Abstract
The repressor element-1 (RE1) silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) silences neuronal genes in neural stem cells (NSCs) and nonneuronal cells through its role as a dynamic modular platform for recruitment of transcriptional and epigenetic regulatory cofactors to RE1-containing promoters. In embryonic stem cells, the REST regulatory network is highly integrated with the transcriptional circuitry governing self-renewal and pluripotency, although its exact functional role is unclear. The C-terminal cofactor for REST, CoREST, also acts as a modular scaffold, but its cell type-specific roles have not been elucidated. We used chromatin immunoprecipitation-on-chip to examine CoREST and REST binding sites in NSCs and their proximate progenitor species. In NSCs, we identified a larger number of CoREST (1,820) compared with REST (322) target genes. The majority of these CoREST targets do not contain known RE1 motifs. Notably, these CoREST target genes do play important roles in pluripotency networks, in modulating NSC identity and fate decisions and in epigenetic processes previously associated with both REST and CoREST. Moreover, we found that NSC-mediated developmental transitions were associated primarily with liberation of CoREST from promoters with transcriptional repression favored in less lineage-restricted radial glia and transcriptional activation favored in more lineage-restricted neuronal-oligodendrocyte precursors. Clonal NSC REST and CoREST gene manipulation paradigms further revealed that CoREST has largely independent and previously uncharacterized roles in promoting NSC multilineage potential and modulating early neural fate decisions.
Keywords: epigenetic, pluripotency
The repressor element-1 (RE1) silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) is a modular transcriptional regulator that orchestrates developmental and homeostatic gene expression programs by recruiting corepressor for element-1–silencing transcription factor (CoREST) and other regulatory cofactors to target gene loci where transcription can be activated or repressed through dynamic epigenetic mechanisms (1–3). REST was initially thought to prevent the expression of neuronal genes in neural stem cells (NSCs) and nonneuronal cells (4–7). More recently, various studies have identified genes that are targets of REST in embryonic stem cells (ESCs) (8) and demonstrated that REST is regulated directly by the core pluripotency factors, Oct4/Sox2/Nanog (9). Moreover, the REST transcriptional regulatory network in ESCs, which includes microRNAs (miRNAs), is highly integrated with these pluripotency factors and their downstream target genes (10, 11). Although these observations suggest that REST is part of the complex molecular circuitry governing pluripotency, REST is not required for pluripotency, and the significance of these findings remains unclear (10, 12). Intriguingly, emerging evidence suggests that these pluripotency factors continue to play instructive roles in NSC maintenance and maturational processes (13–15), although their functional interactions with REST and CoREST in determining early neural cell identity and function have not been explored.
Although CoREST is the C-terminal cofactor for REST, it is likely that these factors have distinct roles in modulating gene expression (16). CoREST is more evolutionarily conserved than REST and thought to subserve more elementary functions in the modulation of neuronal genes (17). Like REST, CoREST is also highly developmentally regulated with distinct temporal and spatial expression patterns. During embryogenesis, REST is expressed in both neural and nonneural tissues, whereas CoREST is preferentially expressed within the nervous system (18, 19). CoREST also acts as a scaffold for recruitment of epigenetic factors (17, 20) and modulates gene expression by binding to REST or independently to promoter regions (1, 16). CoREST and REST therefore may play complementary roles in promoting NSC identity and function. In fact, recent studies by our group suggest that REST and CoREST display both overlapping and unique context-specific roles in mediating neural cell fate decisions, including neuronal and glial subtype specification and oligodendrocyte (OL) lineage maturation (11, 21, 22).
In the present study, we analyzed the functional roles played by CoREST and REST in NSCs and in their proximate neural progenitor species. Our data reveal that, in comparison with REST, CoREST targets a much larger number of genes, including those with seminal roles in promoting NSC maintenance and lineage restriction as well as mediating the functions of chromatin-modifying enzymes that represent core components of the REST-CoREST regulon. Moreover, we observed that CoREST preferentially targets components of pluripotency networks, suggesting previously unrecognized roles for both pluripotency networks and for CoREST in NSC maintenance and maturational functions.
Results
Profiles of CoREST and REST NSC Expression.
We defined CoREST and REST expression and subcellular localization in NSCs by performing immunofluorescence microscopy (Fig. S1A) and Western blot analysis (Fig. S1B). We observed that, along with REST, CoREST is ubiquitously expressed within the nucleus and the cytoplasm of NSCs and other neural cell types (21, 22).
Identification of CoREST and REST Target Genes.
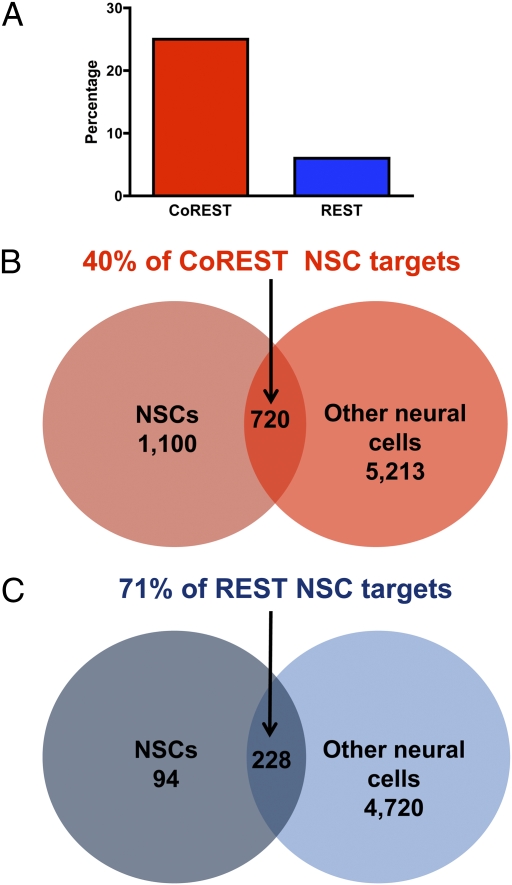
To characterize the functional roles of CoREST and REST, we performed a series of chromatin immunoprecipitation-on-chip (ChIP-chip) experiments in NSCs and other neural cell types (21, 22). We found a total of 7,033 CoREST target genes across all cell types examined, including 1,820 (25%) in NSCs, the most for any cell type (Fig. 1A and Dataset S1). In contrast, for REST, we found a total of 5,042 target genes across all cell types, with only 322 (6%) in NSCs, a statistically significant difference (P < 0.001).
Fig. 1.
CoREST and REST target genes in NSCs and other neural cell types. We identified 7,033 total CoREST targets and 5,042 total REST targets in neural cell species through ChIP-chip experiments. We compared profiles of CoREST and REST target genes in NSCs with those at specific developmental stages and in individual neural cell types, including neuronal subtypes (cholinergic, medium spiny, GABAergic, and glutamatergic neurons), oligodendrocyte (OL) lineage species, neural progenitors/precursors (neuronal-oligodendrocyte progenitors, radial glia, and OL precursors), and astrocytes (21, 22). (A) Percentages of the total number of CoREST or REST target genes present in all neural species that are targets within NSCs. (B and C) Comparative profiles of CoREST (B) and REST (C) target genes in NSCs and other neural cell types.
To examine the potential roles of CoREST and REST in modulating NSC maintenance and maturational functions, we compared profiles of target genes found in NSCs with those present at distinct developmental stages and in specific neural cell types, including neural precursor/progenitor cells, mature neuronal subtypes, glial cell subtypes, and OL developmental species (Fig. 1 B and C) (21, 22). We found that, of CoREST target genes in NSCs, CoREST also binds to 40% of these genes in other neural cell types. In contrast, we found that, of REST target genes in NSCs, REST also binds to 71% of these genes in other neural cell types. Thus, in comparison with the percentage of REST target genes, a higher and statistically significant (P < 0.001) percentage of CoREST target genes are unique targets within NSCs, suggesting a potentially distinct and broader role for CoREST in modulating NSC gene expression programs.
Whereas CoREST functions, in part, by binding to REST at RE1 sites, it can also independently bind to DNA and regulate transcription (1, 16). We therefore compared profiles of CoREST and REST target genes in NSCs with each other and with a previously characterized set of RE1 motif-containing genes (Fig. S2) (4, 6). Among genes targeted by REST, 72% contain known RE1s, suggesting that REST modulates gene expression primarily through these RE1 sites. In contrast, among genes targeted by CoREST, only 41% contain known RE1s, a statistically significant difference (P < 0.001).
Characterization of Diverse Functional Roles for CoREST and REST NSC Target Genes.
To further examine the potentially distinct and overlapping roles for CoREST and REST in NSCs, we analyzed target genes using Ingenuity pathways analysis and found that CoREST targets are enriched in a diverse array of pathways whereas REST targets are enriched for a much more delimited set of biological functions (Table S1). These pathways are consistent with previously characterized functions for RE1-containing genes (4, 6). We did not identify any pathways that were significantly enriched among the target genes bound by both CoREST and REST, suggesting distinct roles for these factors in NSCs.
To further delineate the roles of CoREST and REST in NSCs, we compared their target genes to sets of genes involved in stem cell maintenance, epigenetic regulatory networks, and disease states (Table 1). For genes involved in pluripotency, including PluriNet (23) and individual and composite Oct4/Sox2/Nanog networks (24), we found that a greater and statistically significantly percentage are CoREST as compared with REST target genes. These results suggest that CoREST is preferentially involved in mediating novel functions for pluripotency networks in NSCs. We also examined genes associated with conditions in which aberrant REST and/or stem cell functions are implicated in disease pathogenesis, including glioblastoma multiforme (GBM), a brain tumor composed of morphologically diverse cells including cancer stem cells (25, 26), and Huntington's disease (HD), a neurodegenerative disorder caused by a mutation in the huntingtin gene that results in aberrant nuclear sequestration of REST and transcriptional dysregulation of RE1-containing genes (27). For genes associated with GBM, our results suggest that CoREST is preferentially involved in mediating aspects of the cancer stem cell state. In contrast, for HD-related genes, we found similar percentages are CoREST and REST targets, including three genes that are targets of both (i.e., Fgf12, Syt4, and Trpc7), suggesting linked roles for CoREST and REST in HD pathogenesis. We also examined genes targeted by brain-related miRNAs, including miR-124a, miR-9, and miR-132, that directly modulate both CoREST and REST within a double-negative feedback loop (28). Among these genes, we found that 19% are CoREST targets and none are REST target genes in NSCs, suggesting that CoREST NSC target genes may be associated with this RNA regulatory circuitry.
Table 1.
Genes involved in stem cell maintenance, epigenetic regulatory networks, and disease states are CoREST and REST targets in NSCs
| Plurinet, % | Oct4/Sox2/Nanog, % | Nanog, % | Oct4, % | Sox2, % | Glioblastoma, % | Huntington's disease, % | REST-miRNA network, % | |
| (225) | (408) | (4,085) | (3,309) | (1,113) | (192) | (37) | (16) | |
| CoREST (1,820) | 22 (49)** | 19 (79)* | 16 (671)* | 17 (574)* | 19 (208)* | 15 (29)* | 19 (7) | 19 (3) |
| REST (322) | 2 (4) | 2 (8) | 2 (88) | 3 (86)* | 2 (25) | 2 (3) | 22 (8)* | 0 (0) |
Shown are genes involved in pluripotency, including “Plurinet” (23) and composite and individual Oct4/Sox2/Nanog networks (24), neurological diseases (glioblastoma, ref. 25; and Huntington's, ref. 27), and REST-miRNA networks (28). Percentage refers to the proportion of genes within a particular network that are CoREST or REST NSC targets, and the numbers in parentheses represent the actual number of genes.
*P < 0.001.
Association of Promoter Occupancy with Profiles of Developmental Gene Expression.
To assess the potential roles played by CoREST and REST in the modulation of neural developmental gene expression programs, we correlated changing profiles for CoREST and REST promoter occupancy with differential gene expression patterns found during distinct stages of NSC lineage restriction. Specifically, we evaluated the transitions from NSCs into less lineage-restricted radial glia (RG) species and into more lineage-restricted bipotent neuronal-OL progenitors (N/OPs) (Table S2). In the transition from NSCs into RG, we found a total of 274 up-regulated and 903 down-regulated genes that have CoREST bound in one of the two cell types. In contrast, we identified a total of 100 up-regulated and 208 down-regulated genes that have REST bound in one of the two cell types. In the transition from NSCs into N/OPs, we found a total of 550 up-regulated and 350 down-regulated genes that have CoREST bound in one of the two cell types. In contrast, we identified a total of 74 up-regulated and 88 down-regulated genes that have REST bound in one of the two cell types (Table S2).
The largest subgroups of genes are those in which CoREST is initially bound in NSCs and then liberated during lineage restriction. In the transitions from NSC to RG and from NSC to N/OP, the liberation of CoREST from gene promoters is associated primarily with context-specific gene repression and activation, respectively. Intriguingly, we observed that these specific biases toward gene activation or repression are recapitulated in the subgroups of genes containing RE1 motifs and those associated with pluripotency networks (Table S2). These CoREST target genes display opposite profiles of differential expression in the transitions from NSCs to RG and from NSCs to N/OPs, respectively, and may serve as important nodal points in NSC-mediated neural fate restriction and lineage specification.
Ablation of REST or CoREST Alters NSC Clonal Properties.
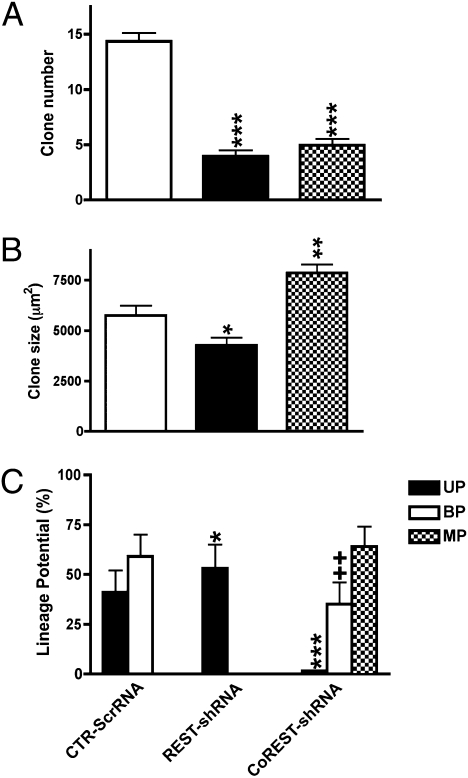
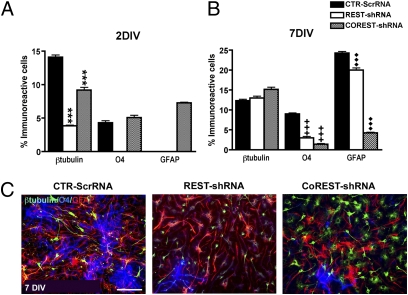
To further define the potential functional roles of REST and CoREST in NSC self-renewal, proliferation, neural lineage restriction, and neuronal and glial lineage specification and maturation, we selectively ablated each gene in proliferating NSCs with lentiviral constructs containing gene-specific shRNAs (Materials and Methods and SI Materials and Methods). Ablation of REST decreased NSC self-renewal as assessed by examination of the generation of secondary clones (Fig. 2A). Compared with controls, these clones contained a smaller complement of β-tubulin+ neuroblasts (Fig. 3A) whose early maturation was delayed (doublecortin+, Fig. S3B), suggesting that REST is required for neurogenesis. Further, contrary to control clones that contain only a small percentage of GLAST+ cells, 100% of REST-depleted clones continued to maintain a significant percentage of intermediate progenitor species colabeled with nestin, RC2, and GLAST (Fig. S4). Moreover, REST ablation resulted in delayed and reduced elaboration of O4+ OL species (Fig. 3 A and B) as well as impaired morphologic maturation (Fig. 3C). Moreover, REST depletion also significantly reduced the elaboration of GFAP+ astrocytes (ASs) (Fig. 3A). These findings suggest that REST has preferential roles in neurogenesis compared with gliogenesis.
Fig. 2.
Effects of selective REST and CoREST depletion on NSC self-renewal, proliferation, and neural fate decisions. (A) REST- and CoREST-depleted secondary clones displayed reduced self-renewal as compared with control clones. (B) Selective ablation of REST resulted in the formation of smaller secondary clones, whereas selective ablation of CoREST resulted in the formation of larger secondary clones compared with control clones. (C) Clonal lineage analysis revealed that in the control condition both neuronal-restricted and bipotent neuronal-oligodendrocyte clones were generated. By contrast, REST ablation resulted in the generation of solely neuronal-restricted clones, whereas CoREST ablation resulted in the generation of a large proportion of clones with multilineage potential, a smaller proportion of bipotent neuronal-astroglial clones and a minimal complement of neuronal-restricted clones. Bars in A–C represent the mean ± SEM of three independent biological replicates. *P < 0.05; ** and ++P < 0.01; and ***P < 0.0001.
Fig. 3.
Effects of selective REST and CoREST depletion on NSC-mediated neurogenesis and gliogenesis. (A) The percentage of β-tubulin+ clones at 2 and 7 d in vitro (DIV) reveals that REST and CoREST are differentially required for neurogenesis. Analysis of O4+ clones reveals that REST and CoREST differentially regulate OL lineage elaboration and maintenance. Analysis of GFAP-immunoreactive clones reveals that REST and CoREST also have selective effects on the generation and maintenance of ASs. Bars in A and B represent the mean ± SEM of three independent biological replicates. ***, +++, and ♦♦♦P < 0.0001. (C) Immunofluorescence microscopic analysis demonstrates the lineage composition of secondary clones derived from NSCs following selective ablation of REST or CoREST compared with the control condition. Neural lineage markers were used to identify neurons (β-tubulin, FITC), oligodendrocytes (O4, DAPI), and astrocytes (GFAP, TRITC). (Scale bar, 200 μm).
Ablation of CoREST also decreased NSC self-renewal (Fig. 2A). However, compared with the normal profile of bipotent neuronal-oligodendrocyte lineage restriction, depletion of CoREST resulted in the generation of clones that preferentially exhibit multilineage potential (Fig. 2C) and bipotent neuronal-astrocyte lineage restriction (Fig. S5). In contrast, depletion of REST resulted in the generation of clones that solely exhibit neuronal restricted lineage potential (Fig. 2C). Clones depleted of CoREST were larger than control clones, whereas REST-depleted clones were significantly smaller than control clones (Fig. 2B), suggesting differences in NSC maintenance. In concert with these observations, further analysis of the lineage potential of CoREST-depleted NSC clones revealed significant increases in the maintenance of distinct intermediate progenitor species exhibiting differential profiles of nestin, RC2, and GLAST immunoreactivity (Fig. S4). These clones also exhibited the precocious elaboration of neuronal species (Fig. S3), impaired maintenance of OL species (Fig. 3B), and precocious elaboration but reduced maintenance of AS species (Fig. 3 A and B). These cumulative findings suggest multiple impairments in fate restriction in CoREST-depleted clones.
These cumulative observations suggest the preferential roles of CoREST in promoting NSC multilineage potential and fate restriction. By contrast, REST collaborates with CoREST in NSC self-renewal but exhibits preferential roles in promoting neurogenesis as previously described (2, 3, 19, 22).
Discussion
We uncovered a broader array of CoREST NSC target genes, in contrast to those of REST, using a ChIP-chip approach and neural developmental paradigms composed of primary neural cell types rather than cell lines. Herein, we consider the potential functions of CoREST revealed through these studies by discussing selected subsets of its NSC transcriptional regulatory network, including genes that play specific roles in modulating ESC and NSC identity, self-renewal, and lineage potential; neural lineage specification; and REST-associated epigenetic regulatory processes.
We identified a number of CoREST NSC target genes with roles in ESC self-renewal and maintenance (e.g., Lin28, Klf5, Tcf3, and c-Myc). Unlike CoREST, we found that REST targets only one gene (i.e., Lin28). These observations suggest that complex crosstalk occurs between CoREST, REST, and pluripotency networks in NSCs, although the significance of these findings is unclear. One possibility is that, because Lin28 is a key mediator of the pluripotent state (29), both CoREST and REST regulate the expression of this factor to maintain NSC identity. Moreover, Klf5, Tcf3, and c-Myc are targets of CoREST in NSCs, but not of REST, suggesting a preferential role for CoREST in maintaining NSC identity via these pluripotency genes. Indeed, Klf5 promotes ESC self-renewal and maintenance of the undifferentiated state through effects on Oct4 and Nanog expression (30). Similarly, Tcf3 limits ESC self-renewal by down-regulating expression of Nanog (31). c-Myc has diverse roles in ESCs and NSCs. It can reprogram neural progenitor cells into pluripotent stem cells in concert with Oct4 alone (32) or in combination with Sox2 and Klf4 (33). In contrast, c-Myc is expressed at higher levels in NSCs than in ESCs (32) and also regulates neural progenitor proliferation (34). We also identified additional CoREST target genes with roles in maintaining NSC multilineage potential (e.g., Tlx, Fgfr2, and Zic1) (35–37). Tlx is a nuclear receptor that mediates NSC proliferation and self-renewal through the recruitment of histone deacetylases (HDACs) to the promoters of the Tlx target genes, p21 and Pten (35). Fgfr2 and Zic1 promote NSC self-renewal and maintenance of the undifferentiated state by inhibiting cell cycle exit (36, 37). These findings further suggest that CoREST may maintain NSC identity, self-renewal, and multilineage potential.
Furthermore, we identified CoREST NSC target genes encoding different classes of neural-specific transcription factors that directly mediate neural lineage commitment and indirectly control lineage commitment by regulating the timing of the expression of other factors. For example, Arx is a homeodomain-containing transcription factor that contributes to the development of GABAergic neurons (38), and Zfp536 is a zinc finger protein that is involved in the intracellular timer mediating OL differentiation (39). In addition, we identified target genes encoding helix–loop–helix (HLH) transcription factors with diverse roles in neural development (e.g., Ngn2, NeuroD, Hes1/3/6, Hey2, and Id2/4). Id proteins promote the maintenance and self-renewal of neural stem and progenitor cells and also control the precise timing of neurogenesis (Id1/2) and oligodendrogliogenesis (Id2/4) by regulating proneural bHLH and other neural differentiation factors (40). For example, Id2 promotes NSC maintenance by sustaining Hes1 expression (41), which actively prevents the differentiation of neuronal and glial lineage species (42, 43). Whereas both Hes1 and Hes3 are essential for the maintenance and proliferation of NSCs (44), Hes6 promotes neuronal differentiation by antagonizing Hes1 activity and inducing the expression of the proneural bHLH transcription factor, Mash1 (45). Further, the Hes-related factor, Hey2, is also critical for NSC maintenance because it negatively regulates neuronal bHLH genes (46). These observations suggest that CoREST mediates the complex interplay between NSC self-renewal and neural differentiation by modulating diverse brain-specific developmental transcription factors.
These neural transcription factors are regulated by signaling pathways that modulate NSC identity, self-renewal, and fate specification (43), and we also found that CoREST NSC target genes encode components of the SHH (e.g., Gas1, Ptch2, and Klf5/9), WNT (e.g., Wnt11 and Fzd 1/3/7), and Notch (e.g., Hes1/3/6, Hey, and Dll1) signaling pathways. SHH signaling plays diverse roles in neural development, including regional patterning of the neuraxis, proliferation and maintenance of various neural stem and progenitors populations, and specification of various classes of neuronal and oligodendroglial cell types (47). WNT signaling also has complex context-specific developmental functions that include directly promoting NSC proliferation and self-renewal through effects on downstream targets including cyclin D1 (43) as well as inducing cell cycle arrest and neuronal lineage commitment and differentiation (48). Notch signaling maintains NSC and neural progenitor pools and prevents precocious neuronal differentiation through downstream effectors, including Hes1 and Hes5 (44). These findings suggest that CoREST may regulate gradient morphogen and cell–cell signaling pathways critical for 3D patterning of the neuraxis and the elaboration of regional NSC species as well as neuronal and glial subtypes.
The potential neural developmental functions for CoREST, suggested by its array of target genes in NSCs, are also supported by complementary evidence derived from our functional clonal analysis paradigms. These observations demonstrated that CoREST modulates NSC self-renewal cooperatively with REST but plays preferential roles in mediating NSC multilineage potential and neural lineage restriction. These are unique functional observations that have begun to identify a role for CoREST, distinct from REST, in mediating NSC cell fate decisions.
Furthermore, our ChIP-chip studies provide additional insights into the distinct but overlapping relationship between CoREST and REST in epigenetic regulation of these NSC fate decisions. We found CoREST target genes that encode epigenetic factors important for establishing cell identity, including histone-modifying and chromatin-remodeling enzymes (e.g., Hdac1, Hdac2, Hdac9, Jmjd1a, Jmjd4, and Brm) (49, 50). Interestingly, Jmjd1a positively regulates pluripotency-associated genes (e.g., Tcl1, Tcfc2l1, and Zfp57) (51) and is a direct target of Oct4 (52), further suggesting crosstalk between CoREST and pluripotency gene programs. We also identified CoREST target genes (e.g., Tcfe2a, E47, Pou3f2, and Creb) encoding components of miRNA circuitry that regulates the expression of factors within the REST macromolecular complex (28). Interestingly, the cAMP response element binding protein (CREB) and several additional regulators control the expression of brain-related miRNAs that form double-negative feedback loops with REST/CoREST/MeCP2 (28, 53, 54).
We further identified CoREST NSC target genes (e.g., Sin3a and Hdac1/2) whose protein products regulate the expression of REST and target genes (e.g., Sin3a, Hdac1/2, Rest, Braf35, and Scp1) encoding factors that serve as epigenetic effectors and components of the modular REST complex, thereby suggesting that CoREST regulates deployment of REST and essential cofactors recruited to the REST macromolecular complex (Fig. S6). Sin3a and HDAC1/2 are components of a distinct regulatory complex that includes the unliganded retinoic acid receptor (RAR) repressor, its corepressor, N-CoR, Sin3a, HDAC1, and CoREST (55). In neural progenitor species, this transcriptional complex represses REST expression and promotes neuronal terminal differentiation (55). We also found that CoREST targets Senp1, a gene encoding a small ubiquitin-like modifier (SUMO)-specific protease. This protease negatively modulates CoREST activity (56), further suggesting that CoREST regulates its own expression and neural developmental functions. Intriguingly, our cumulative observations strongly suggest that in NSCs, CoREST modulates its own as well as REST expression both directly, by binding to the REST promoter, and indirectly, by modulating Sin3a and HDAC1/2 expression as well as other regulatory factors.
We also observed that CoREST NSC target genes encode other important REST cofactors (e.g., Hp1 and Scp1). Hp1 is a nonhistone protein that localizes to heterochromatin and promotes gene silencing and long-range chromatin interactions (57, 58). Hp1 also mediates RE1 gene silencing by functioning as an adaptor between di- and trimethylated histone 3 on lysine 9 and DNA methylation-dependent states and appears to lock genes into a “repressed” state, suggesting that CoREST regulation of Hp1 in NSCs may be important for preventing ectopic expression of REST-regulated genes (59). In addition, Scp1 is an evolutionarily conserved transcriptional regulator that negatively regulates RNA polymerase II (PolII) activity (60). Interestingly, Scp1 may play a role in the prevention of PolII-mediated transcription despite the presence of the preinitiation complex characteristic of the stem cell state (60). In fact, Scp1 cooperates with REST to suppress neuronal differentiation during development, and down-regulation of Scp1 by miR-124 is critical for inducing neurogenesis (60). Our findings suggest that CoREST regulation of Scp1 in NSCs also provides an important additional layer of contextual control for orchestrating neurogenesis. This conclusion is supported by previous studies demonstrating that CoREST, REST, and Scp1 represent nodes within highly integrated transcriptional and posttranscriptional regulatory networks required for mediating neuronal gene expression, including several brain-related miRNA regulatory feedback loops (28, 61).
Future studies using CoREST and REST coimmunoprecipitation along with increasingly high-resolution whole-genome approaches will further expand our understanding of the complex and interrelated roles of CoREST and REST. These strategies are particularly relevant because recent studies suggest that RE1 motifs are widely distributed across the genome well beyond traditional promoter sequences (6) and REST regulatory effects are highly complex, in part, as a result of hierarchical binding affinities (62). In addition, more detailed functional manipulations will help elucidate their distinct and overlapping roles in coordinating NSC identity and function.
Nonetheless, our results support a model in which CoREST preferentially mediates gene networks underlying neural stem cell multilineage potential and fate restriction. Our experimental data imply that CoREST may coordinate these complex NSC functions by targeting critical neural developmental factors and seminal signaling pathways involved in NSC functions distinctly from REST and also by modulating components of the REST macromolecular complex itself. Moreover, CoREST may play previously unanticipated roles in the pathogenesis of disease processes, such as cancer and neurodegeneration. Finally, REST and CoREST are also highly integrated with pluripotency gene networks in ESCs and NSCs, respectively, raising intriguing questions about the potential biological functions of these networks beyond ESCs, the regulatory topology of these networks, and the implications of these relationships for lineage diversity and plasticity in the nervous system.
Materials and Methods
Cell Cultures and Growth Factor Preparations.
Cell culture paradigms were used as previously described with certain modifications (21, 22, 63–68) (SI Materials and Methods).
Specific Antibody Preparations.
All antibodies exhibited selective immunoreactivity for mouse cellular preparations, and each antibody exhibited a complete absence of cross-reactivity with other antigens and epitopes (21, 22) (SI Materials and Methods).
Western Blot Analysis.
Cell cultures were processed for Western blot analysis as previously described (21, 22, 64) (SI Materials and Methods).
Statistical Analysis.
Comparison of proportions was calculated using the χ2 test and mean values were calculated with Student's t test. Statistical differences between the CTR-ScrRNA, REST-shRNA, and CoREST-shRNA conditions were defined using probability values (*, +, and ♦P < 0.05; **, ++, and ♦♦P < 0.01; and ***, +++, and ♦♦♦P < 0.0001).
Chromatin Immunoprecipitation-on-Chip (ChIP-Chip) Assays and Data Analysis.
ChIP-chip assays and data analysis were performed as previously described (21, 22, 69–71) (SI Materials and Methods).
Functional Classification.
Target genes were functionally annotated using Ingenuity pathways analysis (Ingenuity Systems).
Gene Expression Analysis.
Gene expression analysis was performed as previously described (21, 22) (SI Materials and Methods).
Supplementary Material
Acknowledgments
A.B. is supported by the National Institutes of Health (Grants AG028872 and AG027734). M.F.M. is supported by the National Institutes of Health (Grants NS38902 and MH66290) and by the F. M. Kirby, the Alpern Family, the Mildred and Bernard H. Kayden, and the Roslyn and Leslie Goldstein Foundations.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0906917107/-/DCSupplemental.
References
- 1.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 3.Chong JA, et al. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 4.Bruce AW, et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto SJ, et al. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 10.Buckley NJ, Johnson R, Sun YM, Stanton LW. Is REST a regulator of pluripotency? Nature. 2009;457(7233):E5–E6. doi: 10.1038/nature07784. [DOI] [PubMed] [Google Scholar]
- 11.Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system–What's the REST of the story? Neurosci Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimozaki K, Nakashima K, Niwa H, Taga T. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- 15.Molero AE, et al. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc Natl Acad Sci USA. 2009;106:21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenway DJ, Street M, Jeffries A, Buckley NJ. RE1 silencing transcription factor maintains a repressive chromatin environment in embryonic hippocampal neural stem cells. Stem Cells. 2007;25:354–363. doi: 10.1634/stemcells.2006-0207. [DOI] [PubMed] [Google Scholar]
- 17.Lakowski B, Roelens I, Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J Mol Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- 18.Grimes JA, et al. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 19.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 20.Andres ME, et al. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrajano JJ, et al. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE. 2009;4:e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrajano JJ, et al. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS ONE. 2009;4:e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller FJ, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan X, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 27.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 30.Parisi S, et al. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 31.Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Stefano B, Prigione A, Broccoli V. Efficient genetic reprogramming of unmodified somatic neural progenitors uncovers the essential requirement of Oct4 and Klf4. Stem Cells Dev. 2008;18:707–716. doi: 10.1089/scd.2008.0180. [DOI] [PubMed] [Google Scholar]
- 33.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 34.Su X, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol. 2008;28:3995–4003. doi: 10.1128/MCB.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue T, Ota M, Ogawa M, Mikoshiba K, Aruga J. Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors. J Neurosci. 2007;27:5461–5473. doi: 10.1523/JNEUROSCI.4046-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng W, Nowakowski RS, Vaccarino FM. Fibroblast growth factor 2 is required for maintaining the neural stem cell pool in the mouse brain subventricular zone. Dev Neurosci. 2004;26:181–196. doi: 10.1159/000082136. [DOI] [PubMed] [Google Scholar]
- 38.Colasante G, et al. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci. 2008;28:10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 41.Bai G, et al. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Ishibashi M, et al. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 45.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278:44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- 47.Hu BY, Du ZW, Li XJ, Ayala M, Zhang SC. Human oligodendrocytes from embryonic stem cells: Conserved SHH signaling networks and divergent FGF effects. Development. 2009;136:1443–1452. doi: 10.1242/dev.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kan L, et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Lee SK. Crucial roles of histone-modifying enzymes in mediating neural cell-type specification. Curr Opin Neurobiol. 2010;20:29–36. doi: 10.1016/j.conb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Daley GQ. Molecular basis of pluripotency. Hum Mol Genet. 2008;17(R1):R23–R27. doi: 10.1093/hmg/ddn050. [DOI] [PubMed] [Google Scholar]
- 52.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ooi L, Wood IC. Regulation of gene expression in the nervous system. Biochem J. 2008;414:327–341. doi: 10.1042/BJ20080963. [DOI] [PubMed] [Google Scholar]
- 55.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Muraoka A, et al. Sumoylation of CoREST modulates its function as a transcriptional repressor. Biochem Biophys Res Commun. 2008;377:1031–1035. doi: 10.1016/j.bbrc.2008.09.149. [DOI] [PubMed] [Google Scholar]
- 57.Smith E, Shilatifard A. The A, B, Gs of silencing. Genes Dev. 2007;21:1141–1144. doi: 10.1101/gad.1559407. [DOI] [PubMed] [Google Scholar]
- 58.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 59.Brenner C, Fuks F. A methylation rendezvous: Reader meets writers. Dev Cell. 2007;12:843–844. doi: 10.1016/j.devcel.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Yeo M, et al. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 61.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruce AW, et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gokhan S, et al. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marmur R, Kessler JA, Zhu G, Gokhan S, Mehler MF. Differentiation of oligodendroglial progenitors derived from cortical multipotent cells requires extrinsic signals including activation of gp130/LIFbeta receptors. J Neurosci. 1998;18:9800–9811. doi: 10.1523/JNEUROSCI.18-23-09800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marmur R, et al. Isolation and developmental characterization of cerebral cortical multipotent progenitors. Dev Biol. 1998;204:577–591. doi: 10.1006/dbio.1998.9099. [DOI] [PubMed] [Google Scholar]
- 66.Zhu G, Mehler MF, Mabie PC, Kessler JA. Developmental changes in neural progenitor cell lineage commitment do not depend on epidermal growth factor receptor signaling. J Neurosci Res. 2000;59:312–320. [PubMed] [Google Scholar]
- 67.Yung SY, et al. Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species. Proc Natl Acad Sci USA. 2002;99:16273–16278. doi: 10.1073/pnas.232586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu G, Mehler MF, Mabie PC, Kessler JA. Developmental changes in progenitor cell responsiveness to cytokines. J Neurosci Res. 1999;56:131–145. doi: 10.1002/(sici)1097-4547(19990415)56:2<131::aid-jnr3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 69.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 70.Sandoval J, et al. RNAPol-ChIP: A novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oberley MJ, Tsao J, Yau P, Farnham PJ. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.