Abstract
No methods proposed thus far have the capability to measure overall molecular flow in the nucleus of living cells. Here, we apply the pair correlation function analysis (pCF) to measure molecular anisotropic diffusion in the interphase nucleus of live cells. In the pCF method, we cross-correlate fluctuations at several distances and locations within the nucleus, enabling us to define migration paths and barriers to diffusion. We use monomeric EGFP as a prototypical inert molecule and measure flow in and between different nuclear environments. Our results suggest that there are two disconnect molecular flows throughout the nucleus associated with high and low DNA density regions. We show that different density regions of DNA form a networked channel that allows EGFP to diffuse freely throughout, however with restricted ability to traverse the channel. We also observe rare and sudden bursts of molecules traveling across DNA density regions with characteristic time of ≈300 ms, suggesting intrinsic localized change in chromatin structure. This is a unique in vivo demonstration of the intricate chromatin network showing channel directed diffusion of an inert molecule with high spatial and temporal resolution.
Keywords: chromatin organization, fluctuation spectroscopy, nucleus structure
Nuclear architecture is fundamental to the manner in which molecules traverse the nucleus (1). The cell nucleus is a functionally and spatially structured organelle (2) in which diffusion is the mode of motion for inert molecules (3–5). The diffusion of molecules within the nucleus is obstructed by the steric constraints imposed by structural components, such as chromatin (6, 7). Proposed models for 3D arrangement of chromatin vary from defined regions of chromatin compartmentalisation (8) to intermingled chromatin fibers and loops (9). The requirement for biologically significant molecules to reach different destinations within the cell nucleus raises the question, how is the diffusive route directed? Given that diffusion cannot be regulated because it is essentially a default mechanism of motion (10), it has been postulated that structural features of the nucleus must impart retention at particular sites and control flux of movement between compartments (1, 11).
Insights into intranuclear trafficking are predominantly derived from measurement of the accessibility of the nuclear landscape and the effect it has on diffusion of biologically active and inert molecules. Current approaches commonly used for such investigations are fluorescence recovery after photo bleaching (FRAP) (12, 13), single particle tracking (SPT) (14), and fluorescence correlation spectroscopy (FCS) (15, 16). FRAP, however, is invasive because it requires high illumination power on the sample to induce photobleaching, and SPT requires the observation of isolated particles for a long time, which yields poor statistics. FCS, in contrast, provides information at the single molecule level with good statistics by averaging the behavior of many molecules (10), and this technique has emerged as the preferred method for studying the motions of intracellular molecules.
Recently, Dross et al. (17) investigated the impact chromatin compaction has on the diffusion of EGFP oligomers by using single-point FCS. A “diffusion map” based on several localized FCS measurements of EGFP diffusion inside the cell nucleus was compiled, using H2B-mRFP labeling as a reference of chromatin density and position. The chromatin network was found to neither exclude nor impede diffusion of EGFP, resulting in a homogenous distribution of the protein throughout the nucleus (with the exception of the nucleolus). It was concluded that either: (i) nuclear structures different from the chromatin network must affect the mobility of the EGFP, given the lack of correlation between diffusion behavior and chromatin density or (ii) there is a dependence on chromatin structure, but it manifests itself on a length scale smaller than the resolution of the FCS technique used (≈0.3 μm).
The limitation of single-point FCS is that it suffers in terms of the spatial environment tested: Only the local diffusion coefficient of molecules within the point spread function (PSF) is detected. The spatial environment around the PSF and, thus, the route the molecules take before crossing the observation volume is not directly observed in the FCS experiment. Recently, a novel approach to spatiotemporal correlation, based on pair correlation functions (pCF) was developed by Digman and Gratton (18) and applied to the study of the nucleocytoplasmic shuttling of inert and biologically active molecules (19).
The pCF method builds on previous approaches to spatiotemporal correlation such as two foci FCS (20). However, pCF analysis is more comprehensive in that only one laser beam is scanned rapidly across different locations in a repeated pattern (line or a circle) resulting in the measurement of correlation between every possible pair of points in the pattern. By detecting the same molecule at two different locations, we measure the average time a molecule takes to move between these two locations (18). If there is a delay from the expected average time to diffuse the distance between the two points, we can make inferences about the existence of barriers to diffusion between those two points. In this instance, the barrier encountered is impenetrable and absence of correlation between positions on either side is observed.
Here, we apply the pCF analysis to the study of intranuclear diffusion of inert molecules and the possible spatial dependence on chromatin environment. We used CHO-K1 cells stably expressing monomeric EGFP, a well characterized protein known to show minimal interaction with its surroundings and, thus, diffuse freely (21). The nuclei of cells were stained with Hoechst 33342 as a means of having a reference for the local DNA density and position. From pCF, analysis of EGFP diffusion within the nucleus DNA-dependent migration paths and barriers to diffusion were definitively established. Based on our results, we propose a model for the molecular flow of inert molecules through the chromatin network.
Results
ACF Carpet Analysis: Measurement of Local Concentration and Diffusion of EGFP Within the Nucleus.
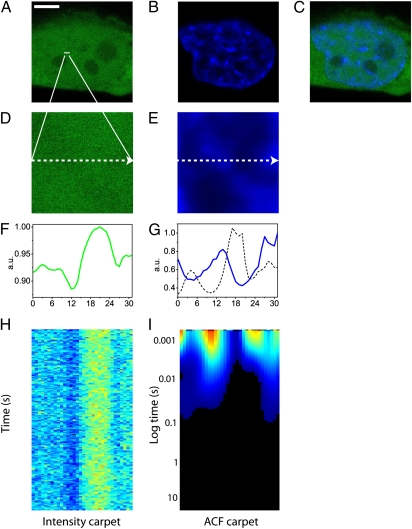
We used CHO-K1 cells stably transfected with EGFP and stained with Hoechst 33342 to measure the impact DNA density has on free diffusion of small molecules within the nucleus. Fig. 1 A–C depicts, respectively, a typical CHO-K1 cell expressing EGFP, its nucleus stained with Hoechst 33342, and their merging. For each cell, five line scans were selected within the nucleus, with each line being deliberately positioned across a region of high DNA density, to test this zone as a barrier to free EGFP diffusion (Fig. 1 D and E). Each line was drawn at maximum zoom, to obtain the pixel dimensions specified under the experimental section. Maximum line scanning speed was selected, with a pixel dwell time of 6.3 μs and a line time of 0.472 ms. For each line scan experiment in the nucleus, it was found that the concentration of EGFP molecules freely diffusing at each point of the line was inversely proportional to DNA density. Comparison of the intensity profile of EGFP across the line measured (Fig. 1F) with the intensity profile of the Hoechst 33342 stain along the same line (Fig. 1G) illustrates this observation.
Fig. 1.
ACF carpet analysis. (A) Free EGFP in a CHOK1 cell. (Scale bar: 5 μm.) (B) Nucleus of a CHOK1 cell stained with Hoechst 33342. (C) Merge of the images shown in A and B. (D) Free EGFP in the plane of the line drawn in the nucleus: 3.3 μm. (E) Hoechst 33342 staining in the plane of the line drawn in the nucleus: 3.3 μm. (F) Intensity profile of free EGFP across the line drawn. (G) Intensity profile of Hoechst 33342 staining across the line drawn (blue line) and an overlay of 1/G(0) (dashed black line), which is equivalent to the intensity profile of free EGFP in F. (H) Fluorescence intensity carpet of the line drawn across freely diffusing EGFP (200,000 lines). (I) ACF carpet of the line drawn across freely diffusing EGFP (90,000 lines analyzed).
After selecting the position of a line within the nucleus, we performed the scan of EGFP intensity along that line several times over many seconds. Over this time, cell movement and photobleaching are likely to occur and files containing either interference were not selected for further analysis (see Materials and Methods for criteria of rejection). The intensity carpet of freely diffusing EGFP shown in Fig. 1H is a construction of 64,000 lines acquired over a distance of 32 pixels. There is a direct relationship between the position of a point in the carpet representation and the time the intensity is acquired at that point. Calculation of the autocorrelation function at each pixel position as a function of time corresponding to a column in the intensity carpet produces what is termed an ACF carpet, shown in Fig. 1I. The concentration and average diffusion coefficient of EGFP at each pixel location is derived by fitting the ACF at every point along the line, as if at each point we were performing a conventional FCS measurement. Careful inspection of Fig. 1I reveals that the ACF carpet has regions where G0 is low (marked in blue) and G0 is high (marked in red). Comparison of the derived 1/G0 (that is directly proportional to the number of molecules) plot with the intensity profile of Hoechst 33342 along the same line (Fig. 1G) further confirms the previous observation that there is an inverse relationship between DNA density and EGFP concentration. As can be seen in Fig. 1I, the maximum of the ACF carpet across the line occurs at time 0 (since the particles correlate inside the PSF) and then correlation is lost at a rate which varies from pixel to pixel as a consequence of the local different environment. It should be noted that unlike single-point FCS measurements where the local diffusion coefficient is derived from a series of points acquired with an interval on the microsecond timescale (corresponding to the pixel dwell time), the ACF carpet here and the local diffusion coefficients derived from it is constructed from acquisition of several lines. The interval between two consecutive time points at the same location is 0.472 ms (corresponding to the line time). This relatively slow sampling at the same point results in our measurements being more sensitive to slower diffusing species. Thus, EGFP diffusion in regions of low and high DNA density were well fitted by using a two-species model. Extraction and fitting of a column corresponding to a region of low DNA density with a two-species model results in D1 = 22.8 μm2/s and D2 = 0.3 μm2/s. Extraction and fitting of a column corresponding to a region of high DNA density results in D1 = 23 μm2/s and D2 = 0.48 μm2/s. Fig. S1 reports the fits these values were derived from and Table S1 summarizes the cumulative results obtained for several experiments. Overall, EGFP diffusion was found to be constant in terms of local diffusion coefficient throughout the nucleus and, therefore, neither impeded nor depended on DNA density, in agreement with reported results (7, 17). However, it was found that the relative contribution of the two species depended on the location in the nucleus. That is the ratio of slow/fast diffusing component was higher in areas of high DNA density.
pCF Analysis: Measurement of the Diffusive Route Taken by EGFP Within the Nucleus.
The ACF-based approach described above does not provide information about the diffusive route taken by free EGFP with respect to DNA position, as it only measures the local diffusion of EGFP in a point. In contrast, pair correlation analysis is based on temporal cross-correlation of a pair of points at a given distance from each other. This analysis can define the diffusive route taken by molecules over many pixels (microns) along the line measured.
pCF analysis was therefore carried out on each line measured, at several different distances determined in each case by the intensity profile of the Hoechst 33342 stain. In general, when the distance is small, the points are within the PSF and the maximum of correlation is observed at a very short time (as with the ACF). As the distance increases, the correlation curve starts at very low amplitudes (even negative) and then increases with a delayed maximum in amplitude, which is due to the average transit time between the two locations (diffusive or not). Given that a molecule cannot be in two places at the same time, those distances that are not possible for the molecule to reach on the timescale of the experiment will never produce a maximum of correlation above zero. The characteristic anticorrelation at a short time and then the increase in the correlation at longer times is the signature that we are detecting the “same molecule” at a later time.
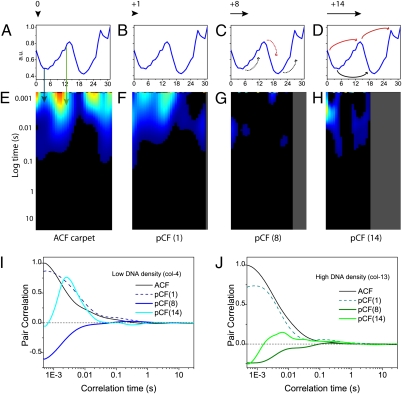
Fig. 2 shows the pCF carpets calculated for EGFP diffusion between the following DNA environments: (i) pCF(0), correlation of each pixel with itself, i.e., autocorrelation (Fig. 2A); (ii) pCF(1), correlation of pixels to adjacent pixels in the same DNA density environment (Fig. 2B); (iii) pCF(8), correlation of pixels in low DNA density to pixels in high DNA density (Fig. 2C); (iv) pCF(14), correlation of pixels in low DNA density to distant pixels in low DNA density, separated by a region of high DNA density (as well as pixels in high DNA density to pixels in high DNA density, separated by a region of low DNA density) (Fig. 2D).
Fig. 2.
pCF carpet analysis of intranuclear diffusion. (A–D) Intensity profile of the Hoechst 33342 stain across the line measured. The black arrow above each profile indicates to scale the distance that columns were cross-correlated in the pCF carpets directly below. The black and red curved arrows indicate respectively the DNA environment that is being cross-correlated from low or high DNA in the pCF carpets directly below. (E) pCF(0) carpet, which corresponds to calculation of an ACF carpet (90,000 lines analyzed). (F) pCF(1) carpet, which corresponds to cross-correlation of adjacent pixels in the same DNA environment within the PSF. (G) pCF(8) carpet which corresponds to cross-correlation of pixels in different density DNA environments. (H) pCF(14) carpet, which corresponds to cross-correlation of pixels in low-low DNA around a high DNA density environment or pixels in high-high DNA around a low DNA density environment. All carpets are depicted on the same color scale: the minimum value was set to 0, and this corresponds to black (no communication). (I) Plot of the amount of correlation for column 4 (low DNA) at each analyzed distance (0, 1, 8, and 14). Curves are normalized to 1 with respect to the ACF. (J) Plot of the amount of correlation for column 13 (high DNA) at each analyzed distance (0, 1, 8, and 14). Curves are normalized to 1 with respect to the ACF.
The pCF(0) carpet shown in Fig. 2E is obviously equivalent to the ACF carpet in Fig. 1H. The pCF(1) carpet shown in Fig. 2F is also similar to the ACF carpet in Fig. 1H. This similarity is because at a distance of one pixel between points, we are still within the PSF and so only local diffusion is measured. At this distance, we can only observe communication within the same DNA environment for both low and high density DNA regions. Fig. 2G shows the pCF(8) carpet, which is no longer a measurement of local diffusion given that we are now cross-correlating points at a distance much larger than the PSF. Inspection of the Hoechst 33342 intensity profile shows that at this distance (800 nm), almost all pixel positions along the line are being cross-correlated from either low-to-high DNA density or high-to-low DNA density. Virtually no positive cross-correlation is observed within the pCF(8) carpet (only anticorrelation). This result suggests that EGFP diffusion from low-to-high or high-to-low DNA density is negligible on the timescale measured.
In contrast, the pCF(14) carpet of Fig. 2H shows strong positive cross-correlation for discrete regions along the line measured. The Hoechst 33342 intensity profile reveals that regions displaying positive correlation are pixel positions that result in cross-correlation of low-to-low DNA density around a region of high DNA density (e.g., columns 3–6, Fig. 2H), or from high-to-high DNA density around a region of low DNA density (e.g., columns 12–15, Fig. 2H). We performed the same analysis across each set of five lines drawn in the nuclei of five different cells, by slightly changing the pixel distance for pCF calculation according to DNA localization: Additional pCF analysis of EGFP intranuclear diffusion is provided in Figs. S2 and S3. Analysis of a line that traversed the nucleolus was also tested (Fig. S4), and it was found to behave like a region of low DNA density that partly excludes EGFP but does not obstruct molecules from reaching the other side.
In general, we observed low-to-low DNA density communication around a high DNA density region in the timescale of 1–30 ms (range of reported cross-correlation peak values, Table 1), and high-to-high DNA density communication around a low DNA density region in the timescale of 10–80 ms (Table 1). As calculated by the normalized curves in Fig. 2 I and J, the fraction of molecules diffusing from low to low DNA density regions around a high DNA density region is much higher compared with that of molecules diffusing from high to high DNA density regions around a low DNA density region: ≈75% versus ≈15%, as given by the ratios of pCF(14) and ACF peaks for columns 4 and 13, respectively. This parameter was defined as “efficiency,” and Table 1 summarizes the cumulative results obtained. Overall we found low-to-low DNA density communication more “efficient” (50–85%, range of reported values in the whole population of cells measured) than high-to-high (10–40%). We also performed two control line experiments, to test the validity of the results attained. We first measured the diffusion of fluorescein (a much smaller molecule than EGFP) across a heterogeneous DNA environment in the CHOK1 cells stained with Hoechst 33342: to test the size dependence of intranuclear diffusion. For a molecule of this size, pCF analysis revealed no barriers to diffusion (Fig. S5). We then measured the diffusion of EGFP in a CHOK1 cell not treated with Hoechst 33342 to test the potential effect this nuclear stain had on the results obtained. In keeping with the results obtained in stained cells, we found the barriers to diffusion at positions that coincided with transition from low to high (or from high to low) EGFP concentration (Fig. S6).
Table 1.
Cumulative statistics derived from pCF analysis
| Measurement | Low-to-low | High-to-high |
| pCF peak, ms | 1–30 | 10–80 |
| Efficiency, % | 50–85 | 10–40 |
The first line reports the range of observed pCF peak positions for the analysis of low-to-low and high-to-high EGFP diffusion in the whole population of cells. The distances at which pCF analysis was carried out were determined on an individual basis by the DNA density variation along the line measured. The second line reports the range of calculated efficiencies for EGFP low-to-low and high-to-high diffusion in the whole population of analyzed cells. The ”efficiency” parameter is defined as the ratio between the amplitude of the pCF (calculated at a distance corresponding to low-to-low or high-to-high communication) and that of the ACF.
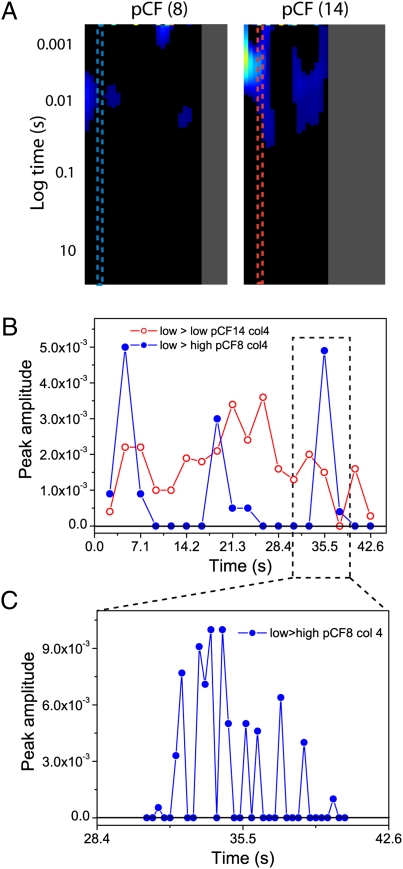
If there is an absence of molecular flow through a region of different DNA density and yet EGFP can be easily detected in both DNA environments, then how did this transit occur? One possible explanation is that the transit through a change in DNA density is statistically a rare event, which upon extensive averaging does not appear in the pCF carpet. It was thus decided to increase the probability of detecting this event (which would give rise to a positive cross-correlation) by sequentially averaging smaller time segments of the original file. The previous pCF(8) analysis depicted in Fig. 2G was calculated by using 90,000 lines (≈42 s). We thus reanalyzed pCF(8) for column 4 (corresponding to almost zero communication in the previous representation) by decomposing the whole time of acquisition into small time increments (5,000 lines, ≈2.36 s) (Fig. 3A). For the same column, pCF(14) was also analyzed, corresponding to positive cross-correlation between low DNA density regions (Fig. 3A). For both pCF distances, a plot was generated of the maximum amplitude of correlation detected for each fragment against the time of acquisition. In the instance a 5,000-line fragment produced a negative cross-correlation, for simplicity we plotted the amplitude of that fragment as zero: because anticorrelation is always indicative of no communication. The pCF(8) analysis of column 4 performed in shorter time intervals detects discrete periods of positive correlation that occur intermittently between longer periods of no communication (blue dots in Fig. 3B). In contrast, for pCF(14) analysis (red dots in Fig. 3B), there is a broad peak of positive correlation that occurs over the entire time of acquisition. One of the rare events of communication detected in pCF (8) was then further decomposed to understand the temporal nature of the periods of communication. The peak observed at ≈35.5 s was analyzed every 600 lines (≈300-ms time intervals), and a plot of correlation amplitudes against time of acquisition was produced (Fig. 3C). Fig. 3C shows that the pCF correlation amplitude arises from several shorter EGFP diffusion events across a low-to-high (or high-to-low) DNA density discontinuity. To prove that the observed bursts did not simply originate from noise fluctuations, we reversed our pCF analysis and cross-correlated points along the same line, but in the opposite direction. By this operation, we found a nearly identical timing of the bursts in the two directions, suggesting that these events are bidirectional (an example is reported in Fig. S7).
Fig. 3.
Decomposition of pCF carpet analysis. (A) Position of column 4 from the experiment depicted in Fig. 2 in the pCF(8) and pCF(14) carpets. As can be seen, column 4 is an area of zero communication in pCF(8) and of positive communication in pCF(14). This column was reanalyzed in shorter time fragments (every 5.0 × 103 lines, 2.36 s) in an attempt to detect communication between different DNA density environments. (B) A plot of the amplitude of the cross-correlation function derived for each 5.0 × 103 line segment against time. In the instance a time segment gave rise to a negative pCF profile (which corresponds to no communication), we reported the amplitude as zero. The plot for pCF(8) analysis of column 4 is depicted in blue, and the plot for pCF(14) analysis of column 4 is depicted in red. (C) Further temporal analysis of a peak of the pCF(8) reported in B, analysis (located at 35.5 s) was then carried out every 6.0 × 102 lines (300 ms) to evaluate the temporal nature of the intermittent periods of communication.
Discussion
The dominant mode of motion of inert molecules throughout the nucleus is by diffusion. How this motion is directed has several significant physiological implications. Here we reveal molecular flow in the nucleus by applying the pair correlation function method (pCF) to the diffusion of monomeric EGFP in live cells. We were able to concomitantly perform ACF and pCF analysis on the line scans acquired. Our ACF analysis, which is local, agrees with the observation by Dross et al. that the local diffusion coefficient of EGFP throughout the nucleus is independent of DNA density and well described by a 2-component model (7, 17). The ACF carpet analysis, however, does not provide information about the translational diffusion of EGFP between two different locations with respect to the DNA environment. Thus, by cross-correlating pairs of points in the nucleus, we found that there was a dependence of the diffusive route taken by EGFP on DNA density, in agreement with the observation by Wachsmuth et al. that EGFP diffusion in the nucleus can be locally impeded by cellular components (7).
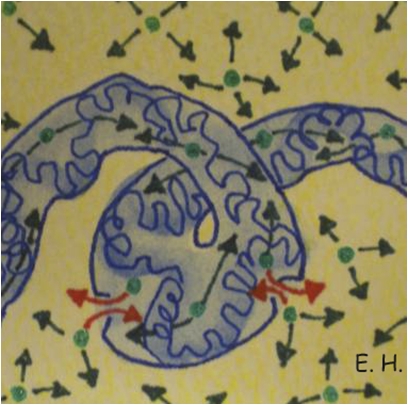
Our analysis of EGFP diffusion between different DNA environments showed migration paths, which allowed for communication between different DNA environments, and established barriers to diffusion resulting in poor or no communication. As schematically shown in Fig. 4, we found the two types of DNA density (high and low) to cause disconnect molecular flow throughout the nucleus, where the high density regions of DNA form a networked channel that allows EGFP to diffuse freely throughout, however with restricted ability to traverse the channel barriers to the low DNA density environment where EGFP diffusion is twice as efficient.
Fig. 4.
Model of diffusion within the nucleus. Schematic of monomeric EGFP (green) diffusing throughout the high DNA density networked channel (blue) as well as the low DNA density surroundings (yellow), with intermittent bursts of EGFP traversing the channel barriers (red).
This conclusion of channeled molecular flow is in agreement with two theoretical chromatin models that have been proposed based on simulation of intranuclear diffusion of inert molecules. The first is the phenomenological ICD (interchromosomal domain) model (8), which postulates a network of channels that enable diffusion of small molecules between chromosomal domains. The second is the MLS (multiloop subcompartment model) (9), which describes the interphase nucleus as being filled with chromosome territories that intermingle only moderately but allow inter- as well as intrachromosomal transport of small molecules via obstructed diffusion. Our measurements lead to a model of two virtually disconnect DNA environments (channels), differentiated by density that allow continuous molecular flow throughout either and restricted movement in between. Occasionally in discrete blocks of time (300 ms), a burst of EGFP molecules can travel across a DNA density barrier. The observed bidirectionality of this process suggests that the bursts may not need metabolic energy. Their confinement in time also indicates that they do not depend on a fixed potential barrier to diffusion, because this fixed barrier would produce a continuous small flow instead of an intermittent one. Local variation of chromatin compaction, due for instance to conformational changes of the DNA, can represent a way to enable communication across regions of different DNA density. Analogously, the role of DNA as a master template has been recently proposed for the regulation of the genetic and molecular flows in bacteria (22).
Overall, our results provide explanation for the presence of EGFP in both DNA environments, yet with no considerable flow being observed between either. EGFP bidirectional diffusion through a region of different DNA density is a statistically rare event that occurs on a short timescale. This event cannot be detected by an ensemble averaging measurement, such as FRAP, because it contains no spatial information about molecular flows. On the contrary, the pCF approach, by tracking single molecules as they travel from a specific location to another, can measure anisotropic diffusion, detect the presence of barriers to diffusion, and produce a map of transport. In conclusion, details of intranuclear diffusion were shown here by the pCF in vivo experiment, which advances our current understanding of molecular flow throughout the nuclear environment and are in agreement with previously proposed theoretical models. Our measurements indicate that chromatin is organized as a networked channel, which directs the diffusion of small molecules throughout and controls communication to the surrounding low DNA density environment. This study proposes the pCF approach as a method to study long-range intracellular molecular diffusion, which could bring a better understanding of the mechanisms of its regulation.
Materials and Methods
Cell Culture and Treatments.
CHO-K1 cells stably transfected with EGFP were grown in Ham's F-12K medium supplemented with 10% of FBS at 37 °C and in 5% CO2. For live imaging, the cells were plated 24 h before experiments onto 35-mm glass bottom dishes (No. 1 0.17-mm thickness). Plated cells were stained with Hoechst 33342 (2 μg/mL for 10 min) for the labeling of double-stranded DNA and then examined by confocal microscopy. All measurements were performed at room temperature.
Two samples were also prepared for control experiments. The first involved plain CHO-K1 cells stained with Hoechst 33342 (2 μg/mL for 10 min) and then with fluorescein (5 μM for 30 min): to measure the size dependence of intranuclear diffusion. The second involved unstained CHO-K1 cells stably transfected with EGFP: to measure the potential effect of Hoechst 33342 treatment on the results attained.
Microscope.
The microscopy measurements were performed on a Zeiss LSM710 META laser scanning microscope, using a 63× water immersion objective 1.2 N.A. (Zeiss). EGFP was excited with the 488-nm emission line of an Argon laser, and Hoechst 33342 was excited with a 405nm diode laser. For each channel, the pinhole was set to 1 Airy Unit. Data were analyzed with SimFCS software developed at the Laboratory for Fluorescence Dynamics (www.lfd.uci.edu). The volume of the point spread function (PSF) was calibrated by measuring the autocorrelation curve for 20 nM fluorescein in 0.01 M NaOH, which has a known diffusion coefficient of 400 μm2/s. The measured values of ω0 (that defines the PSF) varied in the range of 0.26 ± 0.04 μm, for the laser wavelength (488 nm) used in this study. The average power at the sample was maintained at the milliwatt level. EGFP and the Hoechst 33342 stain were measured sequentially, adopting the following collection ranges: 409–480nm (Hoechst 33342) and 500–598nm (EGFP). The potential for cross talk between EGFP and the Hoechst 33342 stain during acquisition was tested and found to be nonexistent through measurement of an unstained EGFP cell, with the detection range adopted for Hoechst 33342.
Experimental.
We measure the diffusion of free monomeric EGFP along several different lines drawn within the same nucleus of several different cells. We use the intensity profile of the Hoechst 33342 stain along the same line in each case as a reference of DNA density and position. We acquire data by rapidly scanning a diffraction limited laser beam (488 nm) along the line drawn. The dimensions of the pixels within the line are defined in such a way that fluorescence intensity is oversampled with respect to the waist of the laser beam. The average waist of the diffraction limited spot calibrated with fluorescein was found to be 260 nm. Therefore, a pixel size of ≈100 nm was required to oversample. Measuring a line of 32 pixels at maximum zoom achieved these pixel dimensions, resulting in a line length of 3.3 μm. The maximum scanning speed for these settings was selected (pixel dwell time 6.3 μs, line time 0.472 ms) so that the EGFP molecules could be correlated in time between lines measured.
These parameters were found to be adequate for studying EGFP-free diffusion in the nucleus, on a spatial scale from ≈200 nm to microns and in a timescale from milliseconds to several seconds. In general for each experiment, 200,000 consecutive lines (with no intervals between lines) were acquired, corresponding to ≈94 s. During this time, cell movements and photobleaching are likely to occur. Thus, the intensity carpets constructed for each of the files acquired were analyzed carefully for both interferences. Time regions within accepted files with no average change in the x, y, and z direction (e.g., cell movement) and no average change in fluorescence intensity (e.g., photo-bleaching) were selected for the correlation analysis. Typically this selection process resulted in the analysis of ≈64,000 lines (corresponding to ≈30 s). Both cell movement and photobleaching would cause lengthening of the correlation profile upon derivation of the ACF, thus preventing further analysis of correlation at earlier times where EGFP diffusion is likely to occur. An image of the Hoechst 33342 stain was also acquired before and after each measurement to check for DNA movement (in those files displaying no cell movement). As reported by others (23, 24), no detectable motion of the DNA was ever observed. The autocorrelation function (ACF) carpet obtained was then analyzed and pair correlation functions derived (mathematical derivation of the pCF is reported in SI Materials and Methods).
Data Analysis.
Calibration of the PSF and calculation of the auto- and pair-correlation functions were done by using the SimFCS software. Details about the mathematical derivation of the pCF for diffusing particles can be found in published papers (18). Intensity data are presented by using a carpet representation in which the x coordinate corresponds to the point along the line (pixels) and the y coordinate corresponds to the time. The autocorrelation function (ACF) and the pair correlation functions (pCF(pixels)) are displayed in pseudo colors in an image in which the x coordinate corresponds to the point along the line and the vertical coordinate corresponds to the autocorrelation time in a log scale. The distances at which pCF analysis was carried out were not fixed across all experiments, but instead determined on an individual basis by the DNA density variation along each line measured. All of the reported pair correlation curves were normalized with respect to the ACF curve. Control experiments to prove the validity of the method were carried out on EGFP diffusion across obvious obstacles: the boundary between two neighbor cells (impenetrable barrier) and the nuclear envelope (barrier introducing a well-known delay in molecular transport) (Fig. S8).
Supplementary Material
Acknowledgments
We thank Milka Stakic for cultivating and transfecting the CHO-K1 cells. This work was supported in part by the Cell Migration Consortium Grant U54 GM064346 (to M.D. and E.G.) and National Institutes of Health Grants P41-RRO3155 and P50-GM076516 (to F.C., E.H., and E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006731107/-/DCSupplemental.
References
- 1.Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 2.Gorski S-A, Dundr M, Misteli T. The road much traveled: Trafficking in the cell nucleus. Curr Opin Cell Biol. 2006;18:284–290. doi: 10.1016/j.ceb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Seksek O, Biwersi J, Verkman A-S. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Politz J-C, Tuft R-A, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14:4805–4812. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phair R-D, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 6.Görisch S-M, Lichter P, Rippe K. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol. 2005;123:217–228. doi: 10.1007/s00418-005-0752-y. [DOI] [PubMed] [Google Scholar]
- 7.Wachsmuth M, Waldeck W, Langowski J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J Mol Biol. 2000;298:677–689. doi: 10.1006/jmbi.2000.3692. [DOI] [PubMed] [Google Scholar]
- 8.Cremer T, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- 9.Münkel C, et al. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 10.Bhorade R, Weissleder R, Nakakoshi T, Moore A, Tung C-H. Macrocyclic chelators with paramagnetic cations are internalized into mammalian cells via a HIV-tat derived membrane translocation peptide. Bioconjug Chem. 2000;11:301–305. doi: 10.1021/bc990168d. [DOI] [PubMed] [Google Scholar]
- 11.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 12.Braga J, Desterro J-M, Carmo-Fonseca M. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol Biol Cell. 2004;15:4749–4760. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sprague B-L, McNally J-G. FRAP analysis of binding: Proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Levi V, Gratton E. Chromatin dynamics during interphase explored by single-particle tracking. Chromosome Res. 2008;16:439–449. doi: 10.1007/s10577-008-1240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Politz J-C, Browne E-S, Wolf D-E, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S-A, Heinze K-G, Schwille P. Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 17.Dross N, et al. Mapping eGFP oligomer mobility in living cell nuclei. PLoS ONE. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Digman M-A, Gratton E. Imaging barriers to diffusion by pair correlation functions. Biophys J. 2009;97:665–673. doi: 10.1016/j.bpj.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardarelli F, Gratton E. In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions. PLoS ONE. 2010;5:e10475. doi: 10.1371/journal.pone.0010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dertinger T, et al. The optics and performance of dual-focus fluorescence correlation spectroscopy. Opt Express. 2008;16:14353–14368. doi: 10.1364/oe.16.014353. [DOI] [PubMed] [Google Scholar]
- 21.Tsien R-Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 22.Llopis P-M, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abney J-R, Cutler B, Fillbach M-L, Axelrod D, Scalettar B-A. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis S-K, Bardeen C-J. The connection between chromatin motion on the 100 nm length scale and core histone dynamics in live XTC-2 cells and isolated nuclei. Biophys J. 2004;86:555–564. doi: 10.1016/S0006-3495(04)74134-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.