Abstract
Both treatment of disease and basic studies of complex tissues can benefit from directing viral vector infection to specific cell types. We have used a unique cell targeting method to direct viral vector transduction to cerebral cortical neurons expressing the neuregulin (NRG) receptor ErbB4; both NRG and ErbB4 have been implicated in schizophrenia, and ErbB4 expression in cerebral cortex is known to be restricted to inhibitory neurons. We find that a bridge protein composed of the avian viral receptor TVB fused to NRG, along with EnvB-psuedotyped virus, is able to direct infection selectively to ErbB4-expressing inhibitory cortical neurons in vivo. Interestingly, although ErbB4 is expressed in a broad range of cortical inhibitory cell types, NRG-dependent infection is restricted to a more selective subset of inhibitory cell types. These results demonstrate a tool that can be used for further studies of NRG and ErbB receptors in brain circuits and demonstrate the feasibility for further development of related bridge proteins to target gene expression to other specific cell types in complex tissues.
Keywords: neuregulin, rabies virus, avian virus receptor for subgroup B, lentivirus, EnvB
In recent years, there has been a significant increase in the development and use of genetic methods for the study of complex tissues. For example, the brain is composed of numerous structures, each of which is populated by multiple neuron types whose axons and dendrites are intimately intertwined. In such tissues, individual cell types each have unique functional roles, which are often difficult to assess independently because of the difficulty of monitoring or manipulating one cell type separately from the others. Genetic methods have proven to be very powerful for the study of such complex tissues, because it has been possible to generate mouse lines that express genes such as Cre-recombinase selectively in a particular cell type. These mice can then be combined with conditional expression of genes that allow the activities of the affected cells to be monitored or manipulated. For example, recently developed genetic tools for studies of the brain allow neuronal activity to be selectively assayed or altered and allow the connectivity of specific cell types to be identified (1).
Despite the great utility of these tools for studies of mice, there is also a need to develop effective genetic strategies for targeting expression to specific cell types in species other than mice. For example, studies of the circuit mechanisms that mediate complex behavior in primates would benefit from cell type targeting in the brain (2), and viral vector-based human gene therapy will have reduced side effects and greater efficacy if specific cell types are targeted. Incorporation of specific promoters into viral vectors has met with limited success, presumably in part due to the difficulty of identifying the relevant regulatory elements. We are not aware of any examples where selective expression has been achieved with a particular promoter element, and it has also been demonstrated that the selectivity did not arise at least in part because of endogenous tropism of the viral vector. For example, the α-CAM kinase promoter was thought to restrict expression to excitatory cortical neurons, but this restriction is only observed in the context of a lentiviral vector (3, 4), which has an extremely strong tropism for excitatory neurons (5); the same promoter fails to restrict expression in the context of an AAV vector, which reliably infects inhibitory neurons (5). Although there is promise for improved methods, there is also the likelihood that many genes are regulated by epigenetic mechanisms and, thus, the regulatory elements that control their expression cannot be exploited to achieve cell type-specific gene expression in the adult. Furthermore, many gene products are subject to posttranslational modifications that alter their functional properties, including binding of receptors to their various ligands. Thus, it might also be useful to target vector delivery to cells based on differential presentation of surface receptors, regardless of the species being studied.
An alternative to transcriptional control of gene expression is “transductional targeting,” which uses engineering of viral vectors to selectively infect cell types of interest. Several such approaches have been developed and demonstrated in cell culture (for review, see refs. 6 and 7), and more recently, to target B lymphocytes in the blood (8) or s.c.-injected tumor cells in vivo (9). However, we are not aware of any successful demonstrations after injections into a complex solid tissue, such as the brain, in vivo. Here, we target gene expression to a specific subset of inhibitory neurons in the cerebral cortex, using a targeting system and vector first described and demonstrated in cultured cells by Snitkovsky et al. (10). They generated a bridge protein composed of the avian virus (ASLV-A) receptor for subgroup A (TVA) fused to the EGF-like region of neuregulin β1 (TVA-NRG1), to target viral infection to cells expressing neuregulin receptors, called ErbBs. We chose to use an ASLV-B receptor for subgroup B (TVB)–NRG1 bridge protein (10) to target ErbB4 expressing neurons in the cerebral cortex because several lines of evidence indicated that it might be possible to target gene expression to a specific subpopulation of neurons expressing ErbB4 receptors and because both ErbB4 and its ligand (NRG) are implicated in schizophrenia (11). TVB-based bridge proteins were used because, unlike TVA-bridge proteins, they can support virus entry when first loaded onto virions as opposed to cell surfaces (12).
Several lines of evidence have implicated disorders of inhibitory cortical circuitry in schizophrenia (13). At the same time, recent evidence has shown that schizophrenia is associated with mutations in both NRG and ErbB4 receptors (11). Visualization of the neocortical pattern of ErbB4 expression in the Allen Brain Atlas (http://mouse.brain-map.org/brain/Erbb4.html), and antibody staining in the hippocampus (14, 15) and cortex (16, 17), indicate that expression of this receptor is restricted to inhibitory neurons, but not necessarily to particular types out of the dozens of distinct inhibitory neocortical cell types (18). Furthermore, functional studies in cortical brain slices show that NRG1 potentiates the release of GABA from parvalbumin (PV)-positive inhibitory neurons, enhancing their effects on postsynaptic pyramidal neurons (19, 20). Although the role of NRG and ErbB4 signaling in other cortical inhibitory cell types remains poorly understood, it appears likely that there are differences in the way that various types of ErbB4-expressing inhibitory neurons respond to NRG (16). Perhaps functional interactions with NRG differ between ErbB4-expressing inhibitory neuron types.
We aimed to target cortical ErbB4-expressing neurons for transgene expression in vivo by using an EnvB-pseudotyped virus and the TVB–NRG1 bridge protein (10). With such a system, it is expected that viral infection might be biased to cells that have particular types of functional interactions with NRG. (See Fig. S1 for a schematic illustration.) We find that the TVB–NRG1 bridge protein successfully targets viral infection selectively to ErbB4-expressing cortical neurons. Subsequent characterization of these cells demonstrates that they are in fact a highly selected subset of cortical inhibitory neurons. These cells have diverse dendritic morphologies, which are both multipolar and bipolar, but are restricted in their expression of characteristic neurochemical markers. Despite the fact that the classical markers PV, somatostatin (SST), and calretinin (CR) all overlap with ErbB4 expression in the cortex (16, 17), cells infected via the TVB–NRG1 bridge protein are negative for SST and only rarely positive for PV, whereas most are positive for CR and/or vasoactive intestinal peptide (VIP). These results therefore demonstrate the successful application of bridge protein targeting of a viral vector to selected cells within a complex tissue in vivo. This targeting strategy should be of general utility for targeting other cell types, and the TVB–NRG1 bridge protein, in particular, provides a unique tool for the further investigation of a selected subset of cortical inhibitory neurons and NRG-sensitive cells in other brain areas.
Results
The results presented here are based on the analysis of mouse or rat cortical tissue 3 d after in vivo cerebral cortical injections of a mixture TVB–NRG1 (10) and EnvB-pseudotyped, G-deleted rabies virus (RV) (see Methods and ref. 21), as well as related control conditions. Identification of infected cells with G-deleted rabies virus was assessed by expression of GFP or mCherry from the rabies genome. The G-deleted rabies was used because it is a highly sensitive method for monitoring infection and results in complete filling of infected cells with GFP or mCherry, allowing detailed morphological observations (22). Importantly, unlike wild-type rabies virus, the G-deleted rabies does not spread from directly infected cells to other nearby or distant cells (22, 23).
Morphology and Distribution of Cortical Cells Infected by TVB–NRG1 and EnvB Pseudotyped RV.
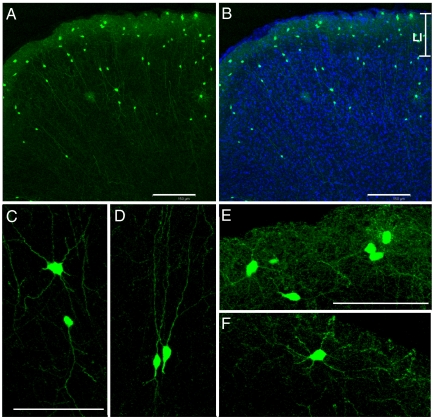
To test whether virus infection mediated by the TVB–NRG1 bridge protein is able to target specific neuron types in vivo, we used EnvB-pseudotyped RV mixed with the TVB–NRG1 bridge protein as an inoculum and injected it into the adult mouse cortex. Three days after injection, animals were perfused and the brains sectioned and stained. We hypothesized that this was likely to result in the selective expression of reporter gene (GFP or mCherry) in a subset of cortical inhibitory neurons. To assess this possibility, our first observations were the distribution and morphologies of neurons infected in the presence of TVB–NRG1 and under control conditions. These experiments used a GFP-expressing virus (EnvB-GFP-RV). In both the immediate vicinity of the injection site and in surrounding regions up to >2 mm away, many GFP-positive cells were detected as shown in Fig. 1 and Fig. S2 A–C. As expected from previous studies in which neurons were infected with GFP-expressing G-deleted rabies virus (22), GFP completely filled the cells’ processes, allowing visual assessment of morphological details (Fig. 1). In the immediate vicinity of the injection (within ≈300 μm), labeled cells sometimes had morphologies characteristic of both glia and neurons (Fig. S2B). We therefore focused more detailed analyses on cells >300 μm and up to 2 mm laterally from the injections where glia-like labeling was rare (e.g., Fig. S2C). In these locations, the GFP-filled cells were distributed throughout the cortical layers and had the morphological features of a diverse group of cortical inhibitory neurons (Fig. 1). In particular, labeled neurons were aspinous and nonpyramidal, strongly suggesting that they were exclusively inhibitory. Such preferential labeling is not expected by chance because only ≈15–20% of cortical neurons in the mouse are inhibitory (5). Outside of layer 1, most labeled cells had a bipolar dendritic morphology (only two primary dendritic processes extending from the cell body) (Fig. 1D). This morphology strongly suggested that these neurons were likely to express calretinin and/or VIP (18). In addition, multipolar neurons were also relatively common (Fig. 1C). Layer 1 neurons were exclusively multipolar (Fig. 1 E and F). A quantitative analysis of GFP-positive neurons from every 24th section revealed that, although cells were located throughout the cortical layers, 46% (189/415 GFP+ neurons in layer 1/GFP positive neurons in all layers) of GFP-positive cells were located in layer 1. Qualitatively similar results were observed 3 d after injections of TVB–NRG1 and EnvB-GFP-RV into the rat cerebral cortex (Fig. S3), indicating that the highly selective transduction of neurons is not unique to mouse cortex.
Fig. 1.
Cortical neurons expressing GFP after infection with EnvB-pseudotyped rabies virus and NRG1-TVB bridge protein. (A) Low power view illustrating overall location and morphologies of infected GFP-labeled neurons. Pial surface is at the top. (B) Same as A with DAPI counterstain (blue) to show cortical layers. The boundaries of layer 1 at the top of the photo are indicated as L1. (C–F) Higher power views illustrating more detailed morphological features of neurons in the cortical plate (C and D) and in layer 1 (E and F). Note that all GFP-expressing cells are aspinous and nonpyramidal. (Scale bars: A and B, 150 μm; C and E, 75 μm.) Bar in C also applies to D. Bar in E also applies to F.
Previous studies using TVA or TVB bridge proteins along with EnvA- or EnvB-pseudotyped retrovirus to infect cultured cells (10, 12) suggest that the utility of this strategy can likely be extended to numerous enveloped viral vectors, including lentiviruses. Because lentiviruses are very useful for long-term and stable gene expression, we also assessed infection of mouse cortical neurons in vivo by using the TVB–NRG1 bridge protein and EnvB-pseudotyped lentivirus (EnvB-LV-GFP). Two weeks after cortical injections, GFP-expressing neurons were seen sparsely scattered throughout the cortical layers in a distribution similar to that observed after rabies injections (Fig. S4). However, as expected from the fact that replication-competent rabies virus generates very high levels of gene expression, and levels of gene expression from lentivirus are likely to be lower and depend on the site of integration into the host cell genome, fewer cells were labeled, GFP expression levels were variable but relatively low, and for most infected cells, detailed morphology could not be visualized. Nevertheless, labeling quality was sufficient to indicate that infected cells were not excitatory pyramidal neurons, and nearly all had small round cell bodies typical of inhibitory cortical neurons. The rare cells that were particularly well-filled with GFP had clear bipolar morphologies similar to those observed after rabies labeling. Although some cells with glial morphology could be seen (e.g., Fig. S4A), such labeling in the immediate vicinity of the injection site was far lower than with rabies virus. Thus, insofar as could be determined by the quality of labeling using these methods, TVB–NRG1 bridge protein-mediated infection with EnvB-LV-GFP was directed predominantly to cortical inhibitory neurons, similar to those infected with EnvB-GFP-RV.
We focus the remainder of our descriptions on infection of neurons by using rabies virus in mouse cortex because this virus allowed better visualization of the detailed morphology of infected cells and more definitive identification of cortical cell types by using mouse lines that express GFP in selected cell types (see below).
NRG1-Mediated Neuronal Infection Selectively Targets Cells Expressing the NRG1 Receptor ErbB4, Requires ErbB4, and Is Mediated by the TVB–NRG1 Bridge Protein.
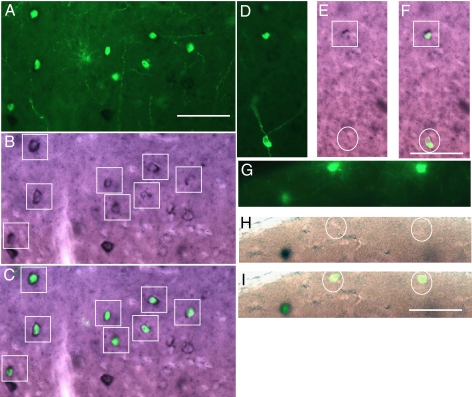
Several further lines of evidence documented in detail in the supporting material and figures indicate that: (i) the observed reporter gene expression is due to virus infection mediated specifically by the TVB–NRG1 bridge protein rather than some other nonspecific interaction; and (ii) the TVB–NRG1-mediated infection requires ErbB4 receptors (it is absent in ErbB4 knockout mice). The supporting material also details a quantitative analysis of the proportion of neurons expressing GFP after TVB–NRG1-mediated rabies virus infection, in which in situ hybridization demonstrates expression of ErbB4 mRNA. Briefly, as illustrated in Fig. 2, TVB–NRG1-mediated infection was highly selective for ErbB4-expressing neurons. Quantification of the percentage of GFP-positive cells in which ErbB4 mRNA could be detected revealed that 72% (331/459) were clearly double-labeled. Because of the stringent criteria adopted and limitations of the labeling methods used, this is a conservative estimate (SI Methods).
Fig. 2.
Double-labeling for GFP expression and ErbB4 in situ hybridization. GFP indicates cells infected with EnvB-pseudotyped virus and TVB–NRG1 bridge protein and stained with a heat-denatured anti-GFP antibody. Black reaction product indicates anti-digoxigenin staining after labeling with an ErbB4 antisense RNA probe. (A–C) Low power views indicating distribution of GFP-positive cells (A), ErbB4 in situ labeling (B), and overlay (C). Many of the GFP-positive cells are also ErbB4-positive. ErbB4-positive cells are marked by boxes (B, C, E, and F), and all of the GFP-positive cells can be seen to be double-labeled in the overlay (C). In some cases, GFP-expressing cells either did not have obvious ErbB4 label (D–F, circled cell) or were clearly not labeled (G–I, circled cells). (Scale bars: A, 50 μm and corresponds to A–C; F, 50 μm and corresponds to D–F; I, 50 μm and corresponds to G–I.)
Characterization of the Neuron Types Selectively Infected by the TVB–NRG1 Bridge Protein.
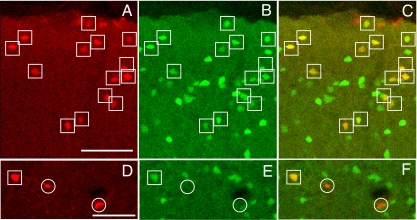
The morphological analyses described above (Fig. 1) strongly suggest that TVB–NRG1-mediated EnvB-RV infection selectively targets inhibitory neurons in the cerebral cortex. To more definitively assess what cell types were infected, EnvB-RV-expressing mCherry (EnvB-mCh-RV) was used instead of EnvB-GFP-RV. This virus facilitated identification of inhibitory neurons by using a line of GAD67 GFP knock-in mice, in which the great majority of inhibitory cortical neurons express GFP (24). After injections of TVB–NRG1 and EnvB-mCh-RV into the cortex of these mice, the distributions and morphologies of mCherry-expressing cells (Fig. 3 A and D) were indistinguishable from that described above by using the EnvB-GFP-RV (Fig. 1). As illustrated in Fig. 3, mCh-expressing cells were detected in layer 1, where all neurons are inhibitory, and cells in deeper cortical layers all had the morphologies typical of inhibitory cortical neurons. Quantitative analyses revealed that 91% (311/343 = mCh+ and GFP+ double-labeled cells/all mCh+ cells) of mCh-expressing cells were also GFP-positive. This proportion is far higher than the value of ≈15% inhibitory neurons expected by chance (5). The majority of mCh-positive/GFP-negative cells were in layer 1 and had the appearance of glial or epithelial cells, which the analyses described above suggest are infected nonselectively. It is also possible that rabies infection and/or mechanical damage might reduce GFP expression, leading to a failure of detection in some infected inhibitory neurons, or that a small population of inhibitory neurons in GAD67 GFP knock-in mice may not express detectable GFP.
Fig. 3.
Cortical neurons expressing mCherry after infection with EnvB-pseudotyped rabies virus and TVB–NRG1 bridge protein in a mouse line that expresses GFP in cortical inhibitory neurons. (A) Low power view illustrating overall location and morphologies of infected mCherry-labeled neurons. Pial surface is at the top. (B) Same section as A, illustrating GFP expressing inhibitory neurons in the GAD-67 GFP knock-in mouse line. (C) Overlay of A and B showing that there are many double-labeled (yellow/orange) cells. Double-labeled cells are marked by boxes, whereas cells expressing mCherry but not GFP are marked by circles (D–F). (D–F) Higher powered view of single- (circled cells) and double-labeled cells (boxed) in the cortical plate (outside layer 1). (Scale bars: A, 75 μm and corresponds to A–C; D, 75 μm and corresponds to D–F.)
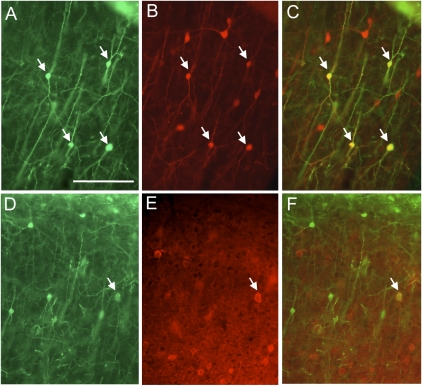
Cortical inhibitory neurons can be further divided into at least a dozen distinct types (18). In the mouse cortex, these types can be separated into three distinct nonoverlapping groups based on immunostaining for PV, SST, and VIP (25). Another very useful marker, CR, largely overlaps with VIP, but also has partial overlap with SST (26). We therefore used double-immunostaining for PV, SST, CR, or VIP and GFP to further characterize the inhibitory neurons infected by TVB–NRG1 and EnvB-GFP-RV. Because the neurons in layer 1 are typically not immunoreactive for the markers we used (27), analyses were restricted to the cortical layers deeper than layer 1. Fig. 4 illustrates double labeling for GFP and CR (Fig. 4 A–C) or PV (Fig. 4 D–F) in cortical tissue sections from animals injected with EnvB-GFP-RV and TVB–NRG1. Although 39% (68/176) of GFP-positive cells were also positive for CR, only 4% (4/112) were positive for PV, and none (0/56) were positive for SST. The percentages for PV and SST are much lower than expected if all inhibitory cell types were infected according to their overall distributions within the cortex. Expected values for PV and SST under identical staining conditions are ≈30% and 20% of all inhibitory neurons, respectively (25). Thus, TVB–NRG1 appears to preferentially infect PV-negative and SST-negative inhibitory neurons. The absence of infection of SST cells also argues that the CR-positive population is unlikely to include cells that express both CR and SST (26). Instead, the CR-positive cells are likely to include those that also express VIP; ≈35% of VIP cells coexpress CR (25).
Fig. 4.
Double-labeling for GFP expression and inhibitory cell type-specific markers. Green label indicates anti-GFP staining of cells infected with EnvB-pseudotyped virus and TVB–NRG1 bridge protein, whereas red corresponds to antibody staining against CR (A–C) or PV (D–F). GFP expression in A and D illustrates overall location and morphologies of infected, GFP-labeled neurons. Pial surface is at the top. B and E correspond to the same sections to their left and illustrate red labeling with anti-CR (B) or PV (E). C and F are overlays of the GFP and antibody-stained images. Some of the double-labeled cells are marked by arrows. Note that GFP-expressing cells colabeled for CR are common, whereas labeling with PV is rare (selected photograph highlights on such cell). (Scale bar: A, 100 μm and corresponds to all images.)
Our quantitative analyses of VIP expression also clearly revealed that many cells infected with TVB–NRG1 and EnvB-GFP-RV express VIP (Fig. S5). Overall, VIP antibody staining was detected in 23% (54/230) of GFP-expressing cells, indicating clear overlap between these populations. Nevertheless, the relatively poor quality of VIP staining relative to other antibodies (compare Fig. 4 with Fig. S5) led to uncertainty about the quantification of antibody/GFP double-staining, suggesting that the true percentage could be higher. In particular, VIP is present not only in cell bodies, but also in axon terminals, giving rise to high levels of “background” (axonal) tissue staining, against which it is sometimes difficult to discern clear cellular labeling (Fig. S5). It is also possible that VIP is not detected at the cell body in some expressing cells because it is instead localized to the axons. Finally, it is also possible that infection with rabies virus decreased VIP immunostaining, leading to false negatives. This interpretation is consistent with previous studies in which neuropeptide expression appears to be decreased after rabies infection (28).
In light of the possibility that decreased neuropeptide expression after rabies infection might have led to our failure to detect SST and PV in infected neurons, we also analyzed the neurons infected in “GIN” mice and “G42” mice, which express GFP selectively in SST-positive and PV-positive inhibitory cortical neurons, respectively (29–31). Methods were similar to those described for GAD-67 GFP knock-in mice (see above). As with the antibody staining against SST or PV, no double-labeled cells were detected in the GIN mice (0/1,047), and only 2% (6/329) were detected in the G42 mice, further supporting the interpretation that TVB–NRG1 does not readily mediate infection of SST- or PV-positive inhibitory neurons.
Discussion
Genetic tools create powerful opportunities for both the study and treatment of complex tissues, such as the cerebral cortex, which contain a mixture of cell types each with unique functional roles. The results presented here demonstrate the effectiveness of TVB–NRG1 bridge protein for selective viral transduction in a complex tissue in vivo. This first demonstration of targeted selective transduction in the brain suggests that this approach is also likely to be successful by using other bridge proteins to target different cell types. Furthermore, the results with TVB–NRG1 bridge protein in the cerebral cortex both provide tools for further studies of the roles of ErbB4 and NRG in neural circuit function and provide unique insight based on the initial observations using these tools.
Relative to the widespread use of transgenic mice for achieving cell-type specific gene expression, the strategy described here has both advantages and limitations. Advantages include the ability to target cell types in species where production of transgenic animals is not practical (such as primates) or for therapeutic purposes in humans. It is also possible that transductional targeting might allow more selective targeting than with methods that are regulated by transcriptional machinery. For example, many mRNAs are subject to alternative splicing, and gene products are subject to posttranslational modification. It might therefore be possible to design bridge proteins that target receptors that are in a particular functional state, such that they will specifically bind to the presented ligand. Such selectivity might have been conferred in our experiments by using the TVB–NRG1 bridge protein. Although infection of cortical neurons was strongly biased against cells expressing PV and SST, there is good evidence that in both the hippocampus and neocortex, these cell types express mRNA for ErbB4, and/or ErbB4 protein (14–17). The more selective infection with the TVB–NRG1 bridge protein might result from targeting of cells that present ErbB4 receptor in a particular functional state or cellular compartment (e.g., exposed at the cell surface). The flip side of this selectivity is that the cell types that are transduced might not be entirely predictable from analyses of mRNA distributions. Although the bridge protein targeting approach shows tremendous promise, relative disadvantages also include the following: variabilities in the numbers or density of cells infected due to variability in injection parameters, mechanical damage that can occur during injections, and the difficulty of vector injection as compared with animal breeding.
In addition to the specific NRG1-mediated infection of inhibitory neurons that was observed, there was also nonspecific infection near the injection sites. The predominant form of nonspecific infection involved cells that had glial morphology. Given the low pH dependence of EnvB-mediated virus entry (32), we speculate that these events are due to a low level of nonspecific uptake of the pseudotyped rabies virus, followed by trafficking to an acidic endosomal compartment where virus-cell membrane fusion occurs.
Because TVB–NRG1 should be able to selectively deliver genetic material to ErbB4 interneurons by using a wide range of enveloped viral vectors, such as lentivirus (10), this method should make the full range of burgeoning genetic technologies available for the study of these neurons (1). For example, it should be possible to identify the sources of synaptic input to these cells (21) and to selectively manipulate their activity with optical and genetic methods (1). And the ability to label these neurons with GFP, as illustrated here, will facilitate their targeting for electrophysiological studies both in brain slices and with two-photon targeted patching in vivo (33). The TVB–NRG1 bridge protein may prove to be particularly valuable in facilitating experiments in nonmouse species, including nonhuman primates. Because the organization of the primate cortex is closer to that of humans than the mouse, studies of ErbB4 interneurons in a primate model is likely to provide useful insight. Such studies in mouse and primate models could also potentially facilitate the development of therapeutic strategies targeting the NRG/ErbB system.
Methods
Preparation of EnvB-Pseudotyped Rabies Virus and Lentivirus.
Production and titer of EnvB-RV-GFP or mCherry was described except the cell line BHK-EnvBGCD was used instead of BHK-EnvAGCD, the plasmid pAB6 instead of pAB7 (12), and 293T-TVBS3 cells (34) instead of 293T-TVA800 (21). The titers of the virus used in these studies were as follows: 3 × 107 transducing units (tu)/mL for EnvB-GFP-RV (assessed by FACscan); and 4 × 107 tu/mL for EnvB-mCh-RV (assessed by manual counting). EnvB-LV-GFP was produced by using the method described (22) except the plasmid of pCI-EnvB instead of pHCMV-RabiesG.
Construction of TVB-Proline Linker and TVB Plasmids.
To construct TVB-proline linker and TVB in pCI plasmid (Promega), PCR products were generated with the primer CACTATAGGCTAGCCTCGAGATGCGCT paired with TTATTACGGTACCTTACCCCGGTCCCCCTAGGAGTT and TAATGCGGTACCTTAGCGGCCGTGAGTGGAGGAGCTG, respectively, digested with NheI and KpnI, and replaced with the gene of TVBS3-NRG1 in pCI (35).
Production of TVB–NRG1, TVB-Proline Linker, and TVB Protein.
Cells (293T) were transfected with 12 μg of pCI plasmids carrying the genes of TVB-proline linker-Herb1 (35) (referred to in here as TVB–NRG1), TVB-proline linker, or TVB using Lipofectamine (Invitrogen). The media was changed 5 h after the transfection and collected 3 d after the transfection. After a brief centrifugation, the supernatants were used as a source of the proteins.
Stereotaxic Animal Injection and Perfusion.
All procedures using live animals were approved by the Salk Institute Animal Care and Use Committee. Adult mice, ICR, GAD67-GFP knock-in, ErbB4−/−ht+ (referred to as “ErbB4 knockout” in the text), GAD67-GFP transgenic (GIN), or G42 mice described and adult Long Evans rat were anesthetized with an intramuscular injection of a mixture of ketamine and xylazine (3.6 mg/kg and 0.36 mg/kg, respectively). Approximately 9 μL of mixture of virus with the supernatant containing TVB–NRG1, TVB-proline linker, or TVB (1:10 volume, virus:supernatant for rabies and 1:1 for lentivirus) was stereotaxically injected to three adjacent locations in the frontal cortex (coordinates: 1–2 mm anterior, 2–3 mm lateral to bregma; depth, 0.7, 0.5, and 0.3 mm below the pia for mice and 2.5 mm anterior, 4 mm lateral to bregma; depth, 1.5, 1.0, and 0.5 mm below the pia) by using a glass pipette. Three days after injection of rabies virus or 2 wk after lentivirus, animals were overdosed with Nembutal (100 mg/kg IP), perfused, and brains were sectioned to a thickness of 40 μm on a freezing microtome (21).
In Situ Hybridization Combined with Immunohistochemistry Against Heat-Denatured GFP.
In situ hybridization using DIG-labeled RNA and immunostaining of free-floating sections were described in ref. 36. Briefly, free-floating brain sections were incubated with denatured antisense DIG-labeled RNA probe (Riboprobe ID:RP_050428_03_A12 in Allen Brain Atlas; http://developingmouse.brain-map.org/data/Erbb4/69672126.html) and then with sheep anti-DIG-AP-Fab (Roche) and rabbit anti-heat–denatured GFP antibodies (36) overnight at 4 °C. After wash, brain sections were incubated with donkey cy2-conjugated anti-rabbit antibody (Jackson Immunoresearch Laboratories) followed by incubation with NBT/BCIP substrate solution. Then the brain sections were mounted on slides and coverslipped with AquaMount (Lerner Laboratories).
Fluorescent Immunohistochemistry and Image Acquisition.
Brain sections were processed for immunochemistry by using the same procedures and antibodies described (25, 26). The images from in situ hybridization combined with immunohistochemistry against heat denatured GFP were acquired with an Olympus BX51 microscope and MicroFire digital camera (Optronics). Images of GFP labeling combined with immunochemistry for inhibitory neuron-specific markers were acquired on a Nikon Optiphot II microscope and MicroFire digital camera (Optronics). The remaining images were acquired with a Leica confocal microscope (Leica TCS SP2 AOBS).
Supplementary Material
Acknowledgments
We thank Keith Roby and Mauricio De La Parra for technical assistance; Dr. Takuma Mori (The Salk Institute, La Jolla, CA) for the supplying G-deleted, mCherry-expressing rabies virus used to grow EnvB-mCh-RV; Drs. Carlos Perez Garcia and Dennis O'Leary (The Salk Institute, La Jolla, CA) for supplying the ErbB4 knockout mice; Drs. Fred De Winter and Kuo-Fen Lee (The Salk Institute, La Jolla, CA) for supplying the ErbB4 in situ probe; and Dr. Takeshi Kaneko (Kyoto University, Kyoto, Japan) for supplying the antibody against heat-denatured GFP. This work was supported by National Institutes of Health Grants MH063912 and EY010742.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006233107/-/DCSupplemental.
References
- 1.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callaway EM. A molecular and genetic arsenal for systems neuroscience. Trends Neurosci. 2005;28:196–201. doi: 10.1016/j.tins.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Dittgen T, et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci USA. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han X, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavillette D, Russell SJ, Cosset FL. Retargeting gene delivery using surface-engineered retroviral vector particles. Curr Opin Biotechnol. 2001;12:461–466. doi: 10.1016/s0958-1669(00)00246-9. [DOI] [PubMed] [Google Scholar]
- 7.Schaffer DV, Koerber JT, Lim KI. Molecular engineering of viral gene delivery vehicles. Annu Rev Biomed Eng. 2008;10:169–194. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang LL, Bailey L, Baltimore D, Wang P. Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci USA. 2006;103:11479–11484. doi: 10.1073/pnas.0604993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler L, et al. Targeting lentiviral vectors to antigen-specific immunoglobulins. Hum Gene Ther. 2008;19:861–872. doi: 10.1089/hum.2007.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snitkovsky S, Young JAT. Targeting retroviral vector infection to cells that express heregulin receptors using a TVA-heregulin bridge protein. Virology. 2002;292:150–155. doi: 10.1006/viro.2001.1314. [DOI] [PubMed] [Google Scholar]
- 11.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boerger AL, Snitkovsky S, Young JAT. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 14.Neddens J, Buonanno A. Selective populations of hippocampal interneurons express ErbB4 and their number and distribution is altered in ErbB4 knockout mice. Hippocampus. 2009;20:724–744. doi: 10.1002/hipo.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vullhorst D, et al. Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci. 2009;29:12255–12264. doi: 10.1523/JNEUROSCI.2454-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazzari P, et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1382. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- 17.Yau HJ, Wang HF, Lai C, Liu FC. Neural development of the neuregulin receptor ErbB4 in the cerebral cortex and the hippocampus: Preferential expression by interneurons tangentially migrating from the ganglionic eminences. Cereb Cortex. 2003;13:252–264. doi: 10.1093/cercor/13.3.252. [DOI] [PubMed] [Google Scholar]
- 18.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 19.Woo RS, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Wen L, et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proc Natl Acad Sci USA. 2010;107:1211–1216. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Etessami R, et al. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: An in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- 24.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: Three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- 27.Uematsu M, et al. Quantitative chemical composition of cortical GABAergic neurons revealed in transgenic venus-expressing rats. Cereb Cortex. 2008;18:315–330. doi: 10.1093/cercor/bhm056. [DOI] [PubMed] [Google Scholar]
- 28.Weihe E, et al. Role of virus-induced neuropeptides in the brain in the pathogenesis of rabies. Dev Biol (Basel) 2008;131:73–81. [PubMed] [Google Scholar]
- 29.Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29:70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyaya B, et al. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 33.Margrie TW, et al. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JA. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 35.Snitkovsky S, Niederman TMJ, Mulligan RC, Young JAT. Targeting avian leukosis virus subgroup A vectors by using a TVA-VEGF bridge protein. J Virol. 2001;75:1571–1575. doi: 10.1128/JVI.75.3.1571-1575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura KC, Kameda H, Koshimizu Y, Yanagawa Y, Kaneko T. Production and histological application of affinity-purified antibodies to heat-denatured green fluorescent protein. J Histochem Cytochem. 2008;56:647–657. doi: 10.1369/jhc.2008.950915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.