Abstract
The mechanism leading to protein-primed DNA replication has been studied extensively in vitro. However, little is known about the in vivo organization of the proteins involved in this fundamental process. Here we show that the terminal proteins (TPs) of phages ϕ29 and PRD1, infecting the distantly related bacteria Bacillus subtilis and Escherichia coli, respectively, associate with the host bacterial nucleoid independently of other viral-encoded proteins. Analyses of phage ϕ29 revealed that the TP N-terminal domain (residues 1–73) possesses sequence-independent DNA-binding capacity and is responsible for its nucleoid association. Importantly, we show that in the absence of the TP N-terminal domain the efficiency of ϕ29 DNA replication is severely affected. Moreover, the TP recruits the phage DNA polymerase to the bacterial nucleoid, and both proteins later are redistributed to enlarged helix-like structures in an MreB cytoskeleton-dependent way. These data disclose a key function for the TP in vivo: organizing the early viral DNA replication machinery at the cell nucleoid.
Keywords: Bacillus subtilis, phage ø29, DNA polymerase, DNA-binding, bacterial cytoskeleton
Protein-primed DNA replication is a mechanism used to initiate DNA synthesis in a variety of prokaryotic and eukaryotic organisms (1). Phages such as ϕ29 (infecting Bacillus subtilis) and PRD1 (infecting Escherichia coli) (1, 2), animal viruses such as adenoviruses (1, 3), bacterial species from the Streptomyces genus (1, 4), and viruses infecting Archaea (5–7) possess replication origins at their linear chromosomes constituted by inverted terminal repetitions with a terminal protein (TP) linked to both 5′ genome ends. TPs also have been reported in linear plasmids isolated from bacteria, yeast, fungi, and higher plants (1) and in animal and plant RNA viruses (1).
The development of an in vitro replication system with purified proteins and DNA from the B. subtilis phage ϕ29 laid the foundations for investigating the protein-primed mechanism of DNA replication and makes ϕ29 a paradigm for studying this process (1, 2). A schematic overview of the in vitro ϕ29 DNA replication mechanism is shown in Fig. S1. The ϕ29 genome consists of a 19,285-bp linear dsDNA, with a TP of 31 kDa (the parental TP) covalently linked to each 5′ end. DNA replication starts with the recognition of the TP-containing DNA ends by a heterodimer formed by the ϕ29 DNA polymerase and a free TP molecule (the primer TP) (8). The DNA polymerase then catalyzes the formation of a covalent bond between deoxyAMP and the hydroxyl group Ser232 of the primer TP. Replication is coupled to strand displacement, and continuous elongation of the DNA polymerase from both DNA ends generates replication intermediates that finally converge in the complete duplication of the parental strands (reviewed in ref. 2). Phage ϕ29 DNA transcription is divided into early and late stages (9). Fig. S1 shows a genetic and a transcriptional map. Genes 2 and 3, encoding phage DNA polymerase and TP, respectively, are located in the left-side early operon.
Although the protein-priming mechanism of DNA replication has been studied extensively in vitro, the underlying mechanisms governing the in vivo organization of the proteins involved in this process are poorly understood. Recently, we provided insights into the in vivo organization of proteins involved in ϕ29 DNA replication by showing that the bacterial actin-like MreB cytoskeleton, playing important roles in several cellular processes (reviewed in 10), is required for efficient DNA replication of the distantly related phages ϕ29, SPP1, and PRD1 (11). Components of the ϕ29 DNA replication machinery, such as the DNA polymerase, are redistributed in peripheral helix-like structures in a cytoskeleton-dependent way.
Here, we analyze the subcellular localization of the TP of phage ϕ29 in live B. subtilis cells and show that it associates with the bacterial nucleoid independently of other phage-encoded proteins. The N-terminal domain of the TP directs its nucleoid association and is required for efficient ϕ29 DNA replication. The TP recruits the phage DNA polymerase to the bacterial nucleoid in early infection, and later both proteins relocalize to peripheral helix-like structures in a process dependent on the bacterial cytoskeleton.
Results
Viral TPs Associate with the Bacterial Nucleoid Independently of Other Phage-Encoded Proteins.
As a first approach to determine the in vivo organization of viral protein primers in bacteria, we engineered a B. subtilis strain containing a xylose-inducible fusion of yfp to the ϕ29 gene 3 cloned at the chromosomal amyE locus. Complementation experiments using the ϕ29 replication-deficient mutant phage sus3(91), unable to synthesize TP, showed that the TP fusion protein YFP-TP was functional (Fig. S2 A and B).
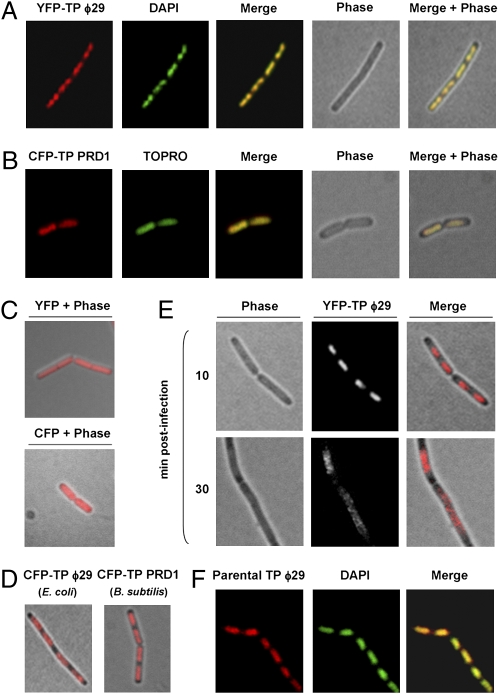
Fig. 1A shows the localization of YFP-TP in B. subtilis noninfected cells. Merged images of both YFP-TP and DAPI fluorescent signals demonstrated that YFP-TP colocalizes with the bacterial nucleoid. Therefore, the nucleoid localization of YFP-TP is independent of other ϕ29-encoded proteins. Importantly, we found that the TP belonging to the distantly related phage PRD1, fused to CFP, similarly colocalizes with the E. coli nucleoid independently of other PRD1 proteins (Fig. 1B), suggesting a conserved functional property of viral TPs in bacteria. Control cells expressing YFP or CFP in B. subtilis and E. coli, respectively, displayed a fluorescent signal uniformly distributed along the entire length of the bacteria (Fig. 1C). We found that ϕ29 and PRD1 TPs also are able to associate with nucleoids of E. coli and B. subtilis, respectively (Fig. 1D).
Fig. 1.
Subcellular localization of ϕ29 and PRD1 TPs. (A) YFP, DAPI staining, phase-contrast, and merged images of typical B. subtilis cells expressing a xylose-induced YFP-ϕ29 TP fusion protein (strain DM-021) analyzed 30 min after xylose addition. (B) CFP, TO-PRO-3 staining, phase-contrast, and merged images of typical E. coli cells expressing an IPTG-induced CFP-PRD1 TP fusion protein (strain DM-051) analyzed 30 min after IPTG addition. (C) Phase-contrast overlay of B. subtilis cells expressing a xylose-induced YFP (strain DM-022) and E. coli cells expressing an IPTG-induced CFP (strain DM-049) 30 min after the addition of the inductor. (D) Phase-contrast overlay of E. coli cells expressing an IPTG-induced CFP-ϕ29 TP (strain DM-050) and B. subtilis cells expressing a xylose-induced CFP-PRD1 TP (strain DM-060) 30 min after the addition of the inductor. (E) Phase-contrast, YFP fluorescence, and merged images of typical ϕ29 sus14(1242)-infected cells expressing a xylose-induced YFP-TP fusion protein (B. subtilis strain DM-021) 10 and 30 min after infection. Fluorescence signals are shown after deconvolution of an image stack, as a max projection. DM-021 cells were grown at 37 °C in LB medium supplemented with 2% glucose to an OD600 of 0.4. Next, xylose was added to a final concentration of 0.5%, and the culture was infected with ϕ29 mutant sus14(1242) at an MOI of 5. (F) Immunofluorescence microscopy assay. B. subtilis 110NA cells were grown at 37 °C in LB medium containing 5 mM MgSO4. At an OD600 of 0.4 the culture was split, and half the culture was infected with phage sus3(91) at a MOI of 25. Samples were harvested 15 min later and processed for immunodetection. Shown are typical unprocessed localization patterns of immunodetected parental TP, DAPI staining, and overlay. For clarity, in A–F YFP fluorescent signals and DAPI or TO-PRO-3 staining are false-colored red and green, respectively.
We next analyzed the localization of the ϕ29 TP fusion in live B. subtilis-infected cells as a function of time. As shown in Fig. 1E, 10 min after infection YFP-TP fluorescent signal occupied the central region of the cell spanning the nucleoid area. Later, 30 min postinfection, corresponding to a period of high ϕ29 DNA accumulation (Fig. S2B), YFP-TP relocalized in helix-like patterns as the cells enlarged before division.
Because the functional YFP-TP fusion, which mimics the role of ϕ29 priming TP, displays nucleoid localization, we studied whether the ϕ29 parental TP, which is covalently linked at each 5′ end of the viral genome, similarly distributes at the bacterial nucleoid independently of priming TP molecules. To do so, we used immunofluorescence techniques, infecting a nonsuppressor B. subtilis strain with the ϕ29 mutant phage sus3(91). Overlay of TP and DAPI fluorescent signals demonstrated that the phage parental TP indeed colocalizes with the B. subtilis nucleoid in the absence of priming TP (Fig. 1F).
N-Terminal Domain of the TP Is Responsible for Its Nucleoid Localization.
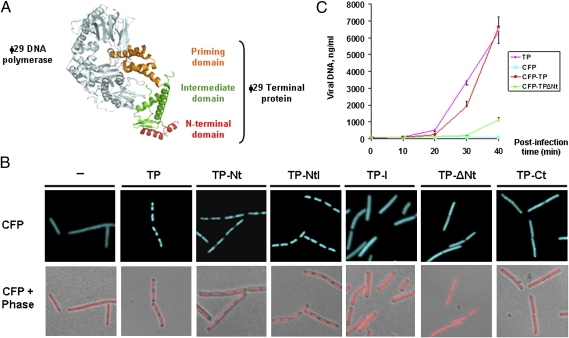
The crystallographic structure of the ϕ29 DNA polymerase/priming TP heterodimer shows that the TP has an elongated three-domain structure (Fig. 2A) (12) with an N-terminal domain spanning residues 1–73 (TP-Nt), an intermediate domain spanning residues 74–173 (TP-I) that makes extensive contacts with the phage DNA polymerase, and a C-terminal priming domain spanning residues 174–266 (TP-Ct) that mimics duplex product DNA in its electrostatic profile and binding site in the polymerase.
Fig. 2.
The TP N-terminal domain localizes at the bacterial nucleoid and is important for efficient ϕ29 DNA replication. (A) Three-dimensional structure of the DNA polymerase/priming TP heterodimer (12). (B) B. subtilis cells expressing CFP (strain DM-024), CFP/TP (strain DM-025), CFP/TP-Nt (strain DM-026), CFP/TP-NtI (strain DM-027), CFP/TP-I (strain DM-028), CFP/TP-ΔNt (strain DM-029), and CFP/TP-Ct (strain DM-030) IPTG-inducible fusions were grown to midexponential phase in LB medium at 37 °C. At an OD600 of 0.4, the cultures were supplemented with 1 mM IPTG. Samples were harvested and analyzed by fluorescence microscopy 30 min after IPTG addition. For clarity, CFP fluorescent signals are false-colored red in merged images. (C) The amount of intracellular accumulated phage ϕ29 DNA was quantified by real-time PCR at different times postinfection with a sus3(91) mutant phage of the following B. subtilis strains: DM-024 (expressing CFP), DM-025 (expressing CFP-TP), DM-029 (expressing CFP-TPΔNt), and DM-032 (expressing wild-type TP). Samples were infected at a MOI of 1 and at different times postinfection were processed as described in Materials and Methods. The amounts of accumulated phage DNA (nanograms of viral DNA per milliliter of culture) are expressed as a function of time after infection. The experiment was carried out in triplicate to calculate SDs.
To determine which TP domain is responsible for its nucleoid localization in live noninfected cells, we engineered a set of B. subtilis strains expressing isopropyl β-d-thiogalactoside (IPTG)-inducible fusions of CFP and a combination of different domains of the TP: TP-Nt, N-terminal/intermediate (TP-NtI), TP-I, intermediate/C-terminal TP (TP-ΔNt), and TP-Ct domains. Experiments using a B. subtilis strain containing a functional IPTG-inducible fusion of cfp to the complete ϕ29 gene 3 at the chromosomal thrC locus (Fig. S2 C and D) confirmed the nucleoid association of the TP in noninfected cells (Fig. 2B). Interestingly, only CFP fusions containing the TP N-terminal domain (i.e., TP-Nt and TP-NtI) displayed nucleoid localization (Fig. 2B). Thus, the N-terminal domain of the ϕ29 TP directs the localization of the TP to the nucleoid.
N-Terminal Domain of the ϕ29 TP Has Sequence-Independent DNA-Binding Capacity.
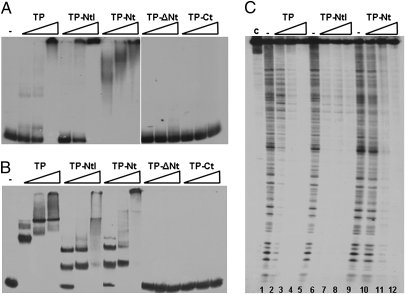
It has been shown that the ϕ29 TP interacts with phage dsDNA in vitro (13). To test whether the N-terminal domain of the TP possesses DNA-binding capacity, we purified different TP truncated proteins: TP-NtI (TP truncated priming domain variant spanning residues 1–172), TP-Nt (TP N-terminal domain spanning residues 1–73), TP-ΔNt (TP truncated N-terminal domain variant spanning residues 74–266), and TP-Ct (TP C-terminal priming domain spanning residues 174–266) and compared their capacity to bind dsDNA with that of wild-type TP. To do so, we used a 297-bp right end fragment of the ϕ29 genome in gel mobility shift assays with increasing amounts of wild-type and mutant TP proteins. Fig. 3A shows that, similar to wild-type TP, the TP-NtI variant and the TP-Nt each retained the ability to bind dsDNA. However, truncated N-terminal mutant proteins TP-ΔNt and TP-Ct lost the ability to bind the dsDNA fragment. Some of the retarded bands in the TP-Nt variant were less discrete than those of the wild-type TP and the TP-NtI variant, probably reflecting a small decrease in DNA-binding capacity because of the absence of the intermediate domain. We next studied whether the ϕ29 TP, and particularly the TP N-terminal domain, are able to interact with a DNA fragment comprising a sequence from the B. subtilis chromosome. Thus, a 216-bp DNA fragment corresponding to the yshC B. subtilis gene was subjected to gel retardation assays (Fig. 3B). As above, only wild-type TP and the truncated mutant proteins bearing the N-terminal region were able to interact with the dsDNA fragment.
Fig. 3.
The N-terminal domain of the TP has sequence-independent dsDNA-binding capacity. Gel mobility shift assays using an end-labeled 297-bp DNA fragment corresponding to the right end of the ϕ29 genome (A) and an end-labeled 216-bp DNA fragment corresponding to the B. subtilis yshC gene (B). The labeled probes were incubated either in the absence (−) or presence of increasing amounts (75, 150, and 300 nM) of the indicated purified protein in a buffer containing 50 mM NaCl. After nondenaturing PAGE, the mobility of the nucleoprotein complexes was detected by autoradiography. (C) DNase I treatment of nucleoprotein complexes formed by ϕ29 wild-type TP and TP-NtI or TP-Nt truncated variants. The 297-bp right end of the ϕ29 DNA fragment was end labeled and used in DNase I footprinting assays in the absence (−) or presence of increasing amounts (0.75, 1.5, and 3 μM) of the indicated proteins. The bottom of the footprints corresponds to the right end of the ϕ29 genome. c, control lane with the dsDNA probe not subjected to DNase I treatment and protein preincubation.
To gain insight into the dsDNA-binding mode of the TP and its N-terminal domain, we performed DNase I footprinting assays with a 297-bp dsDNA fragment from the ϕ29 right end (Fig. 3C). A typical DNase I digestion pattern was observed in the absence of protein (lane 1), but full protection of the dsDNA fragments was seen at the higher TP concentrations tested (lanes 3 and 4). Likewise, hardly any degradation products were observed for the TP-NtI and the TP-Nt variants (lanes 7 and 8 and lanes 11 and 12, respectively), indicating that the dsDNA was almost completely protected from DNase I attack with no obvious sequence preference. Together, these results show that a functional property of the TP, binding to dsDNA, resides in the N-terminal domain of the protein.
Efficient ϕ29 DNA Replication Requires the TP N-Terminal Domain.
To assess the importance of the N-terminal domain for the efficiency of in vivo ϕ29 DNA replication, we performed complementation assays using the sus3(91) mutant phage and measured by real-time PCR the amount of accumulated viral DNA in B. subtilis strains expressing wild-type TP, CFP-TP, CFP-TPΔNt, and CFP, respectively. Fig. 2C shows that similar efficient ϕ29 DNA replication was obtained in sus3(91)-infected cells that were complemented in trans with either TP or CFP-TP, indicating that the CFP-TP fusion is functional. In contrast, the efficiency of DNA replication in the strain expressing CFP-TPΔNt was severely decreased compared with the strain producing CFP-TP. Western blot analysis demonstrated that the amount of proteins expressed in trans was similar to that of TP synthesized in a typical ϕ29 infection cycle (Fig. S3A). Also, analyses of phage DNA accumulation by agarose gel electrophoresis confirmed that the TP N-terminal domain is required for efficient intracellular accumulation of ϕ29 DNA (Fig. S3B).
ϕ29 TP Recruits the Phage DNA Polymerase to the Bacterial Nucleoid, and Both Proteins Subsequently Are Redistributed in a Helix-Like Manner.
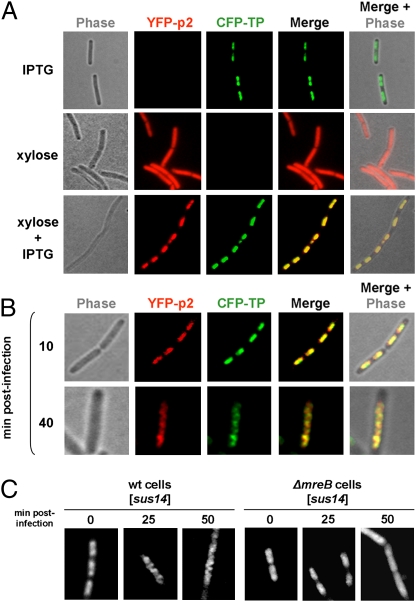
We previously showed that ϕ29 DNA polymerase localizes uniformly in non-infected cells, and that at middle stages of the infection cycle, it localizes into clear helix-like structures in B. subtilis (11). Nevertheless, at 10 min postinfection, the ϕ29 DNA polymerase fused to YFP (YFP-p2) colocalizes with the bacterial nucleoid (Fig. S4). Because ϕ29 priming TP and DNA polymerase interact, forming a heterodimer to initiate phage DNA replication (8), and because the TP is able to localize at the bacterial nucleoid independently of other phage-encoded proteins (Fig. 1A), we analyzed whether the ϕ29 TP has a role in recruiting the DNA polymerase at the bacterial nucleoid. To do so, we constructed a B. subtilis strain able to express simultaneously functional CFP-TP and YFP-p2 fusions (Fig. S2 C–E) from IPTG- and xylose-inducible promoters, respectively. The results show that, in the absence of CFP-TP expression, YFP-p2 distributes uniformly in noninfected cells (Fig. 4A). However, when the growing culture was supplemented with both IPTG and xylose (i.e., when both the CFP-TP and the YFP-p2 fusions were expressed), YFP-p2 relocalized to the bacterial chromosome. Thus, the ϕ29 TP plays a crucial role recruiting the phage DNA polymerase to the bacterial nucleoid.
Fig. 4.
The ϕ29 TP recruits the DNA polymerase to the bacterial nucleoid, and both proteins are redistributed in an MreB-dependent way. (A and B) B. subtilis strain DM-023, expressing YFP-p2 and CFP-TP in a xylose- and IPTG-dependent way, respectively, was grown in LB medium supplemented with 5 mM MgSO4 and 2% glucose. At an OD600 of 0.4, cultures were supplemented with 1 mM IPTG and/or 0.5% xylose, as indicated (A) and simultaneously were infected with a sus14(1242) phage at a MOI of 5 (B). Shown are phase-contrast, YFP and CFP fluorescence, and merged images. For clarity, YFP and CFP fluorescent signals are false-colored red and green, respectively. (C) B. subtilis strains expressing YFP-TP in a xylose-dependent way under wild-type (DM-021) and ΔmreB (DM-031) backgrounds were grown in LB medium supplemented with 25 mM MgSO4 and 2% glucose. At an OD600 of 0.4, cultures were supplemented with 0.5% xylose and infected with a sus14(1242) phage, as indicated. Samples were harvested 25 and 50 min postinfection. In A–C fluorescence signals are shown after deconvolution of an image stack, as a max projection.
To unravel further the in vivo organization of the ϕ29 protein priming machinery, we simultaneously examined the localization of the phage TP and DNA polymerase in B. subtilis-infected cells. As shown in Fig. 4B, 10 min postinfection, CFP-TP and YFP-p2 colocalized at the bacterial nucleoid. Importantly, 40 min postinfection both CFP-TP and YFP-p2 had relocalized and followed similar helical paths. Merging the CFP and YFP signals revealed a substantial colocalization.
Proper Redistribution of ϕ29 TP to Helix-Like Patterns Requires the MreB Cytoskeleton.
We previously demonstrated that the helical localization of the ϕ29 DNA polymerase at middle stages of infection is lost in mreB cytoskeleton mutant strains (11). To investigate whether the helical-like redistribution of ϕ29 TP also depends on MreB, we examined the subcellular localization of YFP-TP in an mreB deletion mutant. As shown in Fig. 4C, and unlike wild-type cells, YFP-TP did not adopt a helical configuration in the MreB cytoskeleton mutant even at late postinfection times. Instead, the fluorescent signals remained associated to the nucleoid area and were not redistributed in a helix-like manner.
Discussion
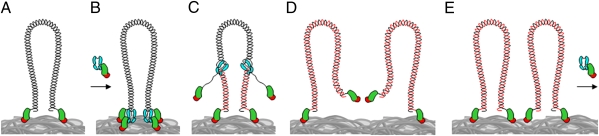
Bacterial viruses produce high numbers of progeny within a narrow time window during their lytic cycle. Following infection, phage DNA replication is expected to be organized rapidly, both spatially and temporally, to allow simultaneous amplification of multiple templates in a short period. Based on the experiments described in this paper, a schematic representation of the initial steps of the ϕ29 infection cycle is shown in Fig. 5.
Fig. 5.
Model of nucleoid-associated early ϕ29 DNA replication organized by the TP. (A) A complete ϕ29 TP-DNA molecule (linear dsDNA shown as a double helix) is shown attached to the bacterial nucleoid surface (gray mass at bottom) by the N-terminal domain (red) of the two parental TPs (red and green). (B) The priming TP interacts with the phage DNA polymerase (cyan), forming a heterodimer that associates with the nucleoid via the TP N-terminal domain and recognizes the origins of replication. (C) After a transition step, the DNA polymerases dissociate and continue processive elongation of the nascent DNA strands (red lines) coupled to strand displacement. (D and E) Once DNA replication is completed, two ϕ29 TP-DNA molecules are ready for another round of replication. For simplicity, other viral proteins involved in DNA replication are not drawn.
Our results show that the parental TP (in green and red) covalently linked to each 5′ end of the ϕ29 genome (linear dsDNA, shown as a double helix) associates with the B. subtilis nucleoid (gray mass at bottom) at early infection times. This binding occurs independently of priming TP, strongly indicating that, after ϕ29 TP-DNA injection takes place, the parental TP directs the viral genome to the bacterial nucleoid (Fig. 5A). In this way, the phage ϕ29 genome gains access to the B. subtilis RNA polymerase, which, like the ϕ29 TP, localizes at the bacterial nucleoid (Fig. S5), initiating transcription of the early genes required for phage DNA replication. Once both ϕ29 TP and DNA polymerase (shown in cyan) are synthesized, they form a heterodimer that associates with the bacterial nucleoid through the N-terminal DNA-binding domain of the priming TP (shown in red) (Fig. 5B) and recognizes the replication origins at both DNA ends by means of specific interactions with the parental TP (14, 15). It seems likely that the association of the ϕ29 TP-DNA to a specific bacterial compartment also might help stabilize the interaction with the heterodimeric complex. The TP N-terminal domain has sequence-independent DNA-binding capacity, and no direct interaction between the ϕ29 TP and nucleoid-associated proteins HBsu, SMC, ScpA, or Noc was detected by yeast two-hybrid assays (Fig. S6). After a transition step, the DNA polymerase dissociates and continues processive elongation (Fig. 5C) until the nascent DNA strand is completed (Fig. 5D). The newly replicated TP-DNA reassociates with the bacterial nucleoid, allowing a new replication cycle to begin (Fig. 5E). The ϕ29 strategy at early infection stages is to anchor its genome to the nucleoid, facilitating the access to the RNA polymerase. In agreement with our model, it was shown previously that the ϕ29 TP-DNA, although not covalently closed, is topologically constrained in vivo (16). Additionally, both novobiocin and nalidixic acid, inhibitors of the DNA gyrase, significantly impair viral DNA replication (16). Hence, the B. subtilis DNA gyrase is required for efficient in vivo ϕ29 DNA replication. This topological constraint might be caused at least in part, as shown here, by the association of the parental TP with the bacterial nucleoid.
After the ϕ29 DNA replication machinery is associated with the nucleoid, both the TP and the DNA polymerase are reorganized, adopting a peripheral helix-like configuration toward the cell poles. Interestingly, it has been reported recently that, as long as the B. subtilis chromosome is being replicated, the two newly synthesized DNA copies are similarly translocated via a peripheral helical structure to the opposite poles (17). Most likely, the TP associates with the newly synthesized bacterial DNA and adopts its morphology during segregation to the future daughter cells. Thus, it is tempting to speculate that ϕ29 DNA uses the motor-like force that provides chromosome segregation in B. subtilis to redistribute its replication machinery at multiple peripheral sites. Conspicuously, convincing evidence has been reported from E. coli and Caulobacter crescentus that MreB proteins, responsible mainly for the regulation of cell shape, also play a leading role in bacterial chromosome partitioning (18–20). Although the notion that MreB has a direct, mitotic-like role in chromosome segregation in B. subtilis remains controversial (21–23), it would explain the observation that essential ϕ29 DNA replication components (i.e., TP and DNA polymerase) redistribute following helical structures in an MreB cytoskeleton-dependent way. The intrinsic dynamic behavior of the replicating bacterial chromosome during its partitioning would enhance efficiency in organizing ϕ29 DNA replication, thereby allowing synchronized replication of multiple templates at various peripheral sites.
The TPs of phage ϕ29 and of eukaryotic adenovirus have remarkably similar roles in vivo. The adenovirus TP associates with the cell nuclear matrix (24, 25). As in ϕ29, this association is postulated to direct replication complexes to the appropriate location within the cell and, additionally, appears to be essential for optimal transcriptional activity of the adenovirus genome. However, although the adenovirus precursor of terminal protein (pTP), like the ϕ29 TP, has DNA-binding capacity (26), the binding of pTP to the nuclear matrix is not dependent on cellular chromosomal DNA (25). Instead, it has been proposed that the DNA-binding ability of pTP stabilizes the adenovirus DNA polymerase on partially unwound DNA origins during the initiation of DNA replication (26). We cannot exclude the possibility that the DNA-binding domain of the ϕ29 priming TP also might play a similar role, locating the DNA polymerase/priming TP heterodimer precisely at the replication origin. Nevertheless, this possibility seems unlikely because hardly any sequence-dependent DNA-binding capacity was observed for the ϕ29 TP, and because the N-terminal DNA-binding domain of the TP is not essential for amplifying ϕ29 DNA in vitro (27). Because (i) the ϕ29 TP localizes at the bacterial nucleoid independently of other phage-encoded proteins; (ii) the ϕ29 DNA polymerase is recruited by the TP to the bacterial nucleoid; and (iii) efficient in vivo ϕ29 DNA replication is compromised in the absence of the TP DNA-binding domain, we propose that the ϕ29 TP plays a significant biological role, attaching the essential ϕ29 DNA replication machinery at the bacterial nucleoid via a sequence-independent DNA-binding domain. Importantly, in addition to ϕ29, the TP of the distantly related E. coli-infecting phage PRD1 also localizes at the bacterial nucleoid independently of other viral-encoded proteins. Moreover, we show that both PRD1 and ϕ29 TP also associate with the nucleoid when expressed in bacteria other than their respective hosts. Taken together, these results reveal a functional property of viral TPs in bacteria that might have been conserved during evolution of a prokaryotic viral ancestor. By playing a key role in the organization of phage DNA replication at the bacterial nucleoid, TPs provide another way for viruses to exploit cell resources to optimize the production of numerous progeny.
Materials and Methods
General Methods.
Phage ϕ29 DNA replication is inhibited by the host protein Spo0A (28). Because the B. subtilis spo0A promoter switch is repressed by an excess of glucose (29), medium was supplemented with 2% glucose. Unless stated otherwise, mid-logarithmically growing B. subtilis cultures were infected at a multiplicity of infection (MOI) of 5, and cell samples were harvested and processed at the indicated times after infection. B. subtilis cells were transformed by standard procedures, as described (23). Selection for B. subtilis transformants was carried out on nutrient agar plates (Oxoid), supplemented with appropriate antibiotics, 0.5% xylose, 1 mM IPTG, or 25 mM MgSO4 when necessary. Details are given in SI Text.
Bacterial and Yeast Strains, Phages, and Growth Conditions.
Bacterial and yeast strains and phages used are listed in Tables S1 and S2, respectively. The delayed-lysis mutant phage ϕ29 sus14(1242) contains a mutation in gene 14 that has no effect on phage DNA replication or phage morphogenesis, thus allowing examination of phage protein and DNA localization at late infection times. Details are given in SI Text.
DNA Techniques and Plasmid Construction.
All DNA manipulations and cloning were carried out by standard methods. Plasmids used are listed in Table S2. Details are given in SI Text. Oligonucleotides used are listed in Table S3.
Protein Purification.
Wild-type ϕ29 TP was expressed in E. coli BL21(DE3) cells harboring gene 3 cloned into plasmid pT7-3 and further purified as described (13). Mutant proteins TP-Nt (ϕ29 TP N-terminal domain, spanning residues 1–73), TP-NtI (ϕ29 TP N-terminal and intermediate domains, spanning residues 1–173), TP-ΔNt (ϕ29 TP lacking the N-terminal domain, spanning residues 74–266), and TP-Ct (ϕ29 TP priming domain, spanning residues 174–266) were expressed in the E. coli strain BL21(DE3). The TP variant TP-NtI was purified essentially as described for the wild-type ϕ29 TP (13). The ϕ29 TP variants TP-Nt, TP-ΔNt, and TP-Ct were purified using Ni2+-NTA resin columns.
Gel Mobility Shift and Footprinting Assays.
Gel retardation and DNase I footprinting assays were performed essentially as described (30, 31). A 297-bp DNA fragment corresponding to the ϕ29 right end was amplified by PCR using genomic ϕ29 DNA as template and primers R-25 and R-OUT SUPER. A 216-bp DNA fragment corresponding to the B. subtilis yshC gene was amplified by PCR using genomic B. subtilis DNA as template and primers yshC_U and yshC_L. The PCR products were labeled at one of the 5′ ends by treating the appropriate primer with polynucleotide kinase and [γ-32P]ATP before the amplification reaction.
Immunofluorescence Microscopy.
Samples were fixed after the indicated times of infection and processed essentially as described (32). Details are given in SI Text.
Epifluorescence Microscopy.
For fluorescence microscopy, overnight cultures were diluted in LB medium containing 5 or 25 mM MgSO4 and grown to early exponential phase at 37 °C. At an OD600 = 0.3–0.6, cells were infected at an MOI of 5 with the indicated ϕ29 phage (Table S2) and supplemented with 0.5% xylose or 1 mM IPTG, as indicated. For live-cell imaging, cells were immobilized on microscope slides covered with a thin film of 1% agarose in water. CFP and YFP fluorescence were detected with a dual CFP/YFP-ET filter (89002; Chroma).
Real-Time PCR.
Cells corresponding to 1-mL aliquots of B. subtilis cultures, withdrawn at different times after infection, were harvested, processed, and analyzed by real-time PCR essentially as described (33). Details are given in SI Text.
Supplementary Material
Acknowledgments
We thank Miguel de Vega and Mario Mencía for critical reading of the manuscript and Jeff Errington and Richard Daniel (Institute for Cell and Molecular Biosciences, Newcastle, UK), David Rudner (Harvard Medical School, Boston), Peter Lewis (The University of Newcastle, Callaghan, Australia), and Etienne Dervyn, Jean-Christophe Meile, and Tatiana Rochat (Institut National de la Recherche Agronomique, Jouy-en-Josas, France) for supplying strains and plasmids. This investigation was supported by Grants BFU2008-00215 and Consolider-Ingenio 2010 24717 from the Spanish Ministry of Science and Innovation to M.S., ANR-08-JCJC-0024-01 from the French National Agency of Research to R.C.-L., and by an Institutional Grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” D.M.-E. was holder of an I3P contract from the Spanish National Research Council. I.H. and D.B.-P. are holders of a Formación de Personal Universitario and a Formación de Personal Investigador fellowship, respectively, from the Spanish Ministries of Education and Science and Innovation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010530107/-/DCSupplemental.
References
- 1.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 2.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet Eng (N Y) 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 3.de Jong RN, van der Vliet PC, Brenkman AB. Adenovirus DNA replication: Protein priming, jumping back and the role of the DNA binding protein DBP. Curr Top Microbiol Immunol. 2003;272:187–211. doi: 10.1007/978-3-662-05597-7_7. [DOI] [PubMed] [Google Scholar]
- 4.Chaconas G, Chen CW. In: The Bacterial Chromosome. Higgins NP, editor. Washington, DC: American Society for Microbiology Press; 2005. pp. 525–539. [Google Scholar]
- 5.Bamford DH, et al. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J Virol. 2005;79:9097–9107. doi: 10.1128/JVI.79.14.9097-9107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Peng X, Basta T, Häring M, Garrett RA, Prangishvili D. Genome of the Acidianus bottle-shaped virus and insights into the replication and packaging mechanisms. Virology. 2007;364:237–243. doi: 10.1016/j.virol.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Blanco L, et al. Effect of NH4+ ions on ϕ29 DNA-protein p3 replication: Formation of a complex between the terminal protein and the DNA polymerase. J Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rojo F, Mencía M, Monsalve M, Salas M. Transcription activation and repression by interaction of a regulator with the α subunit of RNA polymerase: The model of phage ϕ29 protein p4. Prog Nucleic Acid Res Mol Biol. 1998;60:29–46. doi: 10.1016/s0079-6603(08)60888-0. [DOI] [PubMed] [Google Scholar]
- 10.Carballido-López R. The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz-Espín D, et al. The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc Natl Acad Sci USA. 2009;106:13347–13352. doi: 10.1073/pnas.0906465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamtekar S, et al. The ϕ29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaballos A, Salas M. Functional domains in the bacteriophage ϕ29 terminal protein for interaction with the ϕ29 DNA polymerase and with DNA. Nucleic Acids Res. 1989;17:10353–10366. doi: 10.1093/nar/17.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serna-Rico A, Illana B, Salas M, Meijer WJJ. The putative coiled coil domain of the ϕ29 terminal protein is a major determinant involved in recognition of the origin of replication. J Biol Chem. 2000;275:40529–40538. doi: 10.1074/jbc.M007855200. [DOI] [PubMed] [Google Scholar]
- 15.González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 16.González-Huici V, Alcorlo M, Salas M, Hermoso JM. Binding of phage ϕ29 architectural protein p6 to the viral genome: Evidence for topological restriction of the phage linear DNA. Nucleic Acids Res. 2004;32:3493–3502. doi: 10.1093/nar/gkh668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci USA. 2008;105:14136–14140. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse T, Møller-Jensen J, Løbner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse T, et al. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 2006;20:113–124. doi: 10.1101/gad.366606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Soufo HJ, Graumann PL. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr Biol. 2003;13:1916–1920. doi: 10.1016/j.cub.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Defeu Soufo HJ, Graumann PL. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 2005;6:10. doi: 10.1186/1471-2121-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Formstone A, Errington J. A magnesium-dependent mreB null mutant: Implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 24.Schaack J, Ho WY, Freimuth P, Shenk T. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 1990;4:1197–1208. doi: 10.1101/gad.4.7.1197. [DOI] [PubMed] [Google Scholar]
- 25.Fredman JN, Engler JA. Adenovirus precursor to terminal protein interacts with the nuclear matrix in vivo and in vitro. J Virol. 1993;67:3384–3395. doi: 10.1128/jvi.67.6.3384-3395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong RN, Meijer LA, van der Vliet PC. DNA binding properties of the adenovirus DNA replication priming protein pTP. Nucleic Acids Res. 2003;31:3274–3286. doi: 10.1093/nar/gkg405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Arnaiz P, et al. Involvement of phage ϕ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 2007;35:7061–7073. doi: 10.1093/nar/gkm749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castilla-Llorente V, Muñoz-Espín D, Villar L, Salas M, Meijer WJJ. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J. 2006;25:3890–3899. doi: 10.1038/sj.emboj.7601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita S, et al. Dissection of the expression signals of the spoA gene of Bacillus subtilis: Glucose represses sporulation-specific expression. J Gen Microbiol. 1989;135:1335–1345. doi: 10.1099/00221287-135-5-1335. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Espín D, et al. Phage ϕ29 DNA replication organizer membrane protein p16.7 contains a coiled coil and a dimeric, homeodomain-related, functional domain. J Biol Chem. 2004;279:50437–50445. doi: 10.1074/jbc.M403297200. [DOI] [PubMed] [Google Scholar]
- 31.Asensio JL, et al. Structure of the functional domain of ϕ29 replication organizer: Insights into oligomerization and DNA binding. J Biol Chem. 2005;280:20730–20739. doi: 10.1074/jbc.M501687200. [DOI] [PubMed] [Google Scholar]
- 32.Lewis PJ, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 33.González-Huici V, Salas M, Hermoso JM. The push-pull mechanism of bacteriophage ϕ29 DNA injection. Mol Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.