Abstract
Glioblastoma multiforme (GBM) is the most common and aggressive form of primary brain tumor and there is no curative treatment to date. Resistance to conventional therapies and tumor recurrence pose major challenges to treatment and management of this disease, and therefore new therapeutic strategies need to be developed. Previous studies by other investigators have shown that a subpopulation of GBM cells can grow as neurosphere-like cells when cultured in restrictive media, and exhibit enhanced tumor initiating ability and resistance to therapy. We report here the identification of internalizing human single chain antibodies (scFvs) targeting GBM tumor sphere cells. We selected a large naive phage antibody display library on the glycosylation-dependent CD133 epitope-positive subpopulation of GBM cells grown as tumor spheres and identified internalizing scFvs that target tumor sphere cells broadly, as well as scFvs that target the CD133 positive subpopulation. These scFvs were found to be efficiently internalized by GBM tumor sphere cells. One scFv GC4 inhibited self-renewal of GBM tumor sphere cells in vitro. We have further developed a full-length human IgG1 based on this scFv and found that it potently inhibits proliferation of GBM tumor sphere cells and GBM cells grown in regular non-selective media. Taken together, these results show that internalizing human scFvs targeting brain tumor sphere cells can be readily identified from a phage antibody display library, which could be useful for further development of novel therapies that target subpopulations of GBM cells to combat recurrence and resistance to treatment.
Keywords: brain tumor sphere cells, inhibitory human IgG, internalizing scFvs, glioblastoma multiforme
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive form of primary brain tumor, accounting for 52% of all primary brain tumor cases and 20% of all intracranial tumors. The median survival time for a newly diagnosed patient is approximately one year (1). Treatment can involve chemotherapy, radiotherapy, and surgery, all of which are acknowledged as palliative measures, but are not curative (2–5). Even with complete gross surgical resection of the tumor, combined with the best available treatment, recurrence has been nearly impossible to prevent, and consequently the long-term survival rate for GBM patients is extremely low.
It has been hypothesized that tumors are heterogeneous and only some of the tumor cells with stem-like properties are able to initiate and sustain tumor development (6–8). It has been further postulated that current therapies targeting the bulk tumor are unable to completely eliminate these tumor initiating subpopulations, leading to tumor recurrence and treatment failure (9, 10). Therapies optimized for eliminating these subpopulations would therefore be more effective in preventing recurrence than those targeting the bulk tumor (9, 11). Tumor initiating cells have been identified initially in hematological malignancies and later in many types of solid tumors including brain tumors. A subpopulation of cells that are positive for the glycosylation-dependent conformational CD133 epitopes (AC133 and/or AC141) have been identified in malignant gliomas, and this subpopulation has been shown to have significantly enhanced potency for initiating tumors in immunocompromised mice (12–16). In one study, as few as 100 CD133 epitope-positive human glioma cells were shown to be capable of tumor initiation in immunocompromised mice, whereas 100,000 CD133 epitope-negative cells isolated from the same tumor mass were non-tumorigenic (14).
Glioma tumor initiating cells can be enriched by culturing primary tumor cells in serum free medium supplemented with epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) (15). In this serum-free selective culture system, a subpopulation of GBM cells proliferate and form tumor spheres. Moreover, these GBM tumor sphere cells more closely resemble the phenotype of primary tumors than do serum-cultured cell lines (17), and demonstrate enhanced ability to self-renew and to give rise to differentiated progenies. As a result, these GBM tumor sphere cells provide an attractive target for the development of therapies targeting brain tumor initiating cells.
Due to accessibility to externally administered agents, cell surface antigens are attractive targets for the development of therapeutics that require intracellular delivery of payloads (18, 19). We and others have previously selected internalizing phage antibodies against a panel of tumor cell lines (20–25) and tumor cells in situ (26). Here we have derived GBM tumor spheres from human brain tumor specimens, and targeted the glycosylation-dependent CD133 epitope-positive subpopulation (from here on we simply refer this population as “CD133 positive”) for phage antibody selection by FACS to recover internalizing scFvs. We have identified a panel of internalizing human scFvs that bind to CD133 positive GBM tumor sphere cells, and one of these scFvs is capable of inhibiting the growth of GBM tumor sphere cells in vitro. We have further developed a full-length human IgG1 molecule based on this inhibitory scFv and found that the human IgG1 inhibits the proliferation of both GBM tumor sphere cells and primary GBM cells. Thus, we have demonstrated that internalizing scFvs targeting GBM tumor sphere cells can be readily identified from a phage antibody display library. These fully human antibody fragments can be used in the future to develop targeted therapeutics attacking the subpopulation of tumor initiating cells.
Materials and Methods
Culture of primary glioblastoma cells and tumor sphere cells
Human GBM tissues obtained from the operating room were either processed immediately or transplanted and maintained in nude mice as xenografts as described (27). The protocol for human tissue acquisitions was approved by the Institutional Review Board and in accordance with an assurance filed with and approved by the Department of Health and Human Services. Animal studies were approved by the Institutional Review Board and adhered to the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. The procedure for obtaining tumor sphere cell cultures is the same for fresh GBM tissues or GBM xenografts. Briefly, GBM tissues were minced, and enzymatically dissociated by sequential digestion with collagenase (1 mg/ml, Invitrogen) and trypsin (0.05%, Invitrogen), and passed through a cell strainer (BD Biosciences) to obtain monodispersed cells. These cells were divided into two groups and were either (1) resuspended in DMEM/F12 containing 2% fetal calf serum (FCS) and then plated at a density of 1×106 cells per 75 cm2 flask to obtain primary GBM cell monolayer cultures, or (2) seeded into tumor sphere forming media (TSFM) containing serum-free Complete NeuroCult medium (StemCell Technologies) supplemented with 10 ng/ml bFGF (Millipore/Chemicon), 10 ng/ml EGF (Sigma) and 0.2% heparin/PBS (StemCell Technologies). For serial propagation, primary tumor spheres were re-dissociated into monodispersed cells by trypsin digestion and further cultured in TSFM.
Cloning efficiency assay
The tumor spheres were treated with 0.05% trypsin and passed through a cell strainer to obtain monodispersed cells and plated into a 96-well plate (1,000 cells/well) and cultured in 200 μl TSFM for 14 days (28). Cells were fed with 25 μl of TSFM by changing medium every 2 days. Experiments were done in triplicates. The number of tumor spheres were counted and averaged to determine the cloning efficiency (15, 29).
Flow cytometry analysis of CD133 epitope expression
Primary GBM cells and tumor sphere cells were dissociated by 0.05% trypsin, washed with PBS, and 100 μl aliquots of 105 cells in PBS were placed in 96-well V-bottom plates and incubated on ice for 30 min with phycoerythrin (PE)-conjugated anti-CD133 antibody (Miltenyi Biotec) or an isotype control PE-labeled mouse IgG (BD/Pharmingen). Cells were washed twice with PBS and analyzed by FACS (LSRII, Becton Dickinson).
Selection for internalizing scFvs targeting GBM tumor sphere cells
A multivalent fd phage antibody display library containing the Sheets repertoire (30, 31) was used for selection on GBM tumor sphere cells that express high levels of the CD133 epitope. For the first round of selection, the library was counter-selected on a panel of control cells to remove non-specific binders as described (22, 24, 26), and incubated for 2h at 4°C with 105 monodispersed GBM sphere cells, washed three times with ice-cold PBS, then incubated with pre-warmed (37 °C) DMEM/F12 containing 10% FCS at 37°C for 1h to allow receptor-mediated phage antibodyinternalization (22, 24). Non-internalized phage were removed by washing cells with 50 mM glycine/150 mM NaCl, pH 2.8. Internalized phage were recovered and propagated as described previously (22, 24, 26). For the second round of selection, tumor sphere cells were incubated with polyclonal round one phage under internalizing conditions as described above and further incubated with the PE-conjugated anti-CD133 antibody to identify subpopulations expressing high levels of CD133, which were sorted by FACSAria (Becton Dickinson) along with internalized phage. The process was repeated for the third round of selection. Monoclonal phage binders were identified by FACS-based 96-well plate screening using biotin-labeled anti-fd antibody (Sigma) and streptavidin-R-PE (Invitrogen) as described (22, 24, 26).
Double-labeling of the CD133 epitope and antigens bound by phage antibodies
Phage antibodies were fluorescently labeled using 6-(fluorescein-5-[and-6]-carboxamido)hexanoic acid, succinimidyl ester (5(6)-SFX) (Invitrogen) and purified as described (22, 32, 33). Monodispersed GBM sphere cells were incubated with biotin-labeled anti-CD133 mAb (5 μg/ml, Miltenyi Biotec) and FITC-labeled phage antibodies. Streptavidin-647 (Invitrogen) was used to detect bound biotin-labeled anti-CD133 antibody.
Phage antibody binding to antigens other than CD133
Caco-2 cells that express high levels of CD133 were incubated with phage antibodies along with the positive control PE-conjugated anti-CD133 antibody at 4 °C for 1h, washed twice with PBS, and bound phage detected by biotin-labeled anti-fd antibody followed by streptavidin-R-PE, and analyzed by FACS. For immunoprecipitation studies, 105 Caco-2 cells were lysed in 1% NP40, 0.15 M NaCl, and 0.01 M sodium phosphate, pH 7.2, on ice for 2 h. Lysates were precleared and incubated with 50 μg/ml scFvs bearing a hexahistidine tag (see below) at 4 °C for 1h, and captured onto Ni-NTA+ beads (Qiagen). As a positive control, precleared Caco-2 lysates were incubated with rabbit anti-CD133 antibody (Cell Signaling Technology) followed by capturing with protein A beads (Thermo Fisher Scientific). All immunoprecipitation products were analyzed by 4–20% gradient SDS-PAGE, followed by Western blot with mouse anti-CD133 followed by horseradish peroxidase-conjugated goat anti-mouse antibody (Jackson ImmunoResearch) using ECL Plus™ (GE Healthcare).
Expression and Purification of recombinant scFvs
ScFvs were subcloned from the phage vector into a bacteria expressionvector, resulting in the addition of a c-myc epitope and a hexahistidine tag at the C-terminus as described (22). ScFv was harvested from the bacterial periplasm (22) and purified using AKTAprime (GE HealthCare) by immobilized metal affinity chromatography (HiTrap His, GE HealthCare) and gel filtration (PD10, GE HealthCare), and analyzed by reducing 10% SDS-PAGE (22, 23).
ScFv internalization by tumor sphere cells
ScFvs were labeled with Q-dot (655) using a two-step conjugation procedure as described (25), incubated with tumor sphere cells at 37 °C for 1 hr, washed with PBS and fixed in 2% paraformaldehyde (Sigma), and viewed under a fluorescence microscope (Olympus) and a confocal microscope (Zeiss). DAPI (nucleus) and Q-dot labeled phage antibodies were imaged in separate channels and merged to show scFv subcellular location. To broaden applicability, we labeled scFvs with FITC using 5(6)-SFX and repeated the binding and internalization experiments.
Quantification of internalized phage antibodies was performed as described (22). Briefly, 1 × 105 GBM sphere cells were incubated with phage antibodies at 37 °C for 1h and 4h respectively, washed with PBS, and divided evenly into two groups, one for quantification of the total phage bound, and the other for the internalized fraction following surface stripping with 50 mM glycine/150 mM NaCl, pH 2.8.
Inhibition of self-renewal of GBM tumor sphere cells by recombinant scFvs
One thousand monodispersed tumor sphere cells were incubated with GC4 scFv (50 μg/ml) in 200 μl TSFM. Media were changed every 2 days by replacing 25 μl of old media with fresh media containing 50 μg/ml scFvs. Colonies (tumor spheroids) were counted after 14 days.
Generation of a recombinant human GC4 IgG1 molecule
The sequence information of the GC4 scFv was used to develop the huma IgG1 as described previously (34). Briefly, the GC4 VH was amplified using primer pairs with overhangs containing MluI and NheI sites, digested with Mlu1 and NheI, ligated into a mammalian expression vector to create in frame fusion with the human heavy chain constant region. The VL gene was amplified with the primer pairs with overhangs containing DraIII and AvrII, and ligated into DraIII- and KpnI-digested plasmid containing the GC4 heavy chain gene to create a plasmid that expresses both heavy and light chains, which was used to transfect CHO-DG44 cells by electroporation and to establish stable lines by selecting with G418 as described previously (34, 35). GC4 IgG1 was purified on a protein A column using the AKTA protein purification system (GE Healthcare), and analyzed by SDS-PAGE under reducing conditions. IgG oncentration was measured using Nanodrop (Thermal Fisher Scientific).
Affinity measurement
Varying concentrations of GC4 IgG1s were incubated with 105 GBM cells at RT for 1h in PBS containing 0.25% BSA. Bound IgG1s were detected by PE-conjugated goat anti-human antibody (Jackson ImmunoResearch) following incubation at RT for 30 min. Apparent dissociation constant was determined by FACS with mean fluorescence intensity (MFI) data fitted using GraphPad Prism (GraphPad Software). The apparent dissociation constant of GC4 scFv was measured similarly except that bound scFvs were detected by mouse anti-hexahistidine antibody (AbD Serotec/MorphoSys) followed by PE-conjugated goat anti-mouse antibody (Jackson ImmunoResearch).
Inhibition of GMB cell proliferation by human GC4 IgG1
To test the effect of GC4 IgG1 on tumor sphere cells, 1,000 monodispersed tumor sphere cells were incubated with 50 μg/ml GC4 IgG1 in 200 μl TSFM, and placed in a 96-well plate. There were no significant aggregations of tumor sphere cells under this condition. Media were changed every 2 days by replacing 25 μl of old media with fresh media containing 50 μg/ml GC4 IgG1. Tumor spheres were counted after 14 days. The assay was done in parallel with a control non-binding human IgG1 (34). To test the effect of GC4 IgG1 on GBM cells grown in non-selective media, soft agar assays were performed. Briefly, 300 μl melted 0.5% agarose was added to a 24-well plate and allowed to solidify. To prepare the top layer, 2.5% SeaPlaque low-melting temperature agarose (Cambrex Bioscience) was melted and diluted (v/v 1:7, final 0.35%) in RPMI-1640/10% FCS media, and kept at 42 °C in a water bath. Ten thousand monodispersed GBM cells were mixed with human IgG1s (at a final concentration of 50 μg/ml) in 300 μl 0.35% low-melting temperature agarose. The mixture was applied to the top of the solidified agarose in the 24-well plate and kept at RT for 30 min to allow solidification of the top layer. The plate was then incubated at 37 °C and colonies were counted after 14 days (36).
Growth inhibition of established tumor spheres
Monodispersed tumor sphere cells were placed in 6-well plates in TSFM at a density of 200 cells per well and allowed to grow for 2–3 days to form tumor spheroids. The GC4 scFv was added to a final concentration of 50 μg/ml and the incubation continued for an additional 4–5 days. The number of tumor spheres with a diameter greater than 50 μm were counted under an inverted microscopy (Nikon).
Statistics
The Student’s t test was used to analyze a pair of variables, and a P value of <0.05 was considered statistically significant.
Results
Isolation and characterization of GBM tumor sphere cells
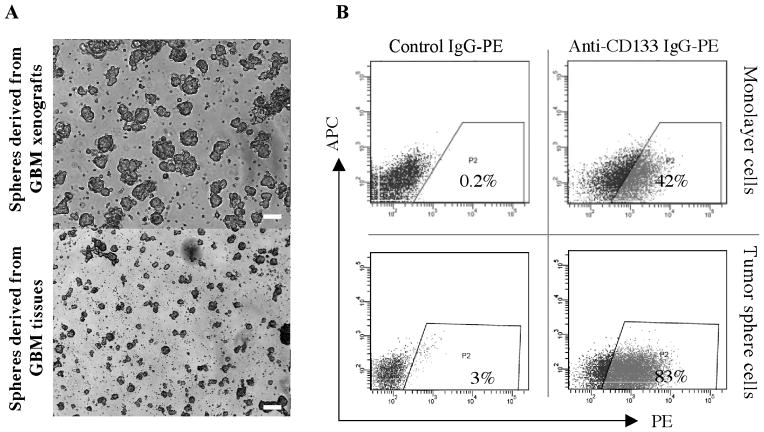
We first isolated GBM tumor spheres from 5 fresh human surgical specimens, as well as from 9 human GBM tissues maintained as xenografts in immunocompromised mice, by growing dissociated GBM cells in restrictive media containing bFGF and EGF. Tumor spheres consisting of a few hundred to a few thousand cells became visible after 7–14 days of culture (Fig. 1). We next compared the cloning efficiency of tumor spheres grown in restrictive media to bulk tumor cells from fresh surgical GBM specimens and GBM xenografts grown in media containing serum. GBM tumor sphere cells have higher cloning (self-renewal) capacity than either of the bulk tumor cells (about 10–20% vs. 1–2%), consistent with reports by others (15). Next, we analyzed the expression of the CD133 epitope (AC141) on GBM tumor sphere cells and GBM cells grown as monolayer cultures in non-selective media containing 2% FCS. We found detectable levels of CD133 epitope expression in 11 out of 14 GBM cultures (grown as spheres or monolayer cells) (Fig. 1 and Table 1a). In most cases (10/11) where epitope expression was evident, tumor sphere cells expressed higher levels of the CD133 epitope than the equivalent cells cultured as a monolayer in non-selective media. In addition, the expression of the CD133 epitope usually remained stable for a longer period of time in tumor sphere cells compared to monolayer primary tumor (4–6 months compared to about 1–2 months).
Fig. 1.
Characterization of GBM tumor spheres. A, Morphology of GBM tumor spheres derived from xenografts and fresh human tissues after 14 days of incubation in serum free restrictive media. Scale bar, 500 μm. B, CD133 epitope expression by GBM cells grown as a monolayer in serum containing media or as tumor spheres in serum free media.
Table 1.
Selection of human scFvs targeting human GBM tumor spheres. (a) Tumor sphere formation and CD133 epitope expression. (b) Summary of binding results for the five unique human scFvs studied. GBMh: human GBM tissues. GBMx: GBM maintained as xenografts in nude mice. ++: large size spheres containing > 100 cells per sphere after 14 days of incubation. +: small size spheres containing < 100 cells per sphere after 14 days of incubation. 0*, expression levels indistinguishable from that of the control (control isotype-matched mouse IgG stain). GBMx-8 and GBMh-2 were used for initial phage antibody library selection. Positive binders were re-tested on additional GBM tissues as indicated. Co-staining with CD133 is shown in Figure 3. Non-sphere GBM cells are derived from sphere cells cultured in differentiation promoting media containing serum. ND, not determined.
| (a) | ||||
|---|---|---|---|---|
| GBM Tissues | Tumor sphere formation | CD133 epitope expression | Target tissues for initial selection | Tissues used for re-test |
| GBMx-1 | ++ | 80% | + | |
| GBMx-2 | ++ | 50% | + | |
| GBMx-3 | ++ | 2% | + | |
| GBMx-4 | ++ | 0* | + | |
| GBMx-5 | ++ | 3% | + | |
| GBMx-6 | ++ | 4% | + | |
| GBMx-7 | ++ | 15% | + | |
| GBMx-8 | ++ | 70% | + | + |
| GBMx-9 | ++ | 0* | ||
| GBMh-1 | + | 0* | ||
| GBMh-2 | ++ | 12% | + | + |
| GBMh-3 | + | 8% | ||
| GBMh-4 | ++ | 3% | + | |
| GBMh-5 | + | 6% | ||
| (b) | |||||
|---|---|---|---|---|---|
| H3 | GC4 | GB1 | GB8 | GH9 | |
| Binding to GBM sphere cells | 100% | 50–99% | Variable# | Variable# | Variable# |
| Binding to non-sphere GBM cells | Unchanged (100%) | Reduced | Reduced concurrently with CD133 | Reduced concurrently with CD133 | Reduced concurrently with CD133 |
| Relationship to CD133+ cells Target antigen | Cover CD133+ CD166 | Cover CD133+ ND | A subset of CD133+ ND | A subset of CD133+ ND | Co-stain w/CD133+ ND |
: variable binding patterns were shown in Table S1.
Selection of internalizing human scFvs targeting the CD133 positive subpopulation of tumor sphere cells
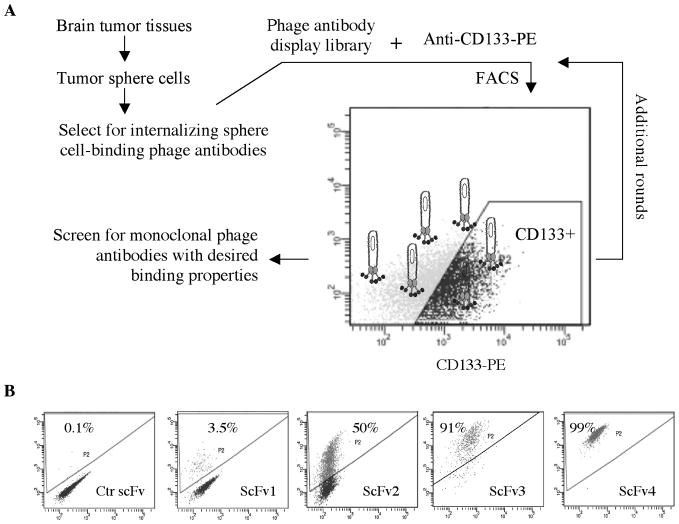
For phage antibody library selection (Fig. 2A), we initially targeted CD133 positive tumor sphere cells derived from fresh human GBM tissues (GBMh2, Table 1b) to increase the likelihood of identifying scFvs that target clinically represented markers. For subsequent rounds of selection we targeted the CD133 positive fraction of tumor sphere cells derived from GBM tissues maintained as xenografts (GBMx-8, Table 1b). To broaden applicability, positive binders were re-tested on additional GBM tissues (Table 1b).
Fig. 2.
Selection of phage antibodies binding to the CD133+ cell population. A, Outline of the selection scheme. Selection was performed by FACS on cells derived from multiple GBM cases. CD133+ cells were gated by comparing to staining results with an isotype-matched PE-conjugated control mouse mAb. Outputs from round-3 selections were screened to identify monoclonal phage antibodies. B, Patterns of phage antibody binding to GBM cells. Percentage of positive cells is indicated. GBMx8 tumor sphere cells were used in the analysis.
To begin, we first counter-selected the 500 million-member naïve phage antibody display library on a panel of non-tumorigenic cell lines to remove non-specific binders, and then selected the library on GBM tumor sphere cells under internalizing conditions to enrich polyclonal phage that target internalizing epitopes. The polyclonal phage antibodies were then used as the input for further selection by FACS to target the CD133 positive cells. After two rounds of FACS-based selection, we analyzed the selection output for binding to GBM sphere cells. About 45% (83/186) of phage antibodies bound to GBM sphere cells but not control cells such as BPH-1. There were twenty unique scFvs among sixty that were sequenced. The phage antibodies exhibit a range of binding patterns, binding from about 3% to 99% of the GBM sphere cell population (Fig. 2B). Five scFvs with the highest frequency of representations in the output were chosen for further studies (Table 1b). They have been re-tested and validated on additional GBM tissues not used in the initial selection (Table 1b). By scanning a sequence database of scFvs previously identified in our laboratory, one scFv that binds to all GBM sphere cells was found to be identical to H3, an scFv that we have previously identified and shown to bind to an internalizing epitope on ALCAM/CD166 (26, 34). To simplify the nomenclature, we used the original name H3 to designate this scFv. The other four scFvs have not been previously identified.
Analysis of co-expression of scFv-targeted antigens and the CD133 epitope
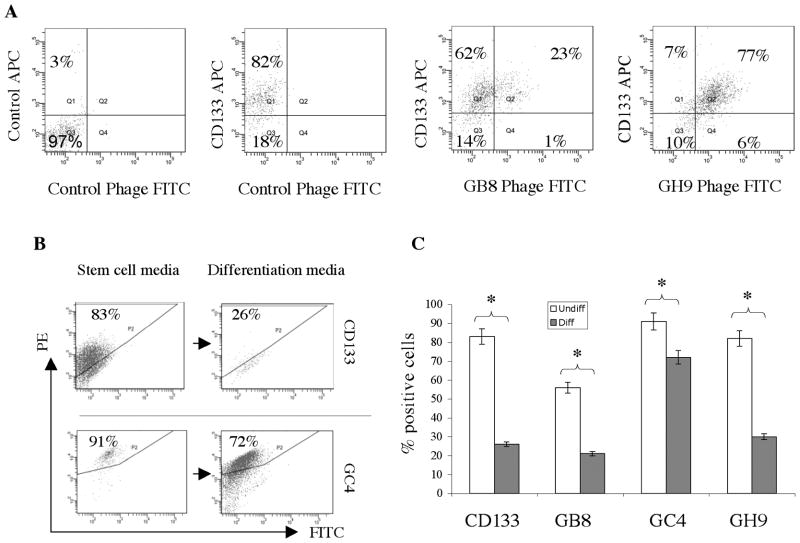
Since the selection scheme was designed to capture binders to the CD133 positive fraction of GBM sphere cells, we used double labeling analysis to determine if the phage antibodies that bind a subset of the cell population bind preferentially to the CD133 positive population. We tested GB8 and GH9 phage antibodies and found that they co-stained with the anti-CD133 epitope mAb (Fig. 3A), whereas very little phage binding to CD133 epitope negative- cells was observed (< 1% for GB8 and < 6% for GH9). Thus, GB8 and GH9 bind to antigens expressed mainly on the CD133+ subpopulation of GBM tumor spheres, indicating that our original selection scheme was successful. Neither GB8 nor GH9 phage antibodies compete with the anti-CD133 epitope mAb for binding (Fig. 3A, no reductions in percentage of CD133-APC positive cells were observed), indicating that these phage antibodies bind to novel epitopes that do not overlap with the widely used CD133 epitopes.
Fig. 3.
Co-expression of the CD133 epitope and phage-bound antigens. A, FITC-labeled phage antibodies and biotin-labeled anti-CD133 mAb were studied on GBM sphere cells. A typical result from studies on GBMx-1 sphere cells is shown. Percentage of total cells is indicated for each quadrant. B, Concurrent down-regulation of the CD133 and the phage-bound epitopes. FACS analysis of the expression of the CD133 epitope (top panels) and the antigen bound by the GC4 phage antibody (bottom panels). Concurrent down-regulations were observed when GBM sphere cells were removed from TSFM and cultured in media that promotes differentiation. C, Quantification of binding status of the anti-CD133 epitope mAb and phage antibodies to GBM tumor sphere cells cultured in TSFM that promote maintenance of the undifferentiated state (Undiff) and non-selective media that promote differentiation (Diff). Experiments were done in triplicates and standard deviations are shown. *, P < 0.05.
To further determine if our scFvs bind to CD133 via a different epitope, we first studied binding of scFvs to Caco-2 cell line that expresses a high level of CD133 (Supplementary Fig. S1-A). As shown in Supplementary Fig. S1-B, none of our scFvs bind to Caco-2 cells, while the positive control anti-CD133 antibody binds strongly to Caco-2 cells. We further performed immunoprecipitation studies and found that our scFvs do not pull down the CD133 molecule while the positive control antibody does (Supplementary Fig. S1-C), consistent with results of the FACS analysis. We therefore conclude that our scFvs do not bind to CD133.
Phage antibodies target antigens preferentially associated with tumor sphere cells grown in selective media
To test if antigens bound by phage antibodies are preferentially expressed by GBM cells growing in restrictive media as tumor sphere cells, we compared expression of the CD133 epitope and scFv-targeted antigens by GBM cells grown in selective vs. non-selective media. We observed concurrent down-regulation of the CD133 epitope and phage antibody-bound epitopes for all the scFvs tested (Fig. 3B & C), suggesting that these phage antibody-targeted antigens are preferentially associated with GBM tumor sphere cells growing in selective media.
Antibody internalization by GBM tumor sphere cells
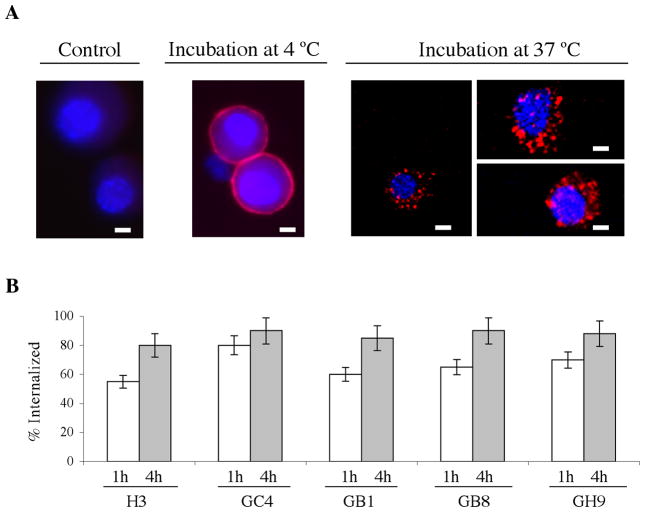
Our phage antibodies were selected for internalizing functions with the goal of developing potential tumor stem cell-targeted therapies based on intracellular delivery strategies. To confirm the internalizing function of selected scFvs, we labeled scFvs with quantum dots (Q-dot 655) and visualized cellular uptake by fluorescence microscopy. An example of the experiment with the GC4 scFv is shown in Fig. 4a. When incubation was at 4 °C, the GC4 scFv was found to bind to the surface of GBM sphere cells. When incubation was at 37 °C, which permits receptor-mediated endocytosis, the GC4 scFv was efficiently internalized (Fig. 4a) into intracellular compartments. To rule out potential Q-dot labeling artifacts, the experiment was repeated with FITC-labeled scFvs and the same results were obtained (data not shown).
Fig. 4. Internalization by GBM tumor sphere cells.
A, Internalization of Q-dot 655-labeled GC4 scFv by GBM tumor sphere cells. Representative images are shown for each indicated incubation condition. Control: GBM sphere cells incubated with Q-dot 655-labeled control non-binding scFv (N3M2). This non-binding scFv was originally picked from the unselected naïve library and has been used as a negative control in our previous studies (25, 50). Scale bar, 2 μm. B, Quantification of percent internalized. Following incubation with phage antibodies, data from two time points (1h and 4h) were collected and analyzed. Surface-bound phage antibodies were removed by washing with a low pH glycine buffer. The percentage of internalized phage is calculated based on the ratio of internalized over total cell associated phage antibodies. The experiments were done in triplicates. Error bars indicate standard deviations.
To quantify the internalization efficiency, we compared the cell-associated phage antibodies before and after treatment with a low pH buffer that strips off surface-bound non-internalized phage antibodies. As shown in Fig. 4b, all four scFvs studied were efficiently internalized. Significant internalization was seen as early as 1h. By 4h most phage antibodies were internalized (Fig. 4b), demonstrating that our selection strategies were effective at enriching internalizing human scFvs that target the GBM tumor sphere cells.
GC4 scFv inhibits GBM tumor sphere cell self-renewal
We next tested the ability of the scFvs to inhibit tumor sphere self-renewal. We incubated affinity-purified recombinant scFvs with dissociated GBM tumor sphere cells for 7 days, and determined the efficiency of clonal expansion. The GC4 scFv caused a significant inhibition of GBM sphere growth in vitro (Fig. 5A) suggesting that it modulates the activity of a cell surface antigen that plays a significant role in GBM tumor sphere cell self-renewal. None of the other scFvs tested showed inhibitory effects under identical conditions (Fig. 5A). To test for non-specific cell toxicity, we studied the effect of the GC4 scFv on cell lines that were not bound by this scFv (SKOV3 and PC3, see Supplementary Table S2). No inhibition was observed in these control experiments.
Fig. 5.
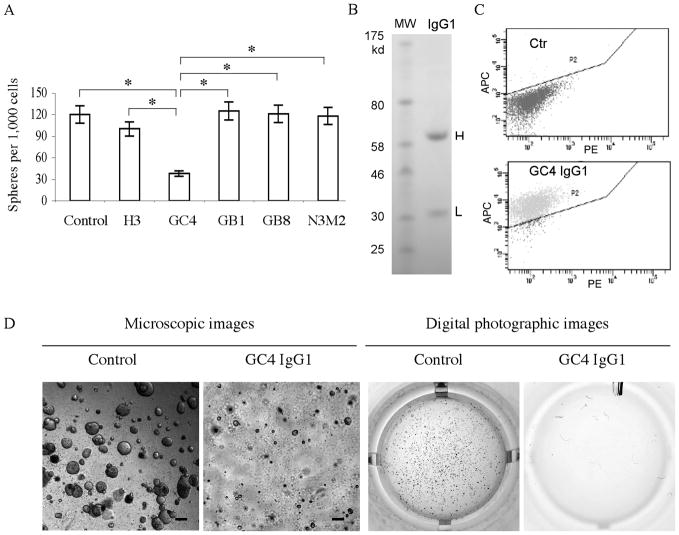
Inhibition of self-renewal of GBM tumor sphere cells. A, The GC4 but not other GBM tumor sphere cell-binding scFvs significantly inhibited GBM sphere cell proliferation. Cloning efficiencies were calculated from tumor spheres formed per 1,000 GBM cells seeded. Control, untreated sphere cells. N3M2, a non-binding scFv control. Experiments were done in triplicates with standard deviations indicated by error bars. *, P < 0.05 for pairs indicated. B, Production of recombinant GC4 IgG. H: heavy chain. L: light chain. MW: molecular weight. C, Quality control study confirming that the GC4 scFv-derived IgG1 retains the ability to bind GBM sphere cells. Ctr: a non-binding human IgG1. D, GC4 IgG1 inhibits colony formation of GBM cells in the soft agar assay. Both microscopic and digital photographic images of the GC4 and control IgG1-treated wells are shown. Scale bar, 250 μm.
To further determine if GC4 scFv is capable of inhibiting the growth of established GBM tumor spheres, we grew monodispersed GBM cells in TSFM to obtain tumor spheroids, and treated them with recombinant GC4 scFv (100 μg/ml). As shown in Supplementary Fig. S2, treatment with the GC4 but not the control scFv significantly reduced the growth of established tumor spheres.
GC4 IgG1 inhibits proliferation of GBM tumor sphere cells
Because the GC4 scFv inhibits proliferation of GBM tumor sphere cells, we sought to develop full-length human GC4 IgG1 for further in vivo applications. Using the VH and VL sequences from the GC4 scFv, we constructed the GC4 IgG1 in a mammalian expression vector, transformed CHO-DG44 cells and produced full-length recombinant human GC4 IgG1. Analysis of purified IgG1 by reducing SDS-PAGE showed heavy and light chains with appropriate sizes (Fig. 5B). For quality control, we tested and confirmed binding of the recombinant GC4 IgG1 to GBM tumor sphere cells (Fig. 5C). In addition, the GC4 IgG competed specifically with the parent GC4 scFv for binding to GBM sphere cells (Supplementary Fig. S3), demonstrating that the IgG and the scFv have the same specificity, consistent with our previous experience with recombinant IgG development (34). We further measured cell binding affinity by FACS and found that the GC4 IgG1 binds with greater affinity than the parental scFv (the binding constant measured at RT is 79 nM for IgG1 (Supplementary Fig. S4) vs. 222 nM for scFv (Supplementary Fig. S5)). We next tested inhibitory effects of the GC4 IgG1 on GBM tumor sphere cells grown in selective media. Like the parental GC4 scFv, recombinant GC4 IgG1 inhibited self-renewal of GBM tumor sphere cells (about 60% inhibition, similar to that of the GC4 scFv).
To determine if this inhibitory effect is also applicable to GBM cells grown under non-selective culture conditions, we treated GBM cells cultured in non-selective media and measured cloning efficiencies using the soft agar assay. As shown in Fig. 5D, the GC4 IgG1 inhibited the clonogenic activity of GBM cells. Compared to controls, the GC4 IgG1-treated GBM cells formed significantly fewer colonies. Moreover, the colonies formed by the GC4 IgG1-treated cells are significantly smaller than those formed by cells treated with the control IgG (Fig. 5D). This experiment was repeated on GBM cells derived from five independent cases. Inhibitory effects were observed in four of these cases. Again, no inhibitory effects were observed on cell lines to which the GC4 antibody does not bind (SKOV3 and PC3; non-binding cells are listed in Supplementary Table S2). These results demonstrated that GC4 human IgG1 is a potent inhibitor of in vitro GBM cell proliferation in non-selective media.
Discussion
It has been hypothesized that bulk tumor is sustained by a subpopulation of tumor initiating cells, and their elimination would promote collapse of the tumor hierarchy, overcoming drug resistance and tumor recurrence associated with current anti-cancer therapies. While conceptually appealing, therapies that target these tumor-initiating cells have yet to be developed for use in clinical practice. To develop human monoclonal antibody-based therapies that target prospective brain tumor initiating cells, we have selected a large naïve phage antibody display library on GBM tumor sphere cells to identify novel internalizing human scFvs that target potential brain tumor initiating cells. We have shown that our antibody library-based approach is effective in identifying novel human antibodies targeting the whole as well as subpopulations of GBM tumor sphere cells. To our knowledge, this is the first time that a phage antibody library selection has been used for this purpose.
GBM tumor initiating cells have been well studied and are generally defined in two ways: by the expression of stem cell markers such as CD133 epitopes, and/or by the ability to grow in restrictive media (tumor sphere formation assay) (14, 15). Both methods have been widely used to enrich brain tumor initiating cells but each by itself may have limitations (16). Thus, in order to more precisely target GBM tumor initiating cells, we have used GBM tumor sphere cells that express high levels of the CD133 epitope as our selection target. The use of two criteria to define prospective GBM tumor initiating cells used for antibody selection helps increase the chances of recovering antibodies targeting these cells.
We have performed the antibody library selection on tumor sphere cells derived from fresh human GBM tissues and GBM tissues maintained in immunocompromised mice. Primary GBM tumors maintained as xenografts are useful for these studies for several reasons. First, these tumors have been well characterized, and shown to have the same gene alterations identified in their corresponding patient tumors (37). Secondly, previous study has shown that brain tumors maintained as xenografts retain many features of the original tumors (27). Finally, these tumors can be propagated indefinitely, and thereby exist as a sustainable source of tissue for extended studies. Xenografts therefore provide a continuous supply of tissues with relatively consistent characteristics, and facilitate experimental reproducibility (27, 38). Similar approaches that aim to identify and characterize tumor initiating cells have been used by others for investigating pancreatic cancer (38).
The GBM tumor sphere cell-targeting scFvs we have identified provide an opportunity to develop novel monoclonal antibody-based therapeutics. For example, we have identified a scFv that inhibits self-renewal of GBM tumor sphere cells. We have further developed a full-length human IgG based on this scFv, and shown that like the parental scFv, this recombinant human IgG inhibits proliferation of GBM tumor sphere cells. The availability of a full-length human IgG with a demonstrated inhibitory activity should allow further testing of anti-tumor efficacy in vivo using appropriate animal models. Even scFvs with no apparent anti-proliferation effects in vitro could nonetheless be useful for therapeutic development. Because our panel of scFvs has been selected for internalizing functions, they can be used to develop therapies based on intracellular delivery strategies. For example, these internalizing scFvs can be conjugated to drugs or drug-loaded nanoparticles, engineered toxin molecules, or appropriate radionuclides to impart a targeted cell killing function (39, 40). These targeted agents could in theory offer an opportunity to determine if in vivo ablation of tumor initiating cells results in improved therapeutic effects such as suppression of recurrence.
In addition to therapeutic development, our panel of GBM tumor sphere cell-targeting scFvs could be useful in identifying additional tumor initiating cell markers other than the CD133 epitopes, which could provide new insights into tumor heterogeneity, facilitate improvement of selection and enrichment protocols for GBM tumor subpopulations, and allow development and optimization of new treatment strategies targeting these antigens.
We would like to point out that although numerous independent studies by others have shown that GBM tumor sphere cells and the CD133 positive GBM cells can initiate tumor formation in vivo and thus are operationally defined as GBM cancer stem cells (16, 41), we have not repeated the in vivo tumor initiation experiment using GBM tumor sphere cells that we have isolated according to published protocols. We have thus refrained from referring to these GBM tumor sphere cells as cancer stem cells, although they showed higher self-renewal ability in vitro, a property commonly associated with cancer stem cells.
The strategy of using antibodies to target tumor subpopulations, while conceptually appealing, is likely to encounter similar challenges associated with antibody targeting of the bulk tumor. For example, issues such as tissue penetration and binding site barrier need to be addressed (42–45). Optimizing antibody-target interactions and pharmacokinetics have been shown to improve efficiencies of antibody-based tumor targeting (44, 46, 47). It should be pointed out that in most cases antibody therapy is most effective in treating solid tumors following volume reduction by surgery, chemotherapy or radiotherapy. In addition, small size lesions such as micrometastasis can be effectively targeted by monoclonal antibodies. Because tumor initiating cells are hypothesized to be the primary force driving recurrence, targeting these cells by antibodies in the above two situations is feasible and should result in improved treatment outcome. Finally, recent studies have shown that at least for some cancers, including GBM, tumor initiating cells exist in a special vascular “niche” (48, 49), which would render them accessible to intravenously injected therapeutics such as monoclonal antibodies.
In summary, our studies show that it is feasible to use the combinatorial antibody library approach to select internalizing scFvs targeting GBM tumor sphere cells, and furthermore a subpopulation of these tumor sphere cells that express high levels of the CD133 epitope. In addition, inhibitory scFvs can be identified and their full-length inhibitory human IgG derivatives can be readily developed. Our methods should be generally applicable to the development of internalizing human monoclonal antibodies targeting subpopulations of heterogeneous tumor cells.
Supplementary Material
Acknowledgments
We thank Dr. James D. Marks for providing the naive phage antibody library, the UCSF Brain Tumor Tissue Core for providing human glioma specimens, Drs. Yong Wang, Weihua Wen, Jianlong Lou, Yongfeng Fan, Fraser Conrad, Eduard B. Dinca and Ramona Voicu for help with experiments.
Financial support: This work is supported in parts by grants from the National Institute of Health (R01 CA118919 to BL, and P50 CA097257 to MSB and CDJ), and by a Developmental Project award from the UCSF Brain Tumor SPORE (to BL).
Abbreviation
- GBM
glioblastoma multiforme
- ScFv
single chain antibody fragment
- EGF
epidermal growth factor
- bFGF
basic fibroblast growth factor
- TSFM
tumor sphere forming media
- FACS
fluorescence activated cell sorting
- FCS
fetal calf serum
- PE
phycoerythrin
- CHO
Chinese hamster ovary
- Q-dot
quantum dot
Footnotes
Potential conflict of interest: No conflict of interest.
References
- 1.Krex D, Klink B, Hartmann C, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 2.Nicholas MK. Glioblastoma multiforme: evidence-based approach to therapy. Expert Rev Anticancer Ther. 2007;7:S23–7. doi: 10.1586/14737140.7.12s.S23. [DOI] [PubMed] [Google Scholar]
- 3.Benitez JA, Dominguez-Monzon G, Segovia J. Conventional and gene therapy strategies for the treatment of brain tumors. Curr Med Chem. 2008;15:729–42. doi: 10.2174/092986708783955491. [DOI] [PubMed] [Google Scholar]
- 4.Chang JE, Khuntia D, Robins HI, Mehta MP. Radiotherapy and radiosensitizers in the treatment of glioblastoma multiforme. Clin Adv Hematol Oncol. 2007;5:894–902. 07–15. [PubMed] [Google Scholar]
- 5.Reardon DA, Desjardins A, Rich JN, Vredenburgh JJ. The emerging role of anti-angiogenic therapy for malignant glioma. Curr Treat Options Oncol. 2008;9:1–22. doi: 10.1007/s11864-008-0052-6. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 8.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–84. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 9.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–45. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Zhao Y, Smith C, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–35. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 11.Sims AH, Howell A, Howell SJ, Clarke RB. Origins of breast cancer subtypes and therapeutic implications. Nat Clin Pract Oncol. 2007;4:516–25. doi: 10.1038/ncponc0908. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 13.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–73. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 14.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 15.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 16.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med. 2008;86:1025–32. doi: 10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen UB, Marks JD. Internalizing antibodies and targeted cancer therapy: direct selection from phage display libraries. Pharm Sci Technol Today. 2000;3:282–91. doi: 10.1016/s1461-5347(00)00280-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu B. Exploring cell type-specific internalizing antibodies for targeted delivery of siRNA. Brief Funct Genomic Proteomic. 2007;6:112–9. doi: 10.1093/bfgp/elm015. [DOI] [PubMed] [Google Scholar]
- 20.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149–61. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 21.Becerril B, Poul MA, Marks JD. Toward selection of internalizing antibodies from phage libraries. Biochem Biophys Res Commun. 1999;255:386–93. doi: 10.1006/bbrc.1999.0177. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Mapping tumor epitope space by direct selection of single-chain Fv antibody libraries on prostate cancer cells. Cancer Res. 2004;64:704–10. doi: 10.1158/0008-5472.can-03-2732. [DOI] [PubMed] [Google Scholar]
- 23.Roth A, Drummond DC, Conrad F, et al. Anti-CD166 single chain antibody-mediated intracellular delivery of liposomal drugs to prostate cancer cells. Mol Cancer Ther. 2007;6:2737–46. doi: 10.1158/1535-7163.MCT-07-0140. [DOI] [PubMed] [Google Scholar]
- 24.An F, Drummond DC, Wilson S, et al. Targeted drug delivery to mesothelioma cells using functionally selected internalizing human single-chain antibodies. Mol Cancer Ther. 2008;7:569–78. doi: 10.1158/1535-7163.MCT-07-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bidlingmaier S, He J, Wang Y, et al. Identification of MCAM/CD146 as the target antigen of a human monoclonal antibody that recognizes both epithelioid and sarcomatoid types of mesothelioma. Cancer Res. 2009;69:1570–7. doi: 10.1158/0008-5472.CAN-08-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan W, Sassoon A, An F, Simko JP, Liu B. Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol Cell Proteomics. 2006;5:2364–73. doi: 10.1074/mcp.M600246-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabos P, Ehtesham M, Kabosova A, Black KL, Yu JS. Generation of neural progenitor cells from whole adult bone marrow. Exp Neurol. 2002;178:288–93. doi: 10.1006/exnr.2002.8039. [DOI] [PubMed] [Google Scholar]
- 29.Westhoff MA, Zhou S, Bachem MG, Debatin KM, Fulda S. Identification of a novel switch in the dominant forms of cell adhesion-mediated drug resistance in glioblastoma cells. Oncogene. 2008 doi: 10.1038/onc.2008.148. [DOI] [PubMed] [Google Scholar]
- 30.O’Connell D, Becerril B, Roy-Burman A, Daws M, Marks JD. Phage versus phagemid libraries for generation of human monoclonal antibodies. J Mol Biol. 2002;321:49–56. doi: 10.1016/s0022-2836(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 31.Sheets MD, Amersdorfer P, Finnern R, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A. 1998;95:6157–62. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B, Marks JD. Applying phage antibodies to proteomics: selecting single chain Fv antibodies to antigens blotted on nitrocellulose. Anal Biochem. 2000;286:119–28. doi: 10.1006/abio.2000.4788. [DOI] [PubMed] [Google Scholar]
- 33.Liu B, Huang L, Sihlbom C, Burlingame A, Marks JD. Towards proteome-wide production of monoclonal antibody by phage display. J Mol Biol. 2002;315:1063–73. doi: 10.1006/jmbi.2001.5276. [DOI] [PubMed] [Google Scholar]
- 34.Liu B, Conrad F, Roth A, et al. Recombinant full-length human IgG1s targeting hormone-refractory prostate cancer. J Mol Med. 2007;85:1113–23. doi: 10.1007/s00109-007-0208-z. [DOI] [PubMed] [Google Scholar]
- 35.Nowakowski A, Wang C, Powers DB, et al. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–50. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–7. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 39.Weiner LM, Dhodapkar MV, Ferrone S. Monoclonal antibodies for cancer immunotherapy. Lancet. 2009;373:1033–40. doi: 10.1016/S0140-6736(09)60251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23:1147–57. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 41.Altaner C. Glioblastoma and stem cells. Neoplasma. 2008;55:369–74. [PubMed] [Google Scholar]
- 42.Thurber GM, Schmidt MM, Wittrup KD. Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Adv Drug Deliv Rev. 2008;60:1421–34. doi: 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams GP, Schier R, McCall AM, et al. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer Res. 2001;61:4750–5. [PubMed] [Google Scholar]
- 44.Adams GP, Tai MS, McCartney JE, et al. Avidity-mediated enhancement of in vivo tumor targeting by single-chain Fv dimers. Clin Cancer Res. 2006;12:1599–605. doi: 10.1158/1078-0432.CCR-05-2217. [DOI] [PubMed] [Google Scholar]
- 45.Juweid M, Neumann R, Paik C, et al. Micropharmacology of monoclonal antibodies in solid tumors: direct experimental evidence for a binding site barrier. Cancer Res. 1992;52:5144–53. [PubMed] [Google Scholar]
- 46.Ackerman ME, Pawlowski D, Wittrup KD. Effect of antigen turnover rate and expression level on antibody penetration into tumor spheroids. Mol Cancer Ther. 2008;7:2233–40. doi: 10.1158/1535-7163.MCT-08-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundaresan G, Yazaki PJ, Shively JE, et al. 124I-labeled engineered anti-CEA minibodies and diabodies allow high-contrast, antigen-specific small-animal PET imaging of xenografts in athymic mice. J Nucl Med. 2003;44:1962–9. [PMC free article] [PubMed] [Google Scholar]
- 48.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Veeravagu A, Bababeygy SR, Kalani MY, Hou LC, Tse V. The cancer stem cell-vascular niche complex in brain tumor formation. Stem Cells Dev. 2008;17:859–67. doi: 10.1089/scd.2008.0047. [DOI] [PubMed] [Google Scholar]
- 50.He J, Wang Y, Feng J, et al. Targeting prostate cancer cells in vivo using a rapidly internalizing novel human single-chain antibody fragment. J Nucl Med. 2010;51:427–32. doi: 10.2967/jnumed.109.069492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.