Cyclotides are a new emerging family of large plant-derived backbone-cyclized polypeptides ( 28–37 amino acids long) that share a disulfide-stabilized core (3 disulfide bonds) characterized by an unusual knotted.[1] Cyclotides contrast with other circular polypeptides in that they have a well-defined three-dimensional structure, and despite their small size, can be considered as micro-proteins. Their unique circular backbone topology and knotted arrangement of three disulfide bonds makes them exceptionally stable to thermal and enzymatic degradation (Scheme 1). Furthermore, their well-defined structures have been associated with wide range of biological functions.[2, 3] Cyclotides MCoTI-I/II are powerful trypsin inhibitors (Ki 20 – 30 pM) which have been recently isolated from the dormant seeds of Momordica cochinchinensis, a plant member of cucurbitaceae family.[4] Although MCoTI cyclotides do not share significant sequence homology with other cyclotides beyond the presence of the three cystine bridges, structural analysis by NMR has shown that they adopt a similar backbone-cyclic cystine-knot topology.[5, 6] MCoTI cyclotides, however, show high sequence homology with related cystine-knot squash trypsin inhibitors,[4] and therefore represent interesting molecular scaffolds for drug design.[7–10]
Scheme 1.
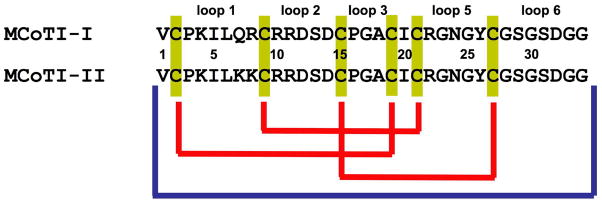
Primary structure and disulfide connectivities of MCoTI cyclotides. Blue and yellow connectors represent peptide and disulfide bonds, respectively.
Determination of the backbone dynamics of these fascinating micro-proteins is key for understanding their physical and biological properties. Internal motions of a protein on different time scales, extending from picoseconds to second, have been suggested to play an important role in its biological function.[11] A better understanding of the backbone dynamics of the cyclotide scaffold will be extremely helpful for evaluating its utility as a scaffold for peptide-based drug discovery. Such insight will help in the design of optimal focused libraries than can be used for the discovery of new cyclotides sequences with novel biological activities.[12, 13] In the current study we report for the first time the determination of internal dynamics of the cyclotide MCoTI-I in free state and complexed with trypsin. Uniformly 15N-labeled natively folded cyclotide MCoTI-I was recombinantly produced in E. coli growing in minimal M9 medium containing 15NH4Cl as only source of nitrogen. Concomitant backbone cyclization and folding was accomplished by using intramolecular native chemical ligation [14, 15] in combination with a modified protein splicing unit (Fig. S1, Supplementary Information).[16–18] Internal dynamics of cyclotide MCoTI-I was obtained from 15N spin-lattice and spin-spin relaxation times and 15N{1H}-heteronuclear Overhauser effect (NOE) enhancements.[11] The backbone flexibility was characterized by the square of the generalized order parameter, S2, which reveal the dynamics of backbone NH groups on the ps-ns time scale.[19, 20] The order parameter satisfies the inequality, 0 ≤ S2 ≤1, in which lower values indicate larger amplitudes of intramolecular motions. Motions on the ms-μs timescales were assessed by the presence of the chemical exchange terms in the spin-spin relaxation.
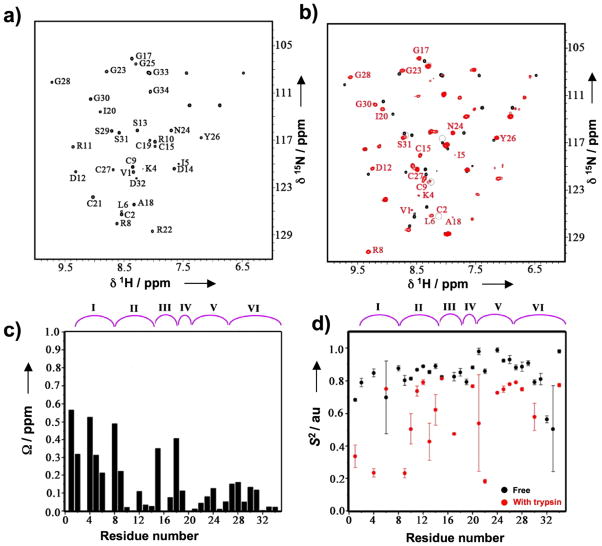
The NMR spectra and S2 values, derived from the 15N relaxation data of free MCoTI-I are shown in Figs. 1a and 1d. Residues Ile5 and Gly23 of free MCoTI-I were excluded from the backbone dynamics analysis since the relaxation data could not be fit to a mono-exponential function, possibly due to chemical exchange.[21] Gln7 was not assigned due to broadening of the NMR signal, presumably caused by fast exchange with water. The S2 values for free MCoTI-I show that most of the NH groups of the cyclotide backbone are highly constrained with S2 values ≥ 0.8 resembling those found in well folded globular proteins (Table 1). The average S2 value, <S2>, for free MCoTI-I was 0.83 ± 0.03. This value similar to that found for the six Cys residues involved in the cystine-knot (<S2> = 0.84 ± 0.02) and is considerably larger than those found for other linear squash trypsin inhibitors (<S2> = 0.71 for trypsin inhibitor from Cucurbita maxima (CMTI-III, 78% homology with MCoTI-I)[22] thus indicating the importance of the backbone cyclization to rigidify the overall structure. Loops 2 through 5 in free MCoTI-I showed <S2> values ≥ 0.8. In particular, loop 5 showed a <S2> value of 0.92 ± 0.02 well above the average for the molecule and the Cys-knot. In contrast, loops 1 and 6 showed <S2> values below the average for the molecule. Thus, loop 6, which is believed to act as a very flexible linker to allow cyclization,[23] had a <S2> value of 0.76 ± 0.17 with only two residues, Asp32 and Gly33 having values below 0.6. Despite this small <S2> value, residues in loop 6 did not require significant chemical exchange terms (Fig. S2 and Table S1, Supplementary Information) suggesting that the mobility observed is mostly due to local vibrations.
Figure 1.
NMR analysis of the backbone dynamic of free and trypsin bound MCoTI-I. (a) NMR {15N,1H}-HSQC spectrum of free MCoTI-I. Chemical shift assignments of the backbone amides are indicated. (b) Overlay of the {15N,1H}-HSQC spectra of free (black) and trypsin bound MCoTI-I (red). Residues with large average amide chemical shift differences between two different states ( > 0.3 ppm) are indicated. Peaks that are broadened in trypsin bound MCoTI-I are indicated by grey circles. (c) Average amide chemical shift difference for all the assigned residues in free and trypsin bound MCoTI-I. Chemical shift difference was calculated as: ΔΩ = [(ΔΩNH2+0.04ΔΩN2)/2]½ , where ΔΩNH and ΔΩN are the changes in the amide proton and nitrogen chemical shifts (ppm), respectively. (d) The order parameter, S2, for the free (black) and the trypsin bound MCoTI-I (red). S2 value is a measure of backbone flexibility and represents the degree of angular restriction of N-H vector in the molecular frame. The MCoTI-I loops are shown on top of panels C and D. Small unassigned peaks in both free and trypsin bound spectra of MCoTI-I are from a minor conformation of the protein due to a known isomerization of the backbone at an Asp-Gly sequence in loop 6 of MCoTI-I.
Table 1.
Average order parameters of structural elements in MCoTI-I in free state and bound to trypsin.
| Structural element | Sequence | <S2>a Free MCoTI-I | <S2>b Trypsin-MCoTI-I |
|---|---|---|---|
| Loop 1 | 3–8 | 0.81 ± 0.01 | 0.49 ± 0.05 |
| Loop 2 | 10–14 | 0.81 ± 0.01 | 0.62 ± 0.07 |
| Loop 3 | 16–18 | 0.84 ± 0.02 | 0.48c |
| Loop 4 | 20 | 0.88c | 0.76c |
| Loop 5 | 22–26 | 0.92 ± 0.02 | 0.61 ± 0.01 |
| Loop 6 | 28–34 | 0.76 ± 0.05 | 0.61 ± 0.05 |
| Cys-knot | 2,10,15,19,21,27 | 0.84 ± 0.02 | 0.60 ± 0.08 |
S2 values for residues 5 and 23 from free MCoTI-I are not included in the average because the relaxation data could not be fitted to a mono-exponential function.
S2 values for residues 2, 5, 8,18,19, 23, 29, 31, 32 and 33 from trypsin bound MCoTI-I are not included in the average because of the lack of signal intensity or the relaxation data could not be fitted to a mono-exponential function.
<S2> contains S2 value for a single residue.
The <S2> value for loop 1, which is responsible for binding trypsin, was 0.81 ± 0.07 This value is 90% of the average value for free MCoTI-I. Residue Leu6 in loop 1 also required chemical exchange terms to be considered indicating the existence of intramolecular conformational exchange on the μs-ms time scale. The mobility observed in loop 1 at both ns-ps and ms time scales have been also described in other trypsin inhibitors[22, 24, 25] and it has been suggested to play an important role in receptor-ligand binding.[11]
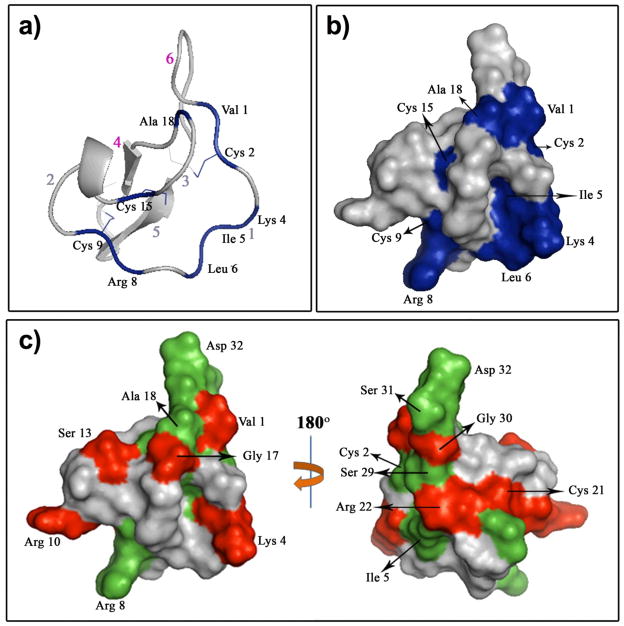
To explore if that was the case in the MCoTI cyclotides, we next studied the effect of ligand-binding on the backbone dynamics of MCoTI-I (Figs. 1 and 2). To exclude the possibility that trypsin could cleave or scramble the disulfide bonds of MCoTI-I upon complex formation, we used a competition experiment of trypsin-[15N]-MCoTI-I with unlabeled MCoTI-I. The results indicated that structure of MCoTI-I is unaltered upon trypsin binding (Fig. S3). Trypsin binding led to large (> 0.3 ppm) and specific changes in the chemical shifts of the residues located in loops 1 (Cys2, Lys4, Ile5, Arg8), 3 (Cys15 and Ala18), and 6 (Val1) (Figs. 1c and 2b and Table S2, Supplementary Information). NMR signals of Cys2, Ile5, Cys19, Ser29, Ser31, Asp32 and Gly33 were significantly broadened presumably due to intramolecular chemical exchanges in the trypsin-MCoTI-I complex. Arg8, Ala18 and Gly23 were excluded from backbone dynamics analysis because their peaks were broadened out in 15N{1H}-NOE spectra. Similar findings have been already reported for other biomolecular interactions.[26] We used these changes to construct the trypsin-MCoTI-I interaction surface. The binding surface is contiguous and spans 46% of the total molecular area of MCoTI-I (Fig. 2b). As expected the major difference in the backbone dynamics was observed in the binding loop (Table 1), where the mobility in the ns-ps time scale was increased in MCoTI once bound to trypsin. Loop 1 showed an <S2> = 0.49 ± 0.02, which is much lower than the value for the rest of the molecule (<S2> = 0.65 ± 0.07). Several residues in loop 2 (Cys9, Arg10, Ser13 and Asp14), loop 3 (Gly17) and loop 5 (Cys21 and Arg22) also showed significantly lower values of S2 upon complex formation (Figs. 1d and 2c). It is likely that the increase in mobility observed in these loops may help to accommodate the increased flexibility of the binding loop (Fig. 2c).
Figure 2.
Trypsin binding to MCoTI-I affects the MCoTI-I backbone dynamics. Ribbon (a) and surface (b) diagrams of the trypsin-MCoTI-I interaction map. Red arabic numbers indicate the positions of the MCoTI-I loops. The MCoTI-I residues with a large chemical shift difference (>0.3 ppm) are in blue. (c) Changes in the MCoTI-I order parameter due to binding to trypsin. Residues with Sf2 - Sb2 > 0.2, where Sf/b2 is the order parameter of the free or trypsin bound MCoTI-I, respectively, are depicted in red. MCoTI-I residues that were broadened in {15N,1H}-HSQC due to binding to trypsin are shown in green. We used a structure of free MCoTI-II (PDB code: 1IB9)[6] to illustrate the changes of MCoTI-I dynamics due to trypsin binding.
Since our data clearly shows that backbone flexibility of MCoTI-I cyclotide increases significantly upon binding to trypsin, we decided to estimate the contribution of these motions to the overall Gibbs free energy of binding (ΔG). The energetic benefit of this increase in backbone flexibility can be estimated from the experimental relaxation data, using the experimentally measured order parameters, S2.[27] The estimated ΔG value was -62 kJ/mol at 298 K. This value is almost identical to the calculated value from the trypsin inhibitory constant of MCoTI-I (Ki ≈ 20 pM,[28] ΔG ≈ −61 kJ/mol ). The calculated entropic contribution (−TΔS) at the same temperature was −46 kJ/mol. These results highlight the importance of the backbone entropic term to the formation of the trypsin-MCoTI-I complex, although a more detailed thermodynamic analysis that also includes the side-chain motions may be required.
In summary, we report the backbone dynamics of the cyclotide MCoTI-I in the free state and complexed to its binding partner trypsin in solution. To our knowledge this is the first time the backbone dynamics of a natively folded cyclotide has been reported in the literature. This has been possible due to the use of modified protein splicing units for the heterologous expression of folded cyclotides using bacterial expression systems[17, 18, 29] to incorporate NMR-active nuclei such as 15N. Our results on the backbone dynamics of free cyclotide MCoTI-I confirm that MCoTI-I adopts a well-folded and highly compact structure with an <S2> value of 0.83. This value is similar to those found in the regions of well-folded proteins with restricted backbone dynamics. The results also indicate that the trypsin-binding loop (loop 1) has smaller S2 value than the averaged value for the whole molecule, indicating a higher mobility of this region in the ps-ns time scale. This region also showed significantly conformational exchange motions in the μs-ms time scale. Loop 6 also posses a higher mobility in the ps-ns time scale than the averaged value for MCoTI-I, although not significant conformational exchange motions were detected in the μs-ms time scale. This result is intriguing since this loop contains a potentially flexible Gly-Ser rich sequence that is mostly absent among other linear trypsin squash inhibitors and therefore it was thought to be a highly flexible linker to allow cyclization. More surprisingly, however, was the fact that the backbone of MCoTI-I, and specially loop 1, increased ps-ns mobility when bound to trypsin. This interesting result has been already observed in other high-affinity protein-protein interactions.[30, 31] The thermodynamic analysis of backbone contribution to the trypsin-MCoTI-I complex formation using measured S2 values also revealed the importance of the backbone entropic term in the formation of trypsin-MCoTI-I complex. Similar findings have been also found in other protease inhibitors.[32] This increment in backbone mobility may help to minimize the entropic penalties required for binding. Hence, we have also observed the appearance in the HSQC spectrum of the trypsin-MCoTI-I complex of a signal corresponding to the ε-NH3+ of Lys4 located in loop 1, which suggests that the amonium group is protected and more rigid when forming the complex (Fig. 3). This is the only Lys residue present in the sequence of MCoTI-I (Scheme 1) and therefore it can be unambiguously assigned. This residue is key for binding to trypsin[29] and is responsible for binding to the specificity pocket of trypsin. This cross-peak was totally absent in the free MCoTI-I sample indicating that the ε-NH+3 of Lys4 is less rigid, and rapidly exchanging with solvent (Fig. 3a). Similarly, the broadening of aliphatic resonances for Arg side-chains with essentially rigid guanidinium groups (i.e. εN-H bond vectors) has been also described in the literature for protein-peptide complexes.[26] Palmer III and co-workers have recently suggested that this dynamic decoupling between the side-chain terminus from the rest of the aliphatic part of the side-chain may be a general biophysical strategy for maximizing residual side-chain and potentially backbone conformational entropy in proteins and their complexes,[33] which is in agreement with our observations regarding the increase in MCoTI-I backbone mobility upon complex formation.
Figure 3.
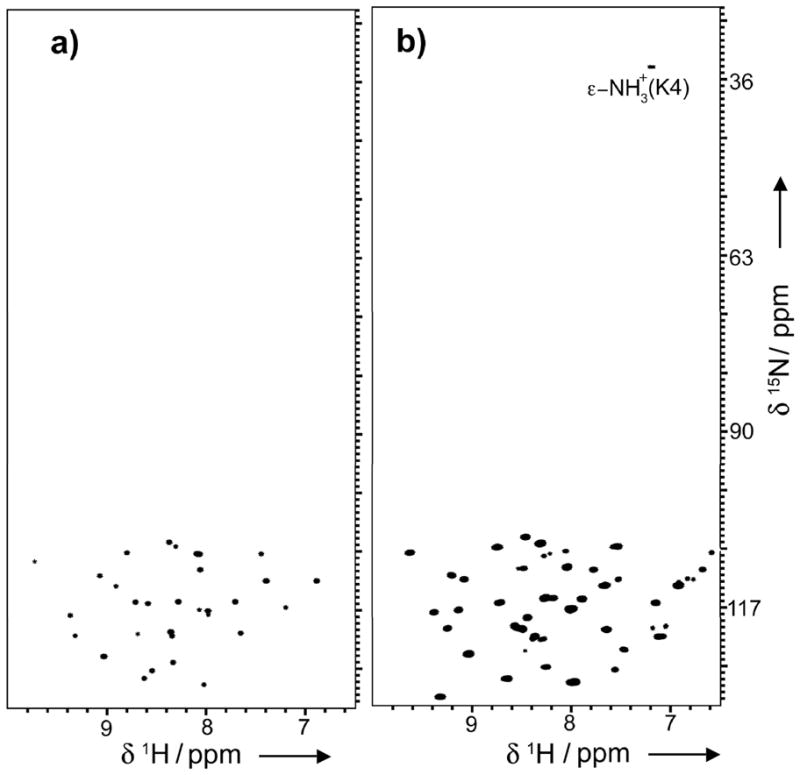
ε-NH3+ of Lys4 is protected from fast exchange with the solvent in trypsin bound MCoTI-I. {15N,1H}-HSQC spectra of free (a) and trypsin-bound MCoTI-I (b) were collected at RT with the 15N-carrier position at 82 ppm and 15N rf field strengths of 5.2 kHz for 90° and 180° pulses and 1.2 kHz for composite decoupling during acquisition.
We have also mapped the binding surface of MCoTI-I once bound to trypsin. The major changes in chemical shifts were observed for the solvent-exposed residues located in loops 1 (Lys4, Ile5, Arg8), 3 (Ala18), and 6 (Val1) (Figs. 1c and 2b). In agreement with these results, we have recently shown that the introduction of non-conservative mutations in these positions has a negative effect on the affinity for trypsin,[29] thus indicating that they may be in close contact with the protease at the binding interface of the molecular complex.
Cyclotides present several characteristics that make them appear as promising leads or frameworks for peptide drug design.[7, 8] Investigation of the backbone dynamics is crucial for a better understanding of the dynamic structural properties for the cyclotide scaffold and how it affects the mode of binding of these interesting molecules. The reported data will help in the design of cyclotide-based libraries for molecular screening and selection of de-novo sequences with new biological activities or developing grafted analogues for use as peptide-based drugs.[9,10]
Supplementary Material
Footnotes
This work was supported by funding from the School of Pharmacy at the University of Southern California and by National Institute of Health award GM090323-01 to J.A.C. and by funding from American Diabetes Association award 1-06-CD-23 to A.S.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Daly NL, Rosengren KJ, Craik DJ. Adv Drug Deliv Rev. 2009;61:918. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Craik DJ, Simonsen S, Daly NL. Curr Opin Drug Discov Devel. 2002;5:251. [PubMed] [Google Scholar]
- 3.Craik DJ, Cemazar M, Wang CK, Daly NL. Biopolymers. 2006;84:250. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Biochemistry. 2000;39:5722. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 5.Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, Le-Nguyen D, Chiche L. Biochemistry. 2001;40:7973. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 6.Felizmenio-Quimio ME, Daly NL, Craik DJ. J Biol Chem. 2001;276:22875. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 7.Clark RJ, Daly NL, Craik DJ. Biochem J. 2006;394:85. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craik DJ, Cemazar M, Daly NL. Curr Opin Drug Discov Devel. 2006;9:251. [PubMed] [Google Scholar]
- 9.Craik DJ, Daly NL, Mulvenna J, Plan MR, Trabi M. Curr Protein Pept Sci. 2004;5:297. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 10.Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Org Biomol Chem. 2008;6:1462. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 11.Jarymowycz VA, Stone MJ. Chem Rev. 2006;106:1624. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 12.Sancheti H, Camarero JA. Adv Drug Deliv Rev. 2009;61:908. doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ireland DC, Colgrave ML, Daly NL, Craik DJ. Adv Exp Med Biol. 2009;611:477. doi: 10.1007/978-0-387-73657-0_205. [DOI] [PubMed] [Google Scholar]
- 14.Camarero JA, Muir TW. J Chem Soc, Chem Comm. 1997;1997:1369. [Google Scholar]
- 15.Camarero JA, Pavel J, Muir TW. Angew Chem Int Ed. 1998;37:347. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Camarero JA, Muir TW. J Am Chem Soc. 1999;121:5597. [Google Scholar]
- 17.Kimura RH, Tran AT, Camarero JA. Angew Chem Int Ed Engl. 2006;45:973. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 18.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Chembiochem. 2007;8:1363. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 19.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4546. [Google Scholar]
- 20.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4559. [Google Scholar]
- 21.Horská A, Horsky J, Spencer RGS. J Magnetic Resonance. 1994;A110:82. [Google Scholar]
- 22.Liu J, Gong Y, Prakash O, Wen L, Lee I, Huang JK, Krishnamoorthi R. Protein Sci. 1998;7:132. doi: 10.1002/pro.5560070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitz A, Avrutina O, Le-Nguyen D, Diederichsen U, Hernandez JF, Gracy J, Kolmar H, Chiche L. BMC Struct Biol. 2008;8:54. doi: 10.1186/1472-6807-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Prakash O, Cai M, Gong Y, Huang Y, Wen L, Wen JJ, Huang JK, Krishnamoorthi R. Biochemistry. 1996;35:1516. doi: 10.1021/bi952466d. [DOI] [PubMed] [Google Scholar]
- 25.Szenthe B, Gaspari Z, Nagy A, Perczel A, Graf L. Biochemistry. 2004;43:3376. doi: 10.1021/bi035689+. [DOI] [PubMed] [Google Scholar]
- 26.Pascal SM, Yamazaki T, Singer AU, Kay LE, Forman-Kay JD. Biochemistry. 1995;34:11353. doi: 10.1021/bi00036a008. [DOI] [PubMed] [Google Scholar]
- 27.Palmer AG., 3rd Annu Rev Biophys Biomol Struct. 2001;30:129. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 28.Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Le Nguyen D, Sommerhoff CP, Diederichsen U, Kolmar H. Biol Chem. 2005;386:1301. doi: 10.1515/BC.2005.148. [DOI] [PubMed] [Google Scholar]
- 29.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Chembiochem. 2009;10:2663. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta K, Shi H, Cruz-Chu ER, Kami K, Ghose R. Biochemistry. 2004;43:8094. doi: 10.1021/bi030268d. [DOI] [PubMed] [Google Scholar]
- 31.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. Nature Str Biol. 2001;8:998. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arumugam S, Gao G, Patton BL, Semenchenko V, Brew K, Van Doren SR. J Mol Biol. 2003;327:719. doi: 10.1016/s0022-2836(03)00180-3. [DOI] [PubMed] [Google Scholar]
- 33.Trbovic N, Cho JH, Abel R, Friesner RA, Rance M, Palmer AG., 3rd J Am Chem Soc. 2009;131:615. doi: 10.1021/ja806475k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.