on Earth, acts as a signal that may trigger development or serve as a repetitive cue that marks the passing of external time. External time cues are used by cellular timers such as circadian clocks to lock their periods to that of the external day. The process of period locking is called “entrainment” and ensures that cellular and behavioral activities happen at times of day when their adaptive value is highest (1–3). Blue light plays a central role in the entrainment of circadian clocks. Indeed blue-light photoreceptors and circadian clocks may have coevolved from a mechanism that originally served to detect (photoreceptor) and avoid (timer) harmful radiation (4–6). Our understanding of the molecular bases of circadian clocks and their responses to light has improved dramatically during the last decade or so, and the eukaryotic model organism Neurospora crassa has become one of the best-studied systems for understanding both processes (7–9).
The key components of the Neurospora circadian clock are the products of the white collar (wc-1 and wc-2), frequency (frq), and frq-interacting helicase (frh) genes (4, 10, 11). The blue-light photoreceptor WC-1, and its interaction partner WC-2, form the transcriptionally and photoactive WHITE COLLAR complex (WCC) that activates frq expression (4, 12). FRQ protein, in turn, complexes with FRH to form an FRQ-FRH complex (FFC) that represses WCC activity (9, 11). Thus, photoreception and temporal organization of gene expression are linked via the WCC (4, 12–14). Hyperphosphorylated WCC is transcriptionally less active, and repression of WCC by FRQ occurs via FRQ-mediated phosphorylation of WCC by Casein kinase 1 and 2 (CK1 and 2) (14, 15).
A second feedback loop that acts to repress WCC activity involves the product of the vivid (vvd) gene (16). Like WC-1, VVD is a PAS/LOV protein and blue-light photoreceptor; however, unlike WC-1, its presence is not essential for circadian rhythmicity in constant darkness (DD) (16–19). Nevertheless, VVD has important roles within the Neurospora circadian system. Without VVD the organism is more sensitive to light, resulting in the rapid breakdown of circadian organization in continuous illumination, whereas in the presence of VVD temporal rhythmicity is maintained in constant light (LL). By influencing clock resetting at both dawn and dusk, VVD affects light resetting and entrainment of the circadian clock (16, 20). Finally, VVD plays a role in maintaining the correct timing of clock-controlled output pathways at different temperatures (21).
Outside the circadian system VVD plays a key role in photoadaptation (18, 22, 23). VVD transiently dimerizes in a light-dependent manner (24–26), and this conformational change may relay light signals to downstream targets. However, despite the significance of VVD for aligning the Neurospora clock with the external day, we still do not know how VVD accomplishes these activities at the molecular level. No interaction partners of VVD (except VVD itself) have been identified, and consequently a mechanistic understanding of VVD's activities is lacking. Here we show that VVD interacts with clock components WC-1 and FRH. Our data suggest that VVD acts in the nucleus as a FRH-dependent corepressor of WCC.
Results
VVD Regulates frq Transcript Levels at Dusk.
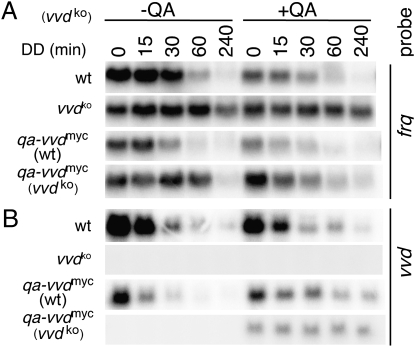
Molecular and physiological data have shown that VVD influences clock resetting at dawn and dusk. At dusk VVD's impact on molecular events is evident when comparing frq mRNA levels of WT and vvd-knockout (vvdko) strains. The frq transcript remains elevated longer in vvdko strains than in the WT, with a delay of about 4 h in reaching basal levels (top two lanes in Fig. 1A) (16, 20). To obtain more direct proof of VVD's role in the regulation of frq transcript levels, we created a strain in which a quinic acid (QA)-inducible copy of an myc epitope-tagged vvd gene (qa-vvdmyc) was inserted at the his-3 locus. The qa-vvdmyc construct was integrated into WT or vvdko strains, and these strains are referred henceforth to as qa-vvdmyc (WT) and qa-vvdmyc (vvdKO), respectively. By using this strategy, we were able to uncouple vvd expression from its normal light regulation (Fig. 1B and Fig. S1). Indeed, the ectopic expression of VVD induced by the addition of QA in qa-vvdmyc (vvdko) restores the normal decline in frq levels (compare the −QA and +QA samples in Fig. 1A, fourth panel) and accelerates frq degradation in qa-vvdmyc (WT) beyond that seen in a normal WT (compare the −QA and +QA samples in the first and third lanes in Fig. 1A and see quantification of data in Fig. S1A). The observation that frq levels are somewhat lower in QA medium was expected, because full expression of frq is dependent on glucose (27). Taken together, these data illustrate an inverse correlation between frq transcript levels and VVD protein.
Fig. 1.
VVD controls frq RNA levels. (A) Northern blots showing frq transcript levels in vvd+ (WT) or vvdko strains with or without an ectopic insertion of a QA-inducible vvd gene. Cultures were grown in LL for 24 h in the presence or absence of the inducer QA before transfer to DD, and samples were harvested after 24 h in LL (time point 0) or at the indicated times in DD. (B) As in A, but Northern blots were hybridized with a probe detecting the vvd transcript. Loading controls and quantitative analysis are shown in Fig. S1.
VVD Interacts with FRH.
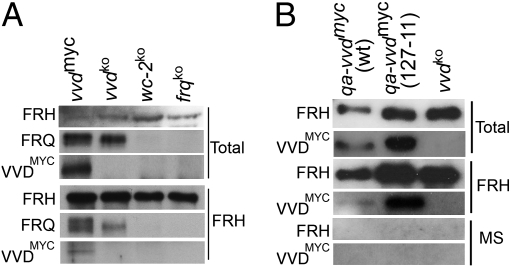
Because the FFC plays a key role in frq negative feedback (11, 28, 29), it was possible that VVD directly modulates the activity of this complex to influence frq levels at dusk. To test whether VVD interacts with the FFC, we performed coimmunoprecipitation (Co-IP) experiments on Neurospora whole-cell lysates using FRH or FRQ antisera, respectively. To facilitate detection of VVD, we used a strain that expresses MYC-tagged VVD in a vvdko background. We have shown previously that this strain rescues all known vvd mutant phenotypes, thus demonstrating that VVDMYC is fully functional (21). Henceforth all references to VVD protein levels are based on VVDMYC expression. Lysates from the vvd myc-tagged strain exposed to 30 min of LL were incubated with either FRH or FRQ antiserum before probing the blotted immunoprecipitates with MYC antiserum to test for the presence of VVD. Unfortunately, the FRQ antiserum proved too unspecific in our Co-IP experiments, so we were unable to judge whether VVD interacts with FRQ. However, when we used FRH antiserum, VVD was specifically immunoprecipitated (Fig. 2A). No signal was detected in frqko and vvdko strains that lacked a tagged copy of the vvd gene, indicating that the identified signal is VVDMYC and not an unspecific signal.
Fig. 2.
VVD interacts with FRH. (A) An antibody against FRH immunoprecipitates VVD in a Co-IP assay. Protein extracts from vvdmyc, vvdko, wc-2ko, and frqko strains grown for 30 min in LL were incubated with FRH antibody. An FRH antiserum immunoprecipitates VVD (bottom lane). As expected, FRH immunoprecipitates FRQ in WT and vvdko strains but not in the frqko and wc-2ko control strains. (B) VVD can interact with FRH in a strain (127-11) that lacks a functional WCC and FFC (SI Materials and Methods) but contains an ectopic qa-2–driven copy of the vvd gene [qa-vvdmyc (127-11)]. Western blots of total (input) extracts (top two lanes), extracts immunoprecipitated with FRH antiserum (middle two lanes), or unimmunized mouse serum (MS) (bottom two lanes) were probed with either FRH or MYC antiserum to detect FRH and VVD, respectively.
To test whether a functional WCC or FFC is necessary for the interaction of VVD with FRH, we exploited the QA-inducible system as outlined above and expressed a qa-vvdmyc in a background in which the wc-1, wc-2, frq, and vvd genes were deleted (strain 127–11) (Fig. 2B). In this qa-vvdmyc (127-11) strain, in which only the central clock gene frh remains intact, VVD still interacts with FRH, indicating that neither a functional FFC nor WCC is necessary for the interaction. The interaction occurs at both dawn and dusk transitions (Fig. S2B). As expected, extracts show a significant depletion of FRH after immunodepletion (top two lanes in Fig. S2A). However, no significant depletion of VVD is seen, suggesting that only a small fraction of total VVD interacts with FRH.
VVD Affects the Transcriptional Limb of FFC-Mediated Negative Feedback.
Next, we investigated the mechanism by which VVD mediates frq RNA turnover. Two distinct pathways that regulate levels of frq message have been described. First, a negative feedback loop involving FRQ and FRH is important for rhythmic down-regulation of frq at the level of transcription (10, 11). Second, the FFC also functions at the posttranscriptional level to control frq mRNA degradation via the exosome (28).
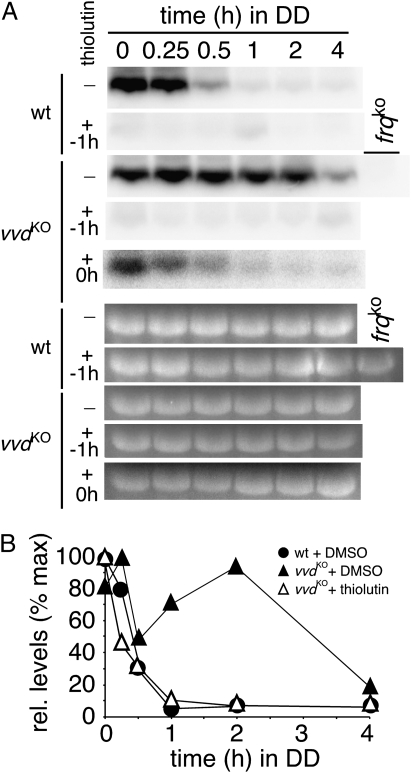
If VVD influences frq transcription, inhibition of transcription should abolish the differences in frq levels that exist between WT and vvdko strains. To test this possibility, we used the transcriptional inhibitor thiolutin (28) to inhibit transcription 1 h before or directly after the transfer of Neurospora cultures from light-to-dark (Fig. 3). When transcription was inhibited before the light-to-dark transition, we saw no or very little frq transcript present in either WT or vvdko strains, suggesting effective repression of transcription by the drug (second and fourth lanes in Fig. 3A). When frq transcription was allowed to proceed to the light–dark boundary before thiolutin was added, the kinetics of frq transcript decline in a vvdko strain were no longer slowed and resembled that of an untreated WT strain (compare fifth lane with top and third lanes of Fig. 3A). These data show that VVD targets frq transcription.
Fig. 3.
VVD represses frq transcription. (A) Northern blots showing frq transcript levels in WT (top two lanes) and vvdko strains (lanes three, four, and five from top) in cultures grown in LL for 24 h (time point 0) or for different lengths of time (h) in DD. Cultures were grown in the presence (+) or absence (−) of the transcriptional inhibitor thiolutin. Thiolutin was added either 1 h before (−1 h) or immediately after (+0 h) liquid cultures were transferred from light to dark. Ethidium bromide–stained ribosomal RNA was used to control for loading of Northern blots. (B) Quantitative analysis of Northern blots (shown in A) of untreated WT (●) and untreated (▲) and thiolutin-treated (immediately after the light-to-dark transfer) (△) vvdko strains. Within each experiment maximum frq RNA levels were set to 100%.
This conclusion was confirmed by an experiment in which we placed frq under the control of the QA-inducible promoter. The qa-frq construct was integrated into a frqko strain or a frqko vvdko double-mutant strain to generate qa-frq (frqko) and qa-frq (frqko vvdKO) strains, respectively. In analogy to our QA-inducible VVDMYC expression system described above, this experiment allowed us to uncouple frq expression from its normal light-induced transcriptional regulation and study the reduction in frq transcript levels in a controlled manner after release from the inducer. If VVD targets frq transcription, replacing the frq promoter with the QA promoter should result in a similar drop in frq transcript levels in both the qa-frq (frqko) and qa-frq (frqko vvdKO) strains after release from the inducer. On the other hand, a posttranscriptional action of VVD on frq levels should result in a difference (i.e., delay) in frq mRNA turnover in qa-frq (frqko vvdKO) strains as compared with qa-frq (frqko). After release from the inducer, frq levels in qa-frq (frqko) and qa-frq (frqko vvdKO) strains were followed for a period of 8 h in DD, and we observed no significant difference in the decline of frq transcript kinetics in the two strains. There is some variability in QA-induced frq levels immediately following release from the inducer, but the kinetics of the decline of frq are similar in all strains and conditions tested (Fig. S3C). These data therefore support the conclusion that VVD influences frq RNA at the level of transcription and not via the exosome-mediated function of the FFC. Interestingly, the presence or absence of light had no significant influence on frq turnover, suggesting that light was not required for the activation of VVD (compare Fig. S3 A and B).
VVD Localizes to Both the Cytoplasm and the Nucleus.
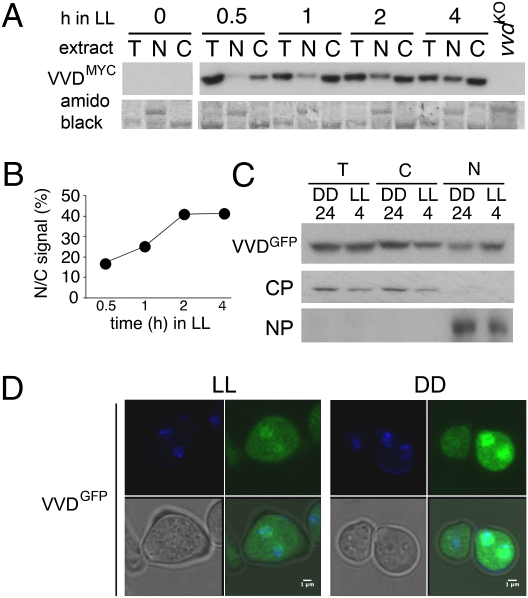
Because our data showed that VVD represses transcription, we explored VVD's localization. VVD is reported to localize to the cytoplasm with no evidence for nuclear localization (18). However, the use of an MYC antibody to detect myc-tagged VVD reduces the threshold at which VVD can be detected (21). We prepared nuclear and cytoplasmic extracts from Neurospora cultures grown in DD and different time points following transfer to LL (Fig. 4). In agreement with previous results, we did not detect VVD in extracts from Neurospora tissue grown in extensive periods of darkness. In contrast, after 30 min in LL, significant amounts of VVD were seen in both the total extracts and the cytoplasmic fraction (Fig. 4A). Upon light exposure a faint signal also is detectable in nuclear extracts, and this signal becomes stronger with increasing time in LL. (Fig. 4A). We could not rule out the possibility that the increase in nuclear VVD is simply a consequence of the drastic increase in VVD levels during exposure to light. However, when expressed as the ratio of nuclear to cytoplasmic VVD levels (Fig. 4B), our data suggested that nuclear entry of VVD may be light regulated.
Fig. 4.
VVD is both a cytoplasmic and nuclear protein. (A) Total (T), nuclear (N), or cytoplasmic (C) extracts were prepared from Neurospora tissue grown for 24 h in DD (0) or with exposure to LL for the indicated times (h). Western blots were probed with an MYC antibody to detect VVDMYC. The amido black-stained membrane serves as a loading control. (B) Graph showing the percent ratio of nuclear to cytoplasmic signal using the Western blot data shown in A. (C) Neurospora extracts of strains expressing GFP-tagged VVD (under ccg-1 promoter control) grown for 24 h in DD or 4 h in LL. Western blots were probed with a GFP antibody to detect VVDGFP. CP, cytoplasmic protein; NP, nuclear protein. (D) Subcellular localization of VVDGFP (under ccg-1 promoter control) in Neurospora conidiospores fixed at time points DD24 and LL4. Each subpanel shows confocal images of fluorescence from DAPI-stained spores (Upper Left), fluorescence from the GFP signal (Upper Right), an overlay of both (Lower Right), and corresponding bright-field image (Lower Left). (Scale bar, 1 μm.)
To gather independent information on VVD's localization within the cell, we generated carboxyl-terminal fusions of GFP to VVD using plasmids (30) in which gene expression is controlled by the strong Neurospora ccg-1 promoter. The ccg-1–driven vvd-gfp construct is capable of rescuing the carotenoid and clock phenotypes of a vvdko strain (Fig. S4 A and C), and levels of VVDGFP are somewhat higher than in a strain that expresses vvd under its own promoter (Fig. S4B). Cell fractionation of the GFP-tagged VVD strains confirmed results obtained earlier with myc-tagged VVD (Fig. 4A) that VVD is localized both in the cytoplasm and nucleus (Fig. 4C). For confocal microscopy, we harvested conidia (that usually contain one to three nuclei) from strains kept in DD for 24 h and from strains kept in LL for 4 h. In a small number of conidia the nuclear localization of VVDGFP can be seen clearly (Fig. 4D), again confirming the results we obtained with our cellular extracts. As expected, conidia obtained from the 1H-GFP fusion strain also show nuclear localization for 1HGFP (Fig. S4D, Bottom). In contrast, we never saw any nuclear signal above background in nuclei from WT strains that were not tagged with GFP (Fig. S4D, Top and Middle). Because published data suggest that GFP alone is cytoplasmic and does not localize to the nucleus (31) the low percentage of nuclei that fluoresce in vvd gfp-tagged strains suggests that nuclear accumulation or entry of VVD may be highly dynamic. In conclusion, our combined data confirmed that a substantial proportion of VVD localizes to the nucleus.
VVD Interacts with the WCC.
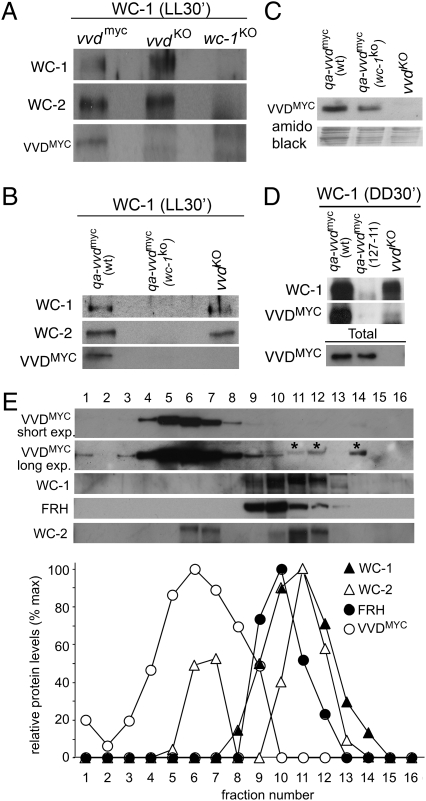
The nuclear localization of VVD and VVD's interaction with FRH led us to test whether VVD represses frq transcription by binding to the WCC. Such an activity would be consistent with the observation that VVD interacts with FRH, because FRH mediates the phosphorylation and inactivation of WCC (15, 32, 33). Moreover, VVD has been shown to influence WC-1 phosphorylation and thus WCC activity (16, 23). We performed Co-IP experiments in which Neurospora whole-cell lysates were incubated with WC-1 antiserum before probing for the presence of VVD. As can be seen in Fig. 5, VVD was immunoprecipitated in both LL (Fig. 5 A and B) and DD (Fig. 5D).
Fig. 5.
VVD interacts with the WCC in the light and in the dark. (A) Protein extracts from vvdmyc, vvdko, and wc-1ko strains were incubated with WC-1 antibody. WC-1 immunoprecipitates VVD in the vvdmyc strain but not in a vvdko or wc-1ko strain. (B) WC-1 antibody does not cross-react with VVD. QA-inducible VVD was expressed in a vvdko and a vvdko, wc-1ko background, and protein extracts coimmunoprecipitated with WC-1 antibody. No VVDMYC was immunoprecipitated in a vvdko, wc-1ko background. (C) Control to show that QA-inducible VVDMYC is stably expressed in the absence of WC-1. (D) A WC-1 antiserum immunoprecipitates QA-induced VVDMYC in DD in a wc-1+ strain (qa-vvdmyc). WC-1 antiserum fails to immunoprecipitate VVDMYC in a strain (127-11) that lacks WC-1 but expresses QA-induced VVDMYC at levels similar to the control strain qa-vvdmyc. (E) A small proportion of VVD is found in sucrose gradient fractions containing WC-1, WC-2, and FRH. Western blot (top five lanes) and graph (Lower) showing the densitometric analysis of VVDMYC, WC-1, WC-2, and FRH from protein extracts size-fractionated on a 10–35% sucrose gradient. Fraction 1 corresponds to low molecular weights and fraction 16 to higher molecular weights. Asterisks denote unspecific signals. For densitometry the maximum signal for each protein was set to 100%.
Because the PAS/LOV domains of VVD and WC-1 are similar, it was important to control for the specificity of the WC-1–VVD interaction. Therefore we created strains in which vvdmyc could be expressed in a wc-1ko background. Because vvd expression is dependent on WC-1, a QA-driven vvd gene was expressed to uncouple VVD expression from WC-1 expression. The qa-vvd construct was integrated into a vvdko or a vvdko wc-1ko double mutant to generate qa-vvdmyc (vvdko) and qa-vvdmyc (vvdko wc-1KO) strains, respectively. In these strains VVDMYC is expressed at comparable levels, although VVD levels appeared slightly lower in the qa-vvdmyc (vvdko wc-1KO) strain (Fig. 5C). In these experiments VVD was immunoprecipitated only in strains that express WC-1 protein (Fig. 5B). In another control experiment using qa-vvdmyc (WT) and qa-vvdmyc (127-11) strains, we observed that VVD is immunoprecipitated only in the qa-vvdmyc (WT) strain (Fig. 5D). If VVD interacts with WC-1 via the WCC, we expected VVD also to interact with WC-2. However, our efforts to test this interaction by Co-IP were hampered by unspecific cross-interactions of the WC-2 and MYC antisera with VVDMYC and WC-2, respectively.
To test whether VVD must be signaling active for the interaction with WC-1, we tested two mutant strains that lack light-induced activity: vvdC71S, which is biologically inactive because of its inability to homodimerize (26), and vvdC108A, a strain in which the formation of a cysteinyl adduct that is critical for light signaling is impaired (18) (Fig. S2C). Both mutant proteins interacted with WC-1, suggesting that VVD does not need to be in a signaling-active state to interact with WC-1.
To investigate further whether VVD is part of higher molecular weight complexes, as suggested by our immunoprecipitation experiments, we performed sucrose gradient experiments (Fig. 5E). In agreement with previous results, we detected WC-2 in two peaks reported to be about 60 kDa and 200 kDa in size and found WC-1 to co-migrate with the higher molecular peak identified for WC-2 (33). FRH migrated in a single peak that overlaps both WC-1 and WC-2. The broad peak of VVDMYC spanning the fractions where monomeric WC-2 migrates is consistent with VVD forming a homodimer, as was suggested recently (24–26). The presence of VVD in fractions that contain the much higher molecular weight complexes of WC-1, WC-2, and FRH is consistent with our Co-IP data that showed an interaction of VVD with WC-1 and FRH.
Does VVD interact with DNA-bound WCC? We tested this possibility in EMSA (Fig. S5). Nuclear proteins (LL 4 h) were extracted from vvdko, vvdmyc or vvdgfp strains and incubated with a previously identified proximal light-responsive element (pLRE) located in the frq promoter (12). In line with published results (12, 34, 35), the free probe was caught in a high molecular weight complex when incubated with the nuclear extracts, and the complex was supershifted when incubated with WC-2 antiserum (Fig. S5). However, we observed no supershift when extracts were incubated with either MYC antibody (Fig. S5A) or GFP antibody (Fig. S5B), suggesting that VVD binds free rather than DNA-bound WC-1.
Discussion
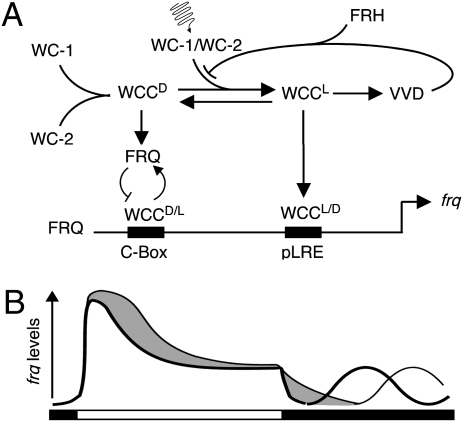
We have shown that VVD interacts with central components of the Neurospora circadian clock and with components of blue-light signaling to inhibit the transcriptional activity of the WCC at dawn and dusk. The model shown in Fig. 6 depicts how VVD may exert its function at the dawn and dusk transitions. We know that WC-1 and WC-2 form multimers that can bind to proximal and distal GATN cis elements in the frq promoter (12). The complex formed in the dark (WCCD) is faster migrating (i.e., smaller) than the one formed in the light (WCCL). The latter mediates transcription of light-induced genes, such as vvd (12, 35). Light induction of frq is regulated mainly through the proximal light-responsive element (LRE), whereas the distal element, although light responsive, is necessary for sustained rhythmicity of frq in DD (12). We have shown here that VVD binds WC-1 both in LL and upon release into DD. It thus seems likely that VVD disrupts the efficient formation of WCCL. This disruption would result in a decrease of light-induced transcription in LL as well as effective disruption in DD of the WCCL that is present at the light–dark transition. In the absence of VVD, the equilibrium between WCCD and WCCL would be shifted toward WCCL, resulting in increased transcription of frq in the light as well as during the first hours in the dark. However, because frq regulates its own transcription via its own gene product, down-regulation of frq in DD is not simply a function of levels and activity of WCC but depends on the level and activity of FRQ.
Fig. 6.
Model of VVD's action in negative feedback regulation of the WCC. (A) In the dark WC-1 and WC-2 heterodimerize to form a WCCD. Upon light exposure a multimeric WCC complex, WCCL, forms (11, 14) that mediates transcription of light-induced genes (e.g., vvd). VVD binds WC-1 to inhibit the efficient formation of WCCL, resulting in a decrease of light-induced transcription. Without VVD the equilibrium between light and dark complexes is shifted toward WCCL, resulting in increased transcription of frq in the light. Upon transfer to DD and without VVD, the increased activity of WCCL results in prolonged activation of frq transcription (possibly via its proximal LRE), whereas in the WT FRQ-mediated phosphorylation of WCCD leads to rapid inactivation of frq transcription. WCCL and WCCD have different preferences for proximal and distal promoter elements, with WCCD preferentially engaged at the distal clock box where negative feedback regulation by FRQ takes place (11). (B) Simplified schematic of the frq RNA profile after light induction and in constant darkness in WT (thick line) and in a vvdKO strain (thin line). The gray area depicts the difference in frq activation between the two strains and is a result of a change in equilibrium between WCCD and WCCL. Prolonged activation in the dark leads to a characteristic phase delay in the onset of frq transcript oscillations and overt circadian rhythmicity.
Because it is known that (i) FRQ cannot repress its own transcription effectively in the light (36), (ii) the light-induction of frq is controlled largely by the proximal LRE (12), and (iii) WCCL is indicative of the light-activated state of WCC (12, 35), it seems plausible that FRQ cannot repress its own transcription effectively via WCCL but exerts its main repressive activity on WCCD. This possibility could provide an alternative explanation of why frq takes longer to decline in a vvdko strain, because the majority of WCC would be predicted to be in the form of WCCL.
Our experiments suggest that only a small amount of VVD is in a complex with WC-1. Because VVD's impact on light resetting of the circadian clock and photoadaptation is quite profound, one would expect that a larger fraction of VVD must complex with the FFC and WCC to achieve its function in these processes. Consequently, we favor the idea that the interaction is short-lived but not necessarily weak. This situation is reminiscent of the observation that only very small amounts of FRQ seem to interact with the WCC (15). Similarly, the observation that FRQ does not appear to be part of the promoter-bound WCC (12) resembles our observation that antisera against VVDMYC or VVDGFP do not supershift the WCC. However, it is possible that these antibodies are less efficient in EMSA experiments or that a DNA-bound interaction is too weak or transient to be detected by EMSA; therefore, we cannot rule out a DNA-bound interaction at this stage. Interestingly, VVD mutants that are thought to be defecive in photosignaling still can interact with WC-1 and FRH, suggesting that the repressive functions of VVD in light signaling may occur as part of a complex, with light-activation of VVD affecting the conformation of the entire complex. VVD's role in repressing light responses aside, there is evidence that VVD promotes some aspects of light signaling (18), and a more recent microarray study suggests that VVD has a role in modulating late light responses in Neurospora (22). It therefore is likely that VVD may have functions that do not require its interaction with the WCC or FFC.
Finally, our data show that VVD can interact with FRH in the absence of a functional FFC or WCC, and it is tempting to speculate that FRH is the primary platform upon which the various complexes assemble.
Materials and Methods
Plasmids and Strains.
Strain 54–3 (bd, a) was used as the WT in all experiments. A detailed description of how strains were created can be found in SI Materials and Methods. All strains were verified using PCR or Southern blot analysis. Homokaryons were generated as previously described (37).
Growth Conditions.
Race tube and liquid culture experiments were carried out in Sanyo MLR-350 light- and temperature-controlled chambers as described in SI Materials and Methods. For transcription inhibitor experiments, thiolutin (Tocris Biosciences) was added at a final concentration of 12 μg/mL. Control samples were treated with DMSO.
Co-IP.
Co-IP experiments and subsequent Western blot analysis were performed as described in SI Materials and Methods.
Cell Fractionation and EMSA.
Cellular fractions were isolated essentially as previously described (38), and details are given in SI Materials and Methods. For EMSA, the frq proximal LRE oligonucleotides (Eurofins MWG Operon) sequences described previously (12) were used to make the dsDNA probe. Binding conditions and gel electrophoresis are described in SI Materials and Methods.
Sucrose Gradients.
Protein extracts were obtained using standard protein extraction buffer as described previously (30), and sucrose gradient experiments were carried out as detailed in S1 Materials and Methods.
RNA Extraction and Northern Blot Analysis.
RNA was extracted using the Qiagen RNeasy mini kit according to the manufacturer's instructions. Northern blot analysis was carried out as previously described (20) and as detailed in S1 Materials and Methods.
Confocal Microscopy.
Conidia were fixed in 4% formaldehyde for 1.5 h, washed in distilled H2O, incubated in 50 μg/mL DAPI (Sigma) for 10 min at room temperature, and then washed and resuspended in 25% glycerol. Details for image collection are given in S1 Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sue Crosthwaite (University of Manchester, Manchester, UK) for helpful suggestions and critical reading of the manuscript and Jay Dunlap, Jennifer Loros (Dartmouth Medical School, Hanover, NH), and Yi Liu (University of Texas Southwestern Medical Center, Dallas, TX) for the generous gift of FRQ WC-1, WC-2, and FRH antisera. We especially thank Jane Kott for help with the microscopy. Microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the University of Manchester Strategic Fund. This work was supported by Grants BB/D00988X/1 and BB/F012055/1 from the Biotechnology and Biological Sciences Research Council to C.H.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009474107/-/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology. Sunderland. MA: Sinauer Associates, Inc.; 2004. [Google Scholar]
- 2.Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol Int. 2003;20:741–774. doi: 10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- 3.Price-Lloyd N, Elvin M, Heintzen C. Synchronizing the Neurospora crassa circadian clock with the rhythmic environment. Biochem Soc Trans. 2005;33:949–952. doi: 10.1042/BST20050949. [DOI] [PubMed] [Google Scholar]
- 4.Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 5.Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57(Suppl 1):S286–S289. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 6.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap JC, et al. A circadian clock in Neurospora: How genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day. Cold Spring Harb Symp Quant Biol. 2007;72:57–68. doi: 10.1101/sqb.2007.72.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 10.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 11.Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 13.Ballario P, et al. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 14.He QY, et al. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 15.Schafmeier T, et al. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 16.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 17.Cheng P, He QY, Yang YH, Wang LX, Liu Y. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc Natl Acad Sci USA. 2003;100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrode LB, Lewis ZA, White LD, Bell-Pedersen D, Ebbole DJ. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet Biol. 2001;32:169–181. doi: 10.1006/fgbi.2001.1264. [DOI] [PubMed] [Google Scholar]
- 20.Elvin M, Loros JJ, Dunlap JC, Heintzen C. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 2005;19:2593–2605. doi: 10.1101/gad.349305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt SM, Elvin M, Crosthwaite SK, Heintzen C. The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev. 2007;21:1964–1974. doi: 10.1101/gad.437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwerdtfeger C, Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol. 2001;39:1080–1087. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 24.Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc. 2008;130:12226–12227. doi: 10.1021/ja804236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoltowski BD, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunlap JC. Genetics and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 28.Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi M, Collett M, Loros JJ, Dunlap JC. FRQ-interacting RNA helicase (FRH) mediates negative and positive feedback in the Neurospora circadian clock. Genetics. 2009;184:351–361. doi: 10.1534/genetics.109.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Folco HD, et al. Histone H1 Is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot Cell. 2003;2:341–350. doi: 10.1128/EC.2.2.341-350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Q, et al. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- 33.Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Froehlich AC, Loros JJ, Dunlap JC. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci USA. 2003;100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crosthwaite SK, Loros JJ, Dunlap JC. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 37.Ebbole D, Sachs MS. A rapid and simple method of isolation of Neurospora crassa homokaryons using microconidia. Fungal Genetics Newsletter. 1990;37:17–18. [Google Scholar]
- 38.Schwerdtfeger C, Linden H. Localization and light-dependent phosphorylation of white collar 1 and 2, the two central components of blue light signaling in Neurospora crassa. Eur J Biochem. 2000;267:414–422. doi: 10.1046/j.1432-1327.2000.01016.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.