Abstract
Photoadaptation, the ability to attenuate a light response on prolonged light exposure while remaining sensitive to escalating changes in light intensity, is essential for organisms to decipher time information appropriately, yet the underlying molecular mechanisms are poorly understood. In Neurospora crassa, VIVID (VVD), a small LOV domain containing blue-light photoreceptor protein, affects photoadaptation for most if not all light-responsive genes. We report that there is a physical interaction between VVD and the white collar complex (WCC), the primary blue-light photoreceptor and the transcription factor complex that initiates light-regulated transcriptional responses in Neurospora. Using two previously characterized VVD mutants, we show that the level of interaction is correlated with the level of WCC repression in constant light and that even light-insensitive VVD is sufficient partly to regulate photoadaptation in vivo. We provide evidence that a functional GFP-VVD fusion protein accumulates in the nucleus on light induction but that nuclear localization of VVD does not require light. Constitutively expressed VVD alone is sufficient to change the dynamics of photoadaptation. Thus, our results demonstrate a direct molecular connection between two of the most essential light signaling components in Neurospora, VVD and WCC, illuminating a previously uncharacterized process for light-sensitive eukaryotic cells.
Keywords: light, white collar-1, white collar-2, photoreceptor, LOV domain
Light is a strong environmental cue for many organisms across different kingdoms. Organisms must not only be aware of the presence or absence of light but be able to detect subtle changes in the intensity of light to elicit proper behavioral and developmental responses. Thus, cells have evolved mechanisms to adapt to prolonged light exposure while retaining sensitivity to escalating changes in light intensity at the same time. Light responses in the filamentous fungus Neurospora crassa have been studied for several decades because of their robustness and easily scored phenotypes (1–6). The WHITE COLLAR (WC)-1 and WC-2 proteins control most light responses in Neurospora (7, 8). WC-1, mainly localized in the nucleus (9–12), is both a FAD-binding photoreceptor and a GATA zinc finger transcription factor (13, 14). Through their PER-ARNT-SIM (PAS) domains (15–17), WC-1 and WC-2, another GATA zinc finger transcription factor, form the white collar complex (WCC). After exposure to light, the WCC binds to the promoter of early light-responsive targets at defined consensus sequences and activates transcription within 15 min or less (14, 18). After prolonged light exposure in Neurospora, induction of both early (average peak in steady-state RNA ∼30 min after exposure to light) and late (average peak in steady-state RNA ∼60 min after exposure to light) light responses exhibit an unambiguous feature of photoadaptation, returning to their basal level of expression after 1–4 h in constant light (7). To remain sensitive to an additional increase in ambient light intensity, Neurospora is able to adapt to various levels of light intensity (9, 19–21).
VIVID (VVD) is an additional blue-light photoreceptor that functions downstream of the WCC to regulate negatively the light responses initiated by the WCC (7, 9, 19,21–22). Loss-of-function vvd mutants were originally identified by the unique appearance of the bright orange conidia while grown in constant light (23), presumably attributable to the persistent activation of carotenoid biosynthesis (7). In addition to its function in attenuating the direct light response in Neurospora, VVD has been shown to take part in regulating various circadian clock properties, most likely through its effects on the WCC, including gating of light input to the clock (21), maintenance of the clock during the light phase (24, 25), and temperature compensation of the circadian phase (26). Despite accumulating genetic and molecular evidence supporting the connection between VVD and WCC, no mechanistic explanation for the relationship has been proposed.
VVD is a small 21-kDa FAD-binding protein consisting of a LOV domain and an N-terminal cap (21, 27), capable of undergoing a photocycle in vitro (9, 27). Light induction of vvd gene expression is robust and fast, with the peak in RNA expression around 15 min and that in protein around 30 min after exposure to light, and it is WCC-dependent (7, 9, 21). Once activated by blue light, the formation of a covalent cysteinyl-flavin adduct within the LOV domain induces an N-terminal conformational change thought to be important for repressor function: A point mutation in the hinge region renders the protein unable to change conformation and causes excessive accumulation of carotenoid in constant light (27). In light, VVD forms a rapidly exchanging dimer in solution, suggesting the possibility that the LOV domain of VVD could interact with other PAS domains, including those found in the WCC proteins (28–30). Cell fractionation studies have shown VVD to be exclusively localized in the cytosol (9), whereas most of the WCC is nuclear (9–12), raising the question of how VVD communicates with the WCC to affect various WCC-mediated activities.
Here, using the protein cross-linker 3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester) (DSP), we show that the WCC coimmunoprecipitates with VVD under both low- and high-light conditions and demonstrate that the level of interaction between VVD and WCC is correlated with the level of WCC repression in constant light. Unexpectedly, two previously characterized vvd mutant alleles, vvdC71S and vvdC108A, are not null for photoadaptation but are partially functional in repressing light responses and, moreover, are sufficient to regulate reinitiation of the light response in constant light with an increase in light intensity. In contrast to previous biochemical evidence, we show by live-cell microscopy that a functional GFP-VVD protein appears in the cytosol on light induction but additionally accumulates in the nucleus, where most of the WCC resides. However, nuclear localization of VVD is independent of the presence of light, as demonstrated by a constitutively expressed GFP-VVD. Manipulating the dynamics of VVD expression offers considerable support for the hypothesis that the function of VVD is WCC-dependent and that VVD protein expression alone is sufficient to repress light responses in vivo.
Results
Physical Interaction Between VVD-V5 and WCC Revealed by Coimmunoprecipitation After DSP Treatment.
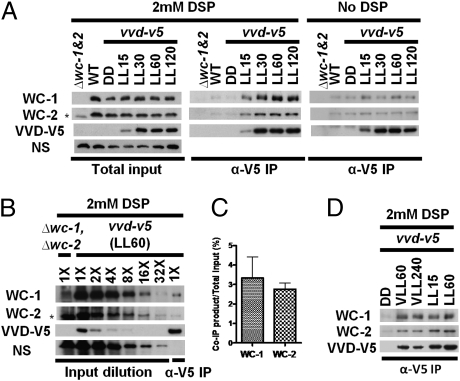
We hypothesized that a central mechanism of photoadaptation could involve association of VVD with other proteins, and therefore made strains with a tagged VVD to facilitate biochemical approaches. In Fig. S1, our data demonstrate that the ectopically expressed VVD appears fully functional and the C-terminal V5 tag is not interfering with biological functions of VVD in vivo. Several attempts using large-scale preparations of purified VVD-V5 to identify the potential interacting partners of VVD were not successful. Therefore, we predicted that if VVD does interact with another protein to repress WCC activity, the interaction might be either weak or transient. Both VVD and WC-1 contain LOV domains, a variant PAS domain with flavin-binding capabilities. WC-1 and WC-2 are PAS domain-containing transcription factors, and formation of the WCC heterodimer is mediated mainly through their shared PAS domains (15–17). Given the similarity between LOV and PAS domains, we hypothesized that VVD might be able to interact directly with the WCC to repress its transcriptional activity and that application of protein cross-linkers might be able to reveal their relationship. We performed a coimmunoprecipitation (co-IP) assay, with DSP treatment before cell lysis. DSP, a reversible protein cross-linker, is widely used to “capture” weak or transient protein-protein interactions (31–34). After the DSP treatment, anti-V5 antibody-coated agarose was used to isolate VVD-V5 and the coprecipitates were hybridized with either anti–WC-1 or anti–WC-2 antibodies to look for interaction. As shown in Fig. 1A, when we performed the co-IP assay following a dark-to-light transfer, either with or without DSP treatment, the level of VVD-V5 in the dark was too low to be detected even in the input lane. Following exposure to light, VVD expression reached an intermediate level 15 min after exposure to white light (LL15) and peaked at LL30. With DSP treatment, the amount of coimmunoprecipitated WC-1 and WC-2 appears to be proportional to the amount of VVD-V5 isolated in the solution, thereby substantiating the specificity of the interaction. Notably, weak bands of WC-1 and WC-2, clearly absent in the wc-1, wc-2 double-knockout strain, can be seen on the gel from a WT (no tag) strain in darkness (DD), indicating that there is a low level of nonspecific binding between WCC and the agarose used in the co-IP assay. Co-IP assays in the absence of cross-linking with DSP treatment show a very weak but real interaction that is above background (Fig. 1A). Next, we determined what fraction of total WCC is bound to VVD-V5. As seen in Fig. 1A, a co-IP assay with anti-V5 antibody-coated agarose isolated most of the VVD-V5 from the total input. In contrast, ≈3% of WC-1 and WC-2 in the lysate coimmunoprecipitated with VVD-V5, as determined by comparing a two-factor serial dilution of total input (Fig. 1 B and C) to the amount in the 1× co-IP lane. To confirm the above findings in the photoadaptation process, we performed co-IP assays following a very low light (VLL, 3 μmol·m2·s)-to-LL (20 μmol·m2·s) transfer. Regardless of the changes in light intensity, VVD-V5 retained the interaction with the WCC, indicating a steady interaction between VVD and WCC in the process of photoadaptation (Fig. 1D).
Fig. 1.
Physical interaction between VVD-V5 and WCC revealed by co-IP after DSP treatment. (A) Anti-V5 co-IP assay with or without DSP treatment following a time course up to 2 h. (B) Relative amount of WC-1 and WC-2 interacting with VVD as determined by the anti-V5 co-IP assay. (C) Densitometric measurements of WC-1 and WC-2 coimmunoprecipitated with the VVD-V5 (n = 5). The data were quantified by comparison with the diluted inputs with the closest image intensity, as shown in B. (D) Anti-V5 co-IP assay with DSP treatment following a very low light (VLL, 3 μmol·m2·s) to LL (20 μmol·m2·s) transfer. NS, nonspecific band on the WC-1 blot as a loading control. Asterisks in A and B highlight a nonspecific band close in size to the WC-2 signal. In all figures, LL indicates a constant white light stimulus with a photon flux of 20 μmol·m2·s.
Level of Interaction Between VVD and WCC Is Correlated with the Level of WCC Repression in Constant Light.
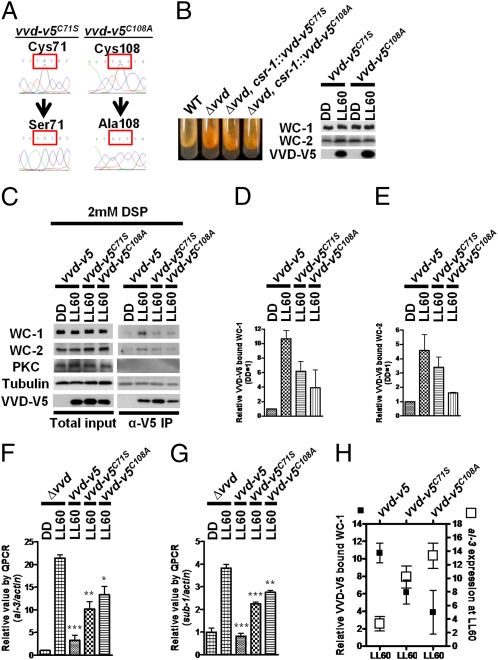
To determine if the physical interaction between VVD and the WCC is required for functional photoadaptation, we asked whether two established loss-of-function vvd mutants, vvd-v5C71S and vvd-v5C108A, still retained their capability to interact with the WCC. Previous studies have shown that Cys71 is essential in controlling movements of the N-terminal cap after light activation (27) and that Cys108 within the LOV domain forms a cysteinyl-flavin adduct in response to blue light and is required for VVD to undergo a complete photocycle in vitro (9). C71S and C108A amino acid changes in VVD caused a vvd-null phenotype as assayed by coloration in constant light (9, 27). After confirming the mutated sequences (Fig. 2A) and transforming the mutated DNA into the Δvvd strain, we found that neither allele could complement the photoadaptation defect of Δvvd as judged from carotenoid levels, despite the robust light induction of their protein levels (Fig. 2B). Consistent with the loss-of-function phenotype, the co-IP data indicate that both mutant proteins display a considerable decrease in their capability to interact with the WCC relative to the WT VVD (Fig. 2 C–E), suggesting that their interaction is important for function and that maximal interaction depends on a fully functional VVD. In addition, the specificity of the interaction is further supported by Western blot analysis with either anti-PKC or antitubulin antibodies (Fig. 2C). In the presence of DSP, PKC is not pulled down in the VVD-V5 co-IP assays in light or dark. Tubulin displays a consistent band, indicating that it binds to the agarose beads independent of VVD-V5 and light conditions.
Fig. 2.
The level of interaction between VVD and the WCC is correlated with the level of WCC repression in constant light. (A) Sequence confirmation of the vvd-v5C71S and vvd-v5C108A alleles. (B) Carotenoid accumulation as a measure of photoadaptation defects in the vvd-v5C71S and vvd-v5C108A strains. Photoadaptation limits the expression of genes whose products are involved in the carotenogenesis pathway. (Right) Light induction of the VVD-V5 protein in either the vvd-v5C71S or the vvd-v5C108A strain. (C) Anti-V5 co-IP assay with the vvd-5, vvd-v5C71S, and vvd-v5C108A strains. PKC and tubulin are controls as described in the text. Densitometric measurements of WC-1 (D) and WC-2 (E) coimmunoprecipitated with VVD-V5 (n = 3). The data were normalized to the background signals in DD. Photoadaptation defects were determined by measuring light induction of al-3 (F) and sub-1 (G) at LL60 using RT quantitative PCR analysis (n = 3, mean values ± SE). Asterisks indicate statistical significance when compared with the Δvvd strain at LL60 as determined by an unpaired t test. ***P < 0.001; **P < 0.01; *P < 0.05. (H) Relative amount of WC-1 coimmunoprecipitated with VVD-V5 (filled squares) vs. al-3 expression at LL60 (open squares) in respective strains (n = 3). LL indicates constant white light stimulus with a photon flux of 20 μmol·m2·s.
Notably, both vvd-v5C71S and vvd- v5C108A mutant proteins still retain a reduced but significant level of WCC interaction (Fig. 2 D and E). If the physical interaction between VVD and WCC is indeed essential for regulating photoadaptation, these two mutant alleles might not be complete loss-of-function alleles, as suggested by their inability to complement repression of carotenogenesis. Therefore, we examined their photoadaptation defects at the molecular level to determine how the level of interaction correlated with the ability to repress WCC-dependent light responses in vivo. The expression of the early light response genes al-3 and sub-1 was followed by quantitative PCR at time 0 and LL60 in both mutants (Fig. 2 F and G). Consistent with our hypothesis that VVD-WCC interaction is necessary for photoadaptation, we found that both vvd-v5C71S and vvd- v5C108A mutant proteins were partially functional in their ability to repress light responses (Fig. 2 F and G) and carotenoid induction (Fig. S2 A and B). We conclude that both transgenic strains displayed mutant photoadaptation color phenotypes resulting from excessive accumulation of carotenoid in constant light over time (Fig. 2B). Intriguingly, the level of interaction between VVD-V5 and WCC is inversely correlated with the functional output of the WCC at LL60 (Fig. 2H), further supporting the hypothesis that the physical interaction between VVD and WCC is specific and determines the level of WCC repression.
Light-Insensitive VVD Is Sufficient to Regulate Photoadaptation in Vivo.
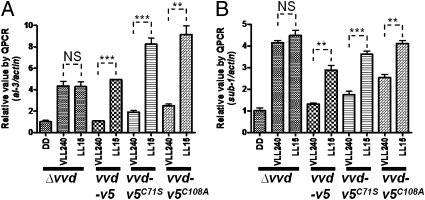
An important feature of photoadaptation is the sensitivity to an increase in light intensity. Although mutant proteins encoded by both vvd-v5C71S and vvd-v5C108A were functional at LL60 to repress the primary light response at least partially, we expected that the ability to respond to increased light might require the VVD protein to act as a photoreceptor and directly sense changes of light intensity independent of the WC-1 photoreceptor. Therefore, we asked whether the vvd mutants were capable of responding to a light increase. Surprisingly, both vvd mutant proteins were still functional, although less so than WT, in regulating photoadaptation in vivo, as indicated by following the reactivation of al-3 and sub-1 expression (Fig. 3) after a low light-to-LL transfer. This suggests that photoadaptation is regulated by VVD protein acting as a negative feedback element on the WCC transcriptional activity, with the positive element (WCC) but not VVD sensitive to a further increase in the amount of light. In the absence of vvd (Fig. 3), there is no reactivation of light-induced gene expression, supporting the absolute requirement for Neurospora VVD in initiating gene expression in response to a change in light intensity.
Fig. 3.
Light-insensitive VVD is sufficient to regulate photoadaptation in vivo. Photoadaptation defects were determined by measuring induction of al-3 (A) and sub-1 (B) following a very low light (VLL, 3 μmol·m2·s) to LL (20 μmol·m2·s) transfer using RT quantitative PCR (QPCR) analysis (n = 3, mean values ± SE). Asterisks indicate statistical significance when compared with the level of expression at VLL240 as determined by an unpaired t test. ***P < 0.001; **P < 0.01. NS, difference between the two groups is not significant.
In the absence of vvd, WC-1 becomes hyperphosphorylated in constant light (19, 21), which correlates with the up-regulated light responses. Although the WCC is inactivated by phosphorylation in response to light (18), the levels of WC-1 phosphorylation may serve as an indicator for the functional output of the WCC. For instance, no noticeable hyperphosphorylated WC-1 was observed at LL60 in either the vvd-v5 or WT strain (Fig. S3), reflecting repressed WCC activity in constant light. On the other hand, consistent with the observation that both vvd-v5C71S and vvd-v5C108A are partially functional (Fig. 2 F and G), WC-1 phosphorylation at LL60 in these two strains appeared intermediate between the Δvvd and WT strains (Fig. S3), supporting their capability to affect WCC activity and repress light response.
GFP-VVD Is Functional in Vivo When Driven by Either the vvd or sod-1 Promoter.
Thus far, the data support a functional and physical interaction between VVD and WCC that is important for photoadaptation, so we next determined when and where this interaction may occur by localization of VVD in living cells. In previous experiments using cell fractionation approaches to separate nuclei from cytoplasm, VVD was seen exclusively in the cytosolic fractions (9), whereas most of the WCC remained in the nucleus (9–12). However, given that the size of VVD (21 kDa) is well below the cutoff for possible passive diffusion through the nuclear pore, which appears to be 60 kDa or above (35–37), we hypothesized that loss of VVD from the nucleus might be a technical artifact attributable to the fact that small protein molecules might simply leak out of the nucleus during the purification process.
To test whether VVD can enter the nucleus and repress WCC activity, we tagged VVD-V5 with a GFP derivative (GFP-S65T, SGPF) (38) at its N terminus and integrated the transgene (Fig. S4A, gfp-vvd-v5), together with 3.5 kbp of its own promoter, into the csr-1 locus in a vvd knockout strain. Additionally, we sought to dissociate the regulation of VVD localization from its light activation. Recent studies on the phototropin photoreceptors in plants have implicated LOV domains in regulating subcellular localization following light activation (39–41). The vvd promoter used to drive the gfp-vvd-v5 allele was replaced with the sod-1 promoter, which constitutively drives gene expression independent of the light conditions (7) (Fig. S4B, gfp-vvd-v5cx). Western blot analysis with anti-V5 and anti-GFP antibodies confirmed the identity and expression kinetics of GFP-conjugated VVD-V5 proteins when driven by either vvd or sod-1 promoters (Fig. S4C). Phenotype analysis indicates that the ectopically expressed GFP-VVD-V5 driven by the vvd promoter is sufficient to restore the photoadaptation defects of Δvvd in vivo, as assayed by coloration (Fig. S4D). In contrast, the transgenic strain with GFP-VVD-V5 driven by the sod-1 promoter displayed a partially complemented phenotype (Fig. S4D), most likely reflecting the relatively lower level of GFP-VVD expression in constant light compared with the endogenous vvd promoter allele (compare LL60 samples in Fig. S4C). Moreover, GFP-VVD is not only phenotypically functional but retains interaction with WC-1 and WC-2 in constant light (Fig. S4E). Consistent with the complemented phenotype on solid medium slants, the photoadaptation defects of Δvvd were complemented by both GFP-VVD alleles at the molecular level as well (Fig. S4 F and G). At the level of our assays, the increased size of VVD (21 kDa + 27 kDa of GFP) to 48 kDa does not appear to affect VVD's interaction with the WCC or its biological functions in vivo, and it behaves like its WT counterpart in the process of photoadaptation.
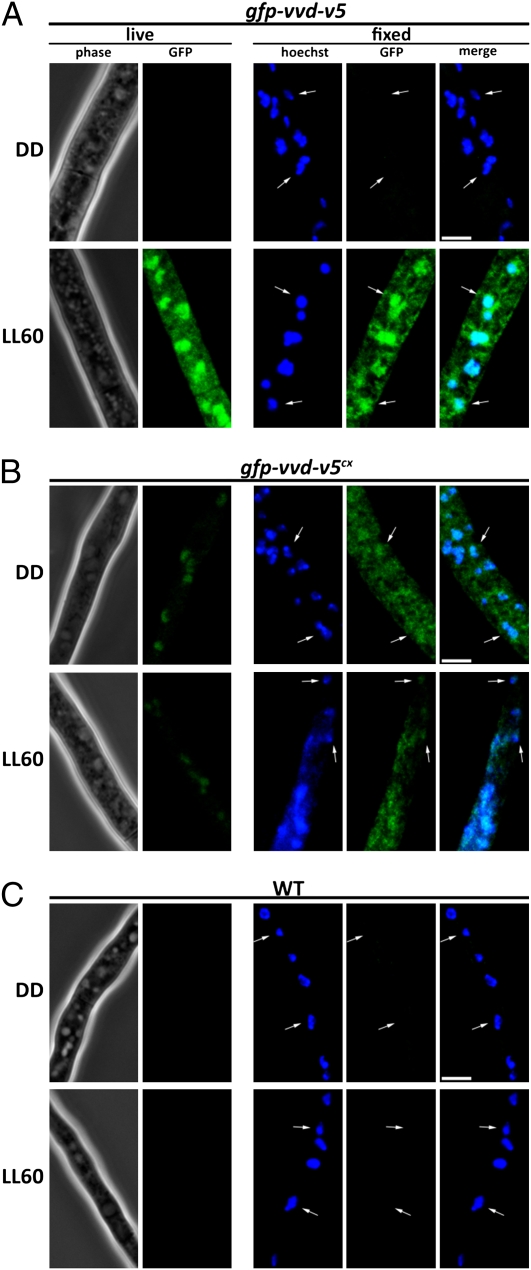
GFP-VVD Accumulates in the Nucleus on Light Induction in Live Cells and Nuclear Localization of GFP-VVD Does Not Require Light Activation.
We followed the dynamics of GFP-VVD-V5 on light induction in vegetative hyphae by live-cell imaging. Consistent with the Western blot analysis (Fig. S4C), light treatment robustly induced GFP signals in the gfp-vvd-v5 strain at LL60 (Fig. 4A). Next, we asked if the expression of GFP-VVD-V5 colocalized with the nuclear Hoechst stain after paraformaldehyde fixation. In agreement with our hypothesis but somewhat surprisingly, given previous work, GFP-VVD-V5 was clearly concentrated in the nucleus on light induction (Fig. 4A and Fig. S5A), although GFP-VVD is also seen in the cytoplasm. Consistent with the Western blot analysis (Fig. S4C), GFP-VVD under sod-1 regulation is seen in both dark- and light-grown hyphae and appears to be expressed at a lower level than under its own promoter in LL. Unlike phototropin proteins in Arabidopsis, nuclear localization of the constitutively expressed GFP-VVD-V5 protein showed no apparent difference in localization before and after light exposure (Fig. 4B and Fig. S5B), thereby suggesting that light-induced conformational changes in VVD protein are not necessary for the nuclear enrichment of VVD. A WT strain without a GFP tag was examined in parallel to control for nonspecific autofluorescent signals in Neurospora (Fig. 4C). Highly expressed GFP alone has been previously shown to be cytoplasmic in Neurospora (42).
Fig. 4.
GFP-VVD accumulates in the nucleus on light induction in live cells, and the nuclear localization of GFP-VVD does not require light activation. (A) Localization of GFP-VVD-V5 before and after light treatment. Mycelia samples for live-cell imaging were collected before (DD) and after light treatment for 60 min. For fixed-cell images, the mycelia were fixed in paraformaldehyde for 7 min and then incubated with Hoechst dye for another 30 min before imaging. (B) Localization of GFP-VVD-V5cx (constitutively expressed) before and after light treatment. (C) WT strain under the same light conditions. LL indicates constant white light (photon flux of 20 μmol·m2·s). White arrows highlight representative nuclei. (Scale bar: 5 μm.) All images were acquired using identical light exposure settings, deconvolved, and displayed using identical linear contrast enhancement.
Constitutively Expressed VVD Is Sufficient to Change the Dynamics of Photoadaptation.
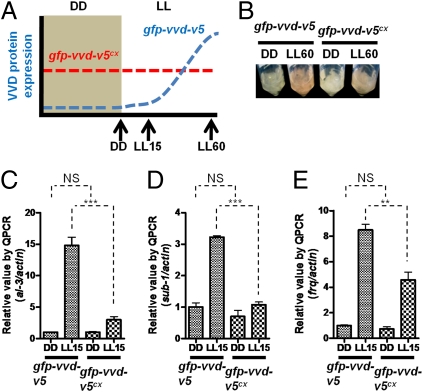
More than 300 genes are light-responsive in Neurospora (7). To test whether the gene repression aspect of photoadaptation is dependent on light-induced genes in addition to vvd, we asked whether constitutively expressed VVD (driven by the sod-1 promoter as shown in Fig. S4C) is sufficient to attenuate WCC-dependent light responses at an early time point when light-responsive genes would not yet be fully translated (Fig. 5A). Compared with VVD driven by its own promoter (gfp-vvd-v5), we found that constitutively expressed VVD (gfp-vvd-v5cx) reduced the level of carotenoid production in hyphal mats in liquid culture (Fig. 5B and Fig. S2B), suggesting that VVD expressed in the dark is repressing WCC-driven carotenogenesis. Dark-expressed VVD effectively repressed acute light responses at LL15 of three early light-responsive genes, al-3, sub-1, and frq (Fig. 5 C–E), suggesting that expression of VVD alone is sufficient to change the dynamics of the photoadaptation response, independent of other light-responsive gene products. Notably, although light-insensitive VVD remained functional in repressing WCC target genes (Fig. 3) in constant light, the constitutively expressed VVD failed to affect the basal expression of al-3, sub-1, and frq (Fig. 5 C–E) in the dark, indicating that the repressor function of VVD is restricted and specific to responses mediated by light-activated WCC.
Fig. 5.
Constitutively expressed VVD is sufficient to change the dynamics of photoadaptation. (A) Schematic depicting the light induction kinetics of GFP-VVD-V5 in either the gfp-vvd-v5 or gfp-vvd-v5cx strain (constitutively expressed). (B) Photoadaptation phenotype of the gfp-vvd-v5 and gfp-vvd-v5cx strains at LL60. Photoadaptation kinetics were determined by measuring light induction of al-3 (C), sub-1 (D), and frq (E) at LL15 using RT quantitative PCR (QPCR) analysis (n = 3, mean values ± SE). Asterisks indicate statistical significance as determined by an unpaired t test. ***P < 0.001; **P < 0.01. NS, difference between two groups is not significant. LL indicates constant white light stimulus with a photon flux of 20 μmol·m2·s.
Discussion
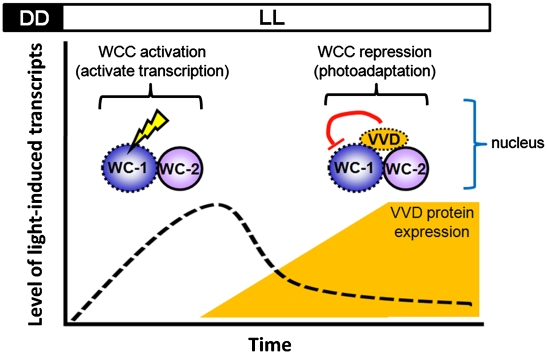
In the filamentous fungus Neurospora, photoadaptation can be described as a series of molecular events that includes the initial attenuation or repression of the wave of gene expression in response to a light stimulus and, if the light remains, a reinitiation of gene expression following an increase in the light intensity. Considerable evidence suggests that VVD regulates the transcriptional activity of the WCC in mediating photoadaptation in constant light as well as several other properties associated with the circadian clock under free-running conditions (7, 21, 24, 26). Despite the genetic relationship between VVD and the WCC, biochemical studies examining their physical interaction and subcellular localization have not shown direct interactions or colocalization. Therefore, the communication between VVD and WCC has been largely viewed as indirect. Through co-IP analysis after protein cross-linking and live-cell imaging of GFP-VVD, we find a specific physical interaction between VVD and WCC and, additionally, that both are present in the nucleus on light induction. Thus, our findings offer a mechanistic explanation for photoadaptation in Neurospora and possibly other light-sensitive eukaryotic cells (Fig. 6).
Fig. 6.
Photoadaptation in Neurospora. After light activation, the WCC transiently binds to the promoter of light-responsive genes to activate transcription, including vvd. The induced VVD protein accumulates in the nucleus and physically interacts with WCC to regulate photoadaptation by repressing WCC activity in constant light. The kinetics of photoadaptation are predominantly regulated by the amount of VVD protein in the system. Components capable of sensing light directly through a chromophore are marked with dashed lines.
In addition to several sequence and functional homologs in other fungi, the LOV/PAS domain found in VVD and WC-1 is widespread and functionally conserved in different kingdoms (27, –48). For instance, LOV domains from phototropins and FKF1 in Arabidopsis are still able to sense blue light when incorporated in Neurospora photoreceptor proteins (49). Interestingly, phototropins (PHOT1 and PHOT2) in Arabidopsis are also involved in sensing and responding to changes of light intensity (50), but their subcellular localization on light activation is complex. A small fraction of PHOT1 is released to the cytoplasm from the plasma membrane (40), whereas PHOT2 becomes associated with the Golgi apparatus (39, 41). How these light-induced changes in localization affect receptor signaling is not completely understood (51). Phototropins may therefore behave in a manner similar to VVD and WCC, whereby communication is mediated by a weak and possibly transient physical interaction after light activation.
The role of other components that VVD might use to repress WCC transcriptional activity remains to be explored. Notably, a few genes other than vvd have been implicated in regulating photoadaptation in Neurospora. For instance, previous work has demonstrated that changes in PKC activity result in altered light responses in constant light (52, 53). However, we were unable to establish a physical interaction between PKC and VVD or WCC (Fig. S6). Additionally, although there is an inverse correlation between WC-1 phosphorylation and VVD activity in constant light, our screenings with a large collection of kinase and phosphatase knockout strains (Table S1) failed to identify any vivid-like strains at the phenotypic level. However, using a genetic screen, Navarro-Sampedro et al. (54) reported several interesting mutants specifically deficient in regulating photoadaptation of con-10 and con-6 (i.e., not all light-responsive genes are affected). Further characterization of these mutants might reveal previously undescribed components involved in mediating photoadaptation in a gene-specific manner. However, given that most if not all light-responsive genes are affected in the Δvvd strain and that, to date, there are no reports of any mutants that display photoadaptation defects close to those of the Δvvd, a more comprehensive genetic or interactome screening may be required to identify any previously undescribed participating components. Meanwhile, our live-cell imaging data demonstrating VVD's presence in the nucleus are in agreement with the nuclear localization of the WCC and the interaction results. A study examining GFP-conjugated WC-2 revealed an unexpected nucleocytoplasmic shuttling that occurs over a very rapid time frame (within minutes) (10). Because the study examined samples in constant conditions, it is unclear how light might affect the shuttling of WCC and whether the shuttling plays any roles in regulating light responses. Taken together, our data reveal a direct molecular connection between two of the most essential light signaling components in Neurospora and provide an important perspective on photoadaptation regulated by proteins containing the widespread LOV/PAS domains.
Materials and Methods
Strains.
The WT strain used here is OR74A. All single-knockout strains came from the Neurospora knockout project (55) and have been deposited in the Fungal Genetics Stock Center (www.fgsc.net). The identity of csr-1 knock-in strains used in this study was examined by PCR analysis (30 cycles) for the absence of the csr-1 ORF in the genome; all appear to be homokaryons, as shown in Fig. S7. NCU (Neurospora crassa unfinished) strain numbers are from Neurospora annotation (www.broad.mit.edu/annotation/genome/neurospora/home.html).
Culture Conditions and Light Treatment.
Frozen conidia were inoculated onto a minimal Vogel slant (56) 1 wk in advance of experiments to generate fresh conidia. On day 0, to form a mycelial layer, 107 conidia suspended in sterile water were inoculated into a 10-cm Petri dish with 20 mL of Bird medium (57) containing 2%(wt/vol) glucose. After 24 h of incubation in DD at 25 °C, a mycelial plug was cut with a no. 4 cork borer (8-mm diameter) and transferred into a 125-mL flask with 50 mL of Bird medium containing 2%(wt/vol) glucose. All procedures were performed in a low red-light environment to avoid any possible light-stimulating effects. After another 24 h of culturing with constant shaking (125 rpm) in DD at 25 °C, the flasks were moved to a shaker at 25 °C with continuous LL covering a wide range of the spectrum from 400 to 700 nm (cool white fluorescent light bulb, F20T12-CW; General Electric; 20 μmol·m2·s) and then harvested before (DD) and after LL treatment. Using vacuum filtration, mycelia were immediately frozen in liquid nitrogen and stored at −80 °C until protein or RNA extraction.
Co-IP Assay with DSP Treatment.
Before harvesting, mycelial plugs were transferred to PBS containing 2 mM freshly prepared DSP (D3669; Sigma). The samples were shaken for 30 min and then quenched with Tris solution (pH 7.5, 20 mM). After shaking for another 15 min, the mycelia were harvested, immediately frozen in liquid nitrogen, and stored at −80 °C until protein extraction. To perform the co-IP assay, 1 mg of total protein was incubated with 30 μL of anti-V5 antibody-coated agarose (A7345; Sigma) overnight at 4 °C. The agarose was washed with protein extraction buffer at least six times before elution. Co-IP samples were eluted and de-cross-linked with 2× Western blot sample buffer (65 °C for 15 min) before loading on the gel.
Supplementary Material
Acknowledgments
We thank laboratory members for extensive discussion, Dhana R. Nair (Dartmouth College) and Pei Zhou (Dartmouth College) for preliminary consultations on imaging, and Randy Lambreghts (Dartmouth Medical School) and Brian Crane (Cornell University) for critical reading of the manuscript. We are deeply grateful to the Fungal Genetics Stock Center at the University of Missouri for supporting our work with Neurospora. This work was supported by grants from the National Institutes of Health to J.J.L. (Grant R01 GM08336) and J.C.D (Grants GM34985 and P01GM68087) and by the Core Grant to the Norris Cotton Cancer Center at Dartmouth Medical School.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011190107/-/DCSupplemental.
References
- 1.Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: Molecular genetics of photoreception. Mol Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- 2.Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 3.Corrochano LM. Fungal photoreceptors: Sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- 4.Chen CH, Loros JJ. Neurospora sees the light: Light signaling components in a model system. Commun Integr Biol. 2009;2:448–451. doi: 10.4161/cib.2.5.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahn YS, et al. Sensing the environment: Lessons from fungi. Nat Rev Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Bell-Pedersen D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linden H, Rodriguez-Franco M, Macino G. Mutants of Neurospora crassa defective in regulation of blue light perception. Mol Gen Genet. 1997;254:111–118. doi: 10.1007/s004380050398. [DOI] [PubMed] [Google Scholar]
- 9.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schafmeier T, et al. Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes Dev. 2008;22:3397–3402. doi: 10.1101/gad.507408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong CI, Ruoff P, Loros JJ, Dunlap JC. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Q, et al. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 14.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 15.Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem. 2003;278:3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- 16.Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballario P, Talora C, Galli D, Linden H, Macino G. Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins. Mol Microbiol. 1998;29:719–729. doi: 10.1046/j.1365-2958.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- 18.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwerdtfeger C, Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol. 2001;39:1080–1087. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 20.Shrode LB, Lewis ZA, White LD, Bell-Pedersen D, Ebbole DJ. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet Biol. 2001;32:169–181. doi: 10.1006/fgbi.2001.1264. [DOI] [PubMed] [Google Scholar]
- 21.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 22.Ballario P, Macino G. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 1997;5:458–462. doi: 10.1016/S0966-842X(97)01144-X. [DOI] [PubMed] [Google Scholar]
- 23.Hall MD, Bennett SN, Krissinger WA. Characterization of a newly isolated pigmentation mutant of Neurospora crassa. Ga J Sci. 1993;51:27. (Abstr) [Google Scholar]
- 24.Elvin M, Loros JJ, Dunlap JC, Heintzen C. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 2005;19:2593–2605. doi: 10.1101/gad.349305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider K, et al. Rhythmic conidiation in constant light in vivid mutants of Neurospora crassa. Genetics. 2009;181:917–931. doi: 10.1534/genetics.108.097808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt SM, Elvin M, Crosthwaite SK, Heintzen C. The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev. 2007;21:1964–1974. doi: 10.1101/gad.437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoltowski BD, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc. 2008;130:12226–12227. doi: 10.1021/ja804236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partch CL, Gardner KH. Coactivator recruitment: A new role for PAS domains in transcriptional regulation by the bHLH-PAS family. J Cell Physiol. 2010;223:553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joshi S, Burrows R. ATP synthase complex from bovine heart mitochondria. Subunit arrangement as revealed by nearest neighbor analysis and susceptibility to trypsin. J Biol Chem. 1990;265:14518–14525. [PubMed] [Google Scholar]
- 32.Park LS, Friend D, Gillis S, Urdal DL. Characterization of the cell surface receptor for a multi-lineage colony-stimulating factor (CSF-2 alpha) J Biol Chem. 1986;261:205–210. [PubMed] [Google Scholar]
- 33.Baskin LS, Yang CS. Cross-linking studies of the protein topography of rat liver microsomes. Biochim Biophys Acta. 1982;684:263–271. doi: 10.1016/0005-2736(82)90015-3. [DOI] [PubMed] [Google Scholar]
- 34.Brenner MB, Trowbridge IS, Strominger JL. Cross-linking of human T cell receptor proteins: Association between the T cell idiotype beta subunit and the T3 glycoprotein heavy subunit. Cell. 1985;40:183–190. doi: 10.1016/0092-8674(85)90321-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang R, Brattain MG. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581:3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 38.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Kong SG, et al. The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses. Plant J. 2007;51:862–873. doi: 10.1111/j.1365-313X.2007.03187.x. [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong SG, et al. Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 2006;45:994–1005. doi: 10.1111/j.1365-313X.2006.02667.x. [DOI] [PubMed] [Google Scholar]
- 42.Folco HD, et al. Histone H1 Is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot Cell. 2003;2:341–350. doi: 10.1128/EC.2.2.341-350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lombardi LM, Brody S. Circadian rhythms in Neurospora crassa: Clock gene homologues in fungi. Fungal Genet Biol. 2005;42:887–892. doi: 10.1016/j.fgb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Briggs WR. The LOV domain: A chromophore module servicing multiple photoreceptors. J Biomed Sci. 2007;14:499–504. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 46.Idnurm A, Crosson S. The photobiology of microbial pathogenesis. PLoS Pathog. 2009;5:e1000470. doi: 10.1371/journal.ppat.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CH, Dunlap JC, Loros JJ. Neurospora illuminates fungal photoreception. Fungal Genet Biol. 2010 doi: 10.1016/j.fgb.2010.07.005. 10.1016/j.fgb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng P, He Q, Yang Y, Wang L, Liu Y. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc Natl Acad Sci USA. 2003;100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demarsy E, Fankhauser C. Higher plants use LOV to perceive blue light. Curr Opin Plant Biol. 2009;12:69–74. doi: 10.1016/j.pbi.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 52.Franchi L, Fulci V, Macino G. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol Microbiol. 2005;56:334–345. doi: 10.1111/j.1365-2958.2005.04545.x. [DOI] [PubMed] [Google Scholar]
- 53.Arpaia G, Cerri F, Baima S, Macino G. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol Gen Genet. 1999;262:314–322. doi: 10.1007/s004380051089. [DOI] [PubMed] [Google Scholar]
- 54.Navarro-Sampedro L, Yanofsky C, Corrochano LM. A genetic selection for Neurospora crassa mutants altered in their light regulation of transcription. Genetics. 2008;178:171–183. doi: 10.1534/genetics.107.079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colot HV, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis RH. Neurospora: Contributions of a Model Organism. New York: Oxford Univ Press; 2000. [Google Scholar]
- 57.Metzenberg RL. Bird medium: An alternative to Vogel medium. Fungal Genet Newsl. 2004;51:19–20. Available at: www.fgsc.net/fgn51/fgn51tofc.htm. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.