Abstract
The human body louse, Pediculus humanus humanus, has one of the smallest insect genomes, containing ~10,775 annotated genes (Kirkness et al. 2010). Annotation of detoxification [cytochrome P450 monooxygenase (P450), glutathione-S-transferase (GST), esterase (Est), and ATP-binding cassette transporter (ABC transporter)] genes revealed that they are dramatically reduced in P. h. humanus compared to other insects except for Apis mellifera. There are 37 P450, 13 GST and 17 Est genes present in P. h. humanus, approximately half of that found in Drosophila melanogaster and Anopheles gambiae. The number of putatively functional ABC transporter genes in P. h. humanus and A. mellifera are the same (36) but both have fewer than An. gambiae (44) or D. melanogaster (65). The reduction of detoxification genes in P. h. humanus may be due to their simple life history, where they do not encounter a wide variety of xenobiotics. Neuronal component genes are highly conserved across different insect species as expected due to their critical function. Although reduced in number, P. h. humanus still retains at least a minimum repertoire of genes known to confer metabolic or toxicokinetic resistance to xenobiotics (e.g., Cyp3 clade P450s, Delta GSTs, B clade Ests and B/C subfamily ABC transporters), suggestive of its high potential for resistance development.
Keywords: Pediculus, body lice, detoxification genes, insecticide resistance
Introduction
The body louse (Pediculus humanus humanus), an obligatory human ectoparasite, is a serious public health threat because it transmits a variety of human diseases, including epidemic typhus, relapsing fever and trench fever (Raoult and Roux, 1999). Unlike other blood-feeding pests such as mosquitoes, blood-sucking hemipterans, and ticks, human lice, including body and head lice (Pediculus humanus capitis), are unique in that they spend virtually their entire life cycle, from egg to adult, either directly on or in close proximity to their human hosts, have a simple life history, and feed exclusively on human blood. Human lice evolved to feed on humans approximately 5.6 million years ago, when the ancestors of chimpanzees and humans diverged (Reed et al., 2004). Body lice differ from conspecific head lice in their choice of habitat on human hosts. Body lice feed mostly on hairless parts of the body but lay their eggs in clothing whereas head lice live exclusively on the head in the hair near the scalp. DNA analysis suggests that body lice originated from head lice not more than approximately 72,000 to 42,000 years ago, when human began to wear clothing (Reed et al., 2004).
The human body louse possesses one of the smallest insect genomes sequenced to date. The size of the P. h. humanus genome (~108 Mb) is significantly smaller than that of Drosophila melanogaster (150 Mb), Anopheles gambiae (220 Mb), Aedes aegypti (800 Mb) and Apis mellifera (236 Mb). The P. h. humanus genome contains approximately 10,775 annotated genes (Kirkness et al. 2010), which is substantially fewer than the number reported for D. melanogaster (13,500 genes) and An. gambiae (14,000 genes) (Holt et al., 2002) but is comparable to that of A. mellifera (11,000 genes) (Honey Bee Genome Sequencing Consortium, 2006). Of the predicted P. h. humanus genes, 90% of the genes share homology and 80% of the genes show orthology to other sequenced insect genes, suggesting that the P. h. humanus genome is almost as complete as other insect genomes in terms of its encoded gene repertoire (Body Louse Genome Sequencing Consortium, unpublished data). Nevertheless, the relatively smaller number of genes may indicate that certain gene families have contracted or been lost in P. h. humanus due, in part, to evolutionary processes leading to its simple life history and its obligate parasitism of a single host species. Thus, gene families involved in responses to environmental variation may have been subject to negative selection in P. h. humanus during the evolutionary processes leading to a parasitic lifestyle.
In this study, we investigated whether families of detoxification genes that play essential roles in environmental interactions and defense against natural and synthetic toxins (Phase I-III xenobiotic metabolism) are expanded or contracted in the P. h. humanus genome. To this end, we have manually annotated and compared the detoxification genes from the P. h. humanus genome to other insect genomes, including the superfamilies of genes encoding cytochrome P450 monooxygenases (P450), glutathione-S-transferases (GST), esterases (Est), and ATP-binding cassette transporters (ABC transporter). We have also annotated and compared some neuronal channel genes that serve as target sites for a variety of insecticides (voltage-dependent sodium channel α-subunits (VDSC), sodium channel auxiliary subunits (TipE) and nicotinic acetylcholine receptors (nAChR)) for use as putatively preserved reference genes that should not have undergone reduction in numbers due to their unique and essential functions. Since pediculicide (insecticide) resistance in body and head lice has been frequently reported and poses a serious threat to public health (Pittendrigh et al., 2006), we have purposely focused our analysis on finding functional repertoires of P. h. humanus detoxification genes that are able to confer metabolic or toxicokinetic resistance to xenobiotics in order to determine their association with the innate potential for resistance development in P. h. humanus.
Results and Discussion
P450 superfamily genes
Annotation efforts revealed 37 putative P450 genes and one P450 pseudogene in the P. h. humanus genome -- the smallest P450 repertoire for any insect genome that has been sequenced (Table 1 and Fig 1). This number is smaller than that of A. mellifera (46 P450s) and substantially smaller than that of D. melanogaster (85 P450s) and An. gambiae (106 P450s) (Claudianos et al., 2006). Amino acid sequences are available for download from http://drnelson.utmem.edu. This reduced set of P450 genes includes obvious louse orthologs to the ecdysteroid biosynthetic genes found in D. melanogaster and other insects: CYP307A1, CYP306A1, CYP302A1, CYP314A1, CYP315A1, and CYP314A1 (Figure 1; Rewitz et al., 2007). An ortholog of the juvenile hormone epoxidase, CYP15A1, identified in the cockroach Diploptera punctata (Helvig et al., 2004), was also annotated in the louse. The louse genome includes a single P450 gene related to the mitochondrial CYP12 family, CYP301C1, a family associated with insecticide resistance in the house fly Musca domestica (Guzov et al., 1998) and in D. melanogaster (Brandt et al., 2002).
Table 1.
Number and class/clade distribution of the cytochrome P450, glutathione-S-transferase (GST), esterase (Est) and ATP-binding cassette (ABC) transporter genes from the genomes of P. h. humanus, A. mellifera, D. melanogaster and An. gambiae.
| Super-family | Class/clade/subfamily | P. h. humanus | A. melliferaa | D. melanogaster* | An. gambiae* |
|---|---|---|---|---|---|
| P450 | CYP4 Clade | 9 | 4 | 32 | 45 |
| CYP3 Clade | 12 | 28 | 36 | 42 | |
| CYP2 Clade | 8 | 8 | 6 | 10 | |
| Mitochondrial Clade | 8 | 6 | 11 | 9 | |
| Subtotal | 37 | 46 | 85 | 106 | |
| GST | Delta | 4 | 1 | 11 | 12 |
| Epsilon | 0 | 0 | 14 | 8 | |
| Omega | 1 | 1 | 5 | 1 | |
| Sigma | 4 | 4 | 1 | 1 | |
| Theta | 1 | 1 | 4 | 2 | |
| Zeta | 1 | 1 | 2 | 1 | |
| Unknown | 0 | 0 | 0 | 3 | |
| Microsomal | 2 | 2 | 1 | 3 | |
| Subtotal | 13 | 10 | 38 | 31 | |
| Est | Dietary/Detoxification | ||||
| A Clade | 3 | 8 | 0 | 0 | |
| B Clade | 0 | 0 | 2 | 14 | |
| C Clade | 0 | 0 | 11 | 2 | |
| Pheromone/Hormone-degrading | |||||
| D Clade | 0 | 1 | 3 | 0 | |
| E Clade | 1 | 3 | 3 | 4 | |
| F Clade | 0 | 0 | 2 | 4 | |
| G Clade | 0 | 1 | 0 | 4 | |
| H Clade | 1 | 0 | 4 | 9 | |
| Neuro/Developmental | |||||
| I Clade | 1 | 2 | 2 | 2 | |
| J Clade | 2 | 2 | 1 | 2 | |
| K Clade | 1 | 1 | 1 | 1 | |
| L Clade | 5 | 5 | 4 | 5 | |
| M Clade | 3 | 1 | 2 | 2 | |
| Subtotal | 17 | 24 | 35 | 51 | |
| ABC transporter | A subfamily | 2 | -c | 19 | 6 |
| B subfamily | 6 | - | 10 | 5 | |
| C subfamily | 5 | - | 12 | 14 | |
| D subfamily | 2 | - | 2 | 1 | |
| E subfamily | 1 | - | 1 | 1 | |
| F subfamily | 3 | - | 3 | 3 | |
| G subfamily | 13 (2)b | - | 15 | 12 | |
| H subfamily | 6 (1) | - | 3 | 2 | |
| uncharacterized | 2 | - | - | - | |
| Subtotal | 40 | 34 d | 65 | 44 | |
The GST and esterase data obtained from either Claudianos et al. (2006) whereas the ABC transporter data from Roth et al. (2001).
The number in parenthesis indicates the number of tentatively non-functional genes due to the lack of critical domains or functional sites.
Data not available.
Subtotal data from the Body Louse Genome Sequencing Consortium.
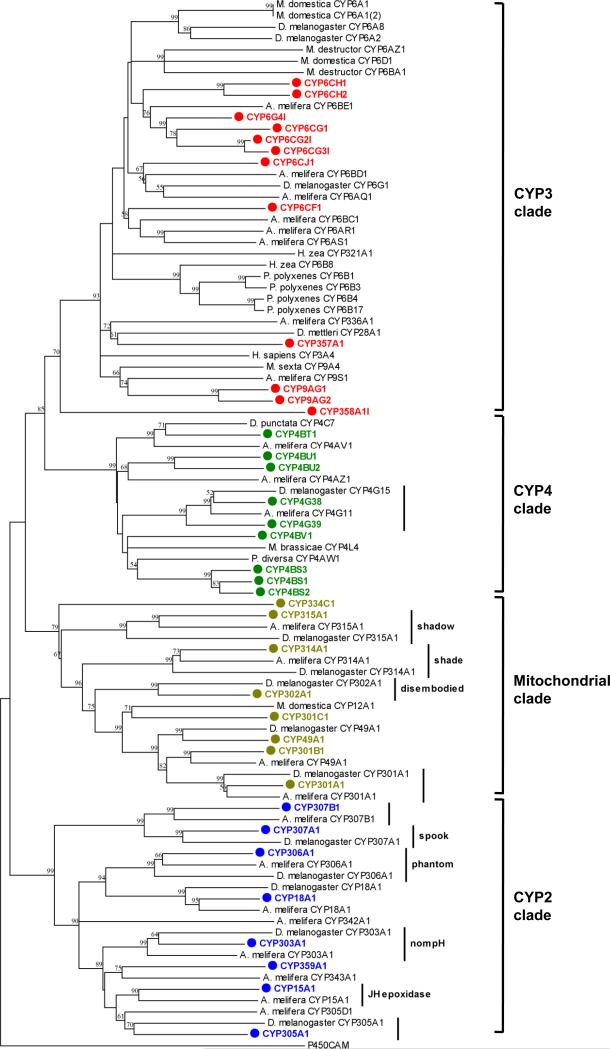
Figure 1.
Rooted, distance-corrected phylogenetic tree of all predicted cytochrome P450 proteins from P. h. humanus (in bold), all complete A. mellifera P450s (highly similar A. mellifera CYP6AS and CYP9 radiations are represented by single sequences), and select P450s from other insects with functions in hormone metabolism or detoxification. Deduced amino acid sequences were aligned using ClustalW (Thompson et al., 1994), and the alignment was used for the generation of tree by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). Putative hormone-related P450 orthologs are indicated with a line and the gene name in D. melanogaster. Groups of apparent orthologs with unknown function are indicated with a line, but no label. Nodes with >50% bootstrap support were only marked with percentage values.
The greatest P450 loss in the P. h. humanus genome, relative to other insects, appears in the CYP3 clade. Only 12 CYP3 genes are present in P. h. humanus compared to 36 in D. melanogaster and 28 in A. mellifera. Additionally, two of these sequences lack key amino acids at highly conserved sites. CYP358A1 lacks the characteristic cysteine residue in the highly conserved heme binding sequence (PfxxGxRxCxG/A) and CYP357A1 lacks the conserved glutamate in the ExLR motif in helix K (Feyereisen, 2005).
Because a single amino acid change can alter the catalytic activity of CYP3 clade P450s (Wen et al., 2005), inference of function for any P450 in this diverse clade, based on amino acid sequence alone, is at best speculative. Nonetheless, a number of CYP3 clade genes in other insects are known to code for P450s involved in detoxification of both natural toxins and pesticides. Both mammals and insects share this detoxification clade (Feyereisen, 2006) and the human liver P450, CYP3A4, is perhaps the most thoroughly characterized representative of this clade (Liu et al., 2007). Among the CYP6 family P450s, which are restricted in insects to the CYP3 clade, CYP6A1 and CYP6D1 contribute to pyrethroid resistance in Musca domestica (Carino et al., 1994; Liu & Scott, 1996), CYP6A2, CYP6A8 and CYP6G1 are important in DDT, malathion and neonicotinoid resistance in D. melanogaster (reviewed in Li et al., 2007), and CYP6Z2 and CYP6M2 are associated with pyrethroid resistance in An. gambiae (Muller et al., 2007). Host plant allelochemicals are detoxified by CYP6B1 and CYP6B3 in the caterpillar Papilio polyxenes (Wen et al., 2006), CYP6B4 and CYP6B17 in Papilio glaucus (Li et al. 2003), CYP6B33 in Papilio multicaudatus (Mao et al. 2007), CYP6B8 and CYP321A1 in the moth Helicoverpa zea (Rupasinghe et al., 2007), CYP6AZ1 and CYP6BA1 in the Hessian fly, Mayetiola destructor (Mittapalli et al., 2005) and CYP6AB3 in the moth Depressaria pastinacella (Mao et al., 2006). Other CYP3 clade member P450s are also reported to participate in detoxification reactions. CYP28A1 detoxifies cactus alkaloids in the cactophilic Drosophila mettleri (Danielson et al., 1997), and CYP9A4 expression is induced in the midgut of Manduca sexta larvae consuming host plant allelochemicals. Despite the low number of CYP3 clade P450s, it is likely that at least some of these 12 genes are involved in metabolic resistance to insecticides in Pediculus lice (Vassena et al., 2003).
The number of encoded CYP4 clade P450s (9 CYP4 genes) is also reduced in P. h. humanus compared with the 32 genes found in D. melanogaster. P450s in this clade are associated with metabolism of both endogenous and xenobiotic substrates (Feyereisen, 2006). For example, CYP4G15 may be involved in ecdysteroid biosynthesis in D. melanogaster (Maïbèche-Coisne et al., 2000) and CYP4C7 may serve to inactivate juvenile hormone in the cockroach, Diploptera punctata, (Sutherland et al., 1998). Other CYP4 genes, such as CYP4AW1 in Phylopertha diversa (Maïbèche-Coisne et al., 2004) and CYP4L4 in Mamestra brassicae (Maïbèche-Coisne et al., 2002), are associated with the breakdown of odorants in chemosensory structures. It is interesting that the louse genome encodes more CYP4 P450s than are found in the A. mellifera genome (with only 4 CYP4 genes), despite the dependence of honey bees on chemical communication (Claudianos et al., 2006). These observations suggest that the large complement of CYP4 P450s in the body louse have functional diversity beyond odorant metabolism.
As in other insect genomes, highly similar groups of CYP3 and CYP4 clade P450s appear to have arisen via recent gene duplication events (Feyereisen, 2005). Louse CYP4BS1, CYP4BS2 and CYP4BS3, which share at least 75% amino acid similarity, are found adjacent to each other on a 9 kb region of the same genomic scaffold. Three additional clusters of similar P450s are similarly found on the same scaffolds: (i) CYP4BU1 and CYP4BU2; (ii) CYP6CH1, CYP6CG2 and CYP6CG3; and, (iii) CYP9AG1 and CYP9AG2. The head-to-head arrangement of CYP306A1 and CYP18A1 is in synteny with the orthologs to these genes in D. melanogaster and A. mellifera, where a similar localization of the genes is evident (Claudianos et al., 2006).
Given the substantial reduction in encoded P450 genes relative to other insects, the louse genome is likely to provide insights into the minimum complement of P450s required for insect adaptation to environmental selection pressures. Comparison with a similarly reduced complement of P450s found in the honey bee may be of particular value in determining the minimum required set of insect P450s, as orthologs between the body louse and the honey bee may participate in as yet uncharacterized hormone metabolism pathways and/or play novel roles in other endogenous functions.
GST superfamily genes
A total of 11 putative cytosolic and two microsomal GST genes were observed in the P. h. humanus genome (Table 1 and Fig. 2). This number is comparable to that found in A. mellifera (10 GSTs), but substantially smaller than that of D. melanogaster (38 GSTs), An. gambiae (31 GSTs), and T castaneum (>34 GSTs) (Claudianos et al., 2006). The eleven cytosolic GSTs in P. h. humanus can be further categorized into five classes: four GSTs in Delta class, four in Sigma, and one each in the Theta, Omega, and Zeta classes (Fig. 2). No GST belonging to Epsilon class has been identified, as is the case for A. mellifera. Sequence identities within the Delta and Sigma classes are 18.2~38.9% and 35.1~45.2%, respectively, indicating that Sigma GSTs have more recently diversified.
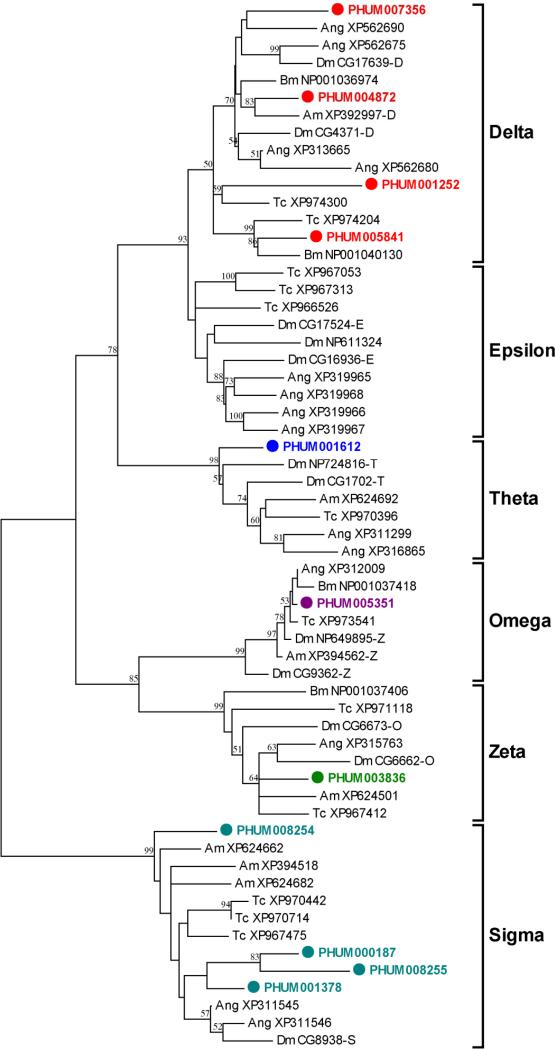
Figure 2.
Unrooted phylogenetic tree of the predicted GST proteins of P. h. humanus, D. melanogaster (Dm), An. gambiae (Ang), A. mellifera (Am) and T. castaneum (Tc). Deduced amino acid sequences were aligned using ClustalW (Thompson et al., 1994), and an unrooted tree was created from the alignment by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). Microsomal GSTs were not included in the tree. Six classes (Delta, Epsilon, Omega, Sigma, Theta and Zeta) of GST are marked. The sequence of PHUM008255 used for the alignment lacks the third exon sequences due to an unresolved nucleotide sequence stretch in the current CDS sequence release (phumanus.CDS-TIGR.PhumU1.0). Sequences from other insects except P. h. humanus are given with NCBI accession number. Nodes with >50% bootstrap support were only marked with percentage values.
The Delta and Epsilon classes of GSTs are unique to insects and are thought to contribute to the adaptation of insects to environmental variation (Ranson et al., 2002). Additionally, the Delta and Epsilon classes of GSTs are directly involved in insecticide detoxification, thereby conferring resistance to various insecticides (Ranson et al., 2002; Enayati et al., 2005). These two classes show a substantial expansion in D. melanogaster and An. gambiae but not in P. h. humanus. The lack of expansion of the Delta and Epsilon classes in P. h. humanus may be due to the relatively reduced ecological and environmental selection pressures that occur within the simple habitat of the external surface of the human body. Nevertheless, the relative abundance of Delta class GSTs with respect to total GSTs (30.7%) in P. h. humanus is similar to that reported for D. melanogaster (28.9%) and An. gambiae (37.7%), and is, in fact, substantially higher than that of A. mellifera (10%). This finding suggests that P. h. humanus possesses a higher potential for detoxification of xenobiotics, including insecticides, than A. mellifera. Indeed, GSTs have been implicated as a DDT resistance mechanism in P. h. capitis (Hemingway et al., 1999) and may function in permethrin resistance (Enayati et al., 2005), indicating that GSTs are actively exploited in the evolution of insecticide resistance in Pediculus lice. Two Delta class GSTs (PHUM004872 and PHUM007356), found on the same scaffold adjacent to each other with 38.4% sequence identity, likely result from a relatively recent local gene duplication.
There are four Sigma GSTs in P. h. humanus, which is three more than the number found in D. melanogaster and An. gambiae (Table 1 and Fig. 2). The increased relative abundance of Sigma GSTs in P. h. humanus is due mainly to local gene duplication events, as three out of four genes (PHUM008254, PHUM008255, and PHUM001378) are found in a cluster. Similar arrangements occur in A. mellifera and T. castaneum genomes, both of which contain greater numbers of Sigma GSTs (Claudianos et al., 2006). Moreover, all of the Sigma GSTs in P. h. humanus, as is the case in A. melifera, lack the hydrophobic N-terminal region that is common in most other insect Sigma GSTs. Evidence exists that insect Sigma GSTs play structural roles in flight muscle (Clayton et al., 1998), protective roles against oxidative stress (Singh et al., 2001), and metabolic roles in processing endogenous substrates (Mittapalli et al., 2007) as well as xenobiotics (Yamamoto et al., 2007). Taken together, the Sigma GSTs in P. h. humanus are likely to play general defensive roles in a broad sense. It remains unclear, however, what factors may have led to the greater diversification of Sigma GSTs in some insect species, such as P. h. humanus, A. mellifera and T. castaneum, beyond their obvious differences in life histories and ecological niches.
The relatively similar distribution of genes in the remaining classes of GSTs (Omega, Theta, and Zeta) across different insect species, including P. h. humanus, suggests that they have more common house-keeping functions in conserved physiological processes, including the metabolism of endogeneous substrates and cellular defense against oxidative stress. Only a single gene is present in the Omega and Zeta classes of GSTs in P. h. humanus, A. mellifera and An. gambiae, revealing a tight one-to-one orthology in these insects. In contrast, D. melanogaster has three genes in the Omega class and two genes in the Zeta class, suggesting a possible functional diversification of these GSTs. The Theta class of GSTs also shows moderate expansions in Diptera species (4 GSTs in D. melanogaster and 2 GSTs in An. gambiae) compared to only one in both P. h. humanus and A. mellifera. It still remains to be elucidated, however, why these groups of GSTs are particularly expanded in dipterans, such as D. melanogaster and An. gambiae.
In summary, the GST composition in P. h. humanus genome is very similar to that of A. mellifera, except that P. h. humanus contains three more Delta GSTs, a finding consistent with an enhanced capacity for metabolic resistance against insecticides in P. h. humanus.
Est superfamily genes
Searching Est superfamily genes that contain esterase homology domains identified 17 putative esterase genes, a substantially smaller number than have been identified in other insects, such as D. melanogaster (35 Ests), An. gambiae (51 Ests) and A. mellifera (24 Ests) (Claudianos et al., 2006). The 17 Ests are categorized into three classes: the first class (Clades A-C) contains primarily intracellular esterases with dietary detoxification functions; the second class (Clades D-H) contains secreted and catalytically active esterases, including juvenile hormone esterases (JHEs); and the most ancient third class (Clades I-M) contains esterases with neuro/developmental function, including acetylcholinesterases (AChEs) (Claudianos et al., 2006). In contrast with D. melanogaster and An. gambiae, no apparent large clusters of Ests are found in P. h. humanus, suggestive of little independent expansion of paralogous Ests. Two catalytic Ests (PHUM005821 and PHUM006997) are found adjacent to each other in a genomic scaffold but their relatively low amino acid sequence similarity (23.5%) implies a relatively ancient gene duplication event. Similarly, two non-catalytic Est genes (217.m01901 and 217.m001826 in TIGR annotation) are also located adjacent to each other in a tail-to-tail arrangement and share similar sequence identity (28.3%), indicating another gene duplication event. The highest amino acid sequence identity was found between PHUM005821 and PHUM005138 (53.9%). Nevertheless, they were substantially separated from one another based on their location in separate genomic scaffolds. The relatively low sequence identity among Ests in pair-wise comparison (average 16.3%) and their scattered localization in different genomic scaffolds suggest that there has been no recent gene duplication in the Est family, as is also thought to be the case for A. mellifera (Claudianos et al., 2006).
As shown in Table 1 and Fig. 3, three Est genes (PHUM005821, PHUM005138, and PHUM000301) are placed in the dietary class (Clade A). All of the P. h. humanus dietary Ests are localized in Clade A. The number of dietary class Est in P. h. humanus is substantially smaller than that of other insects, perhaps reflecting the monophagous human blood-feeding nature of the body louse throughout its entire life cycle. The substantially increased number of dietary class Ests in An. gambiae (16) is likely due to the polyphagy it exhibits throughout its holometabolous life cycle.
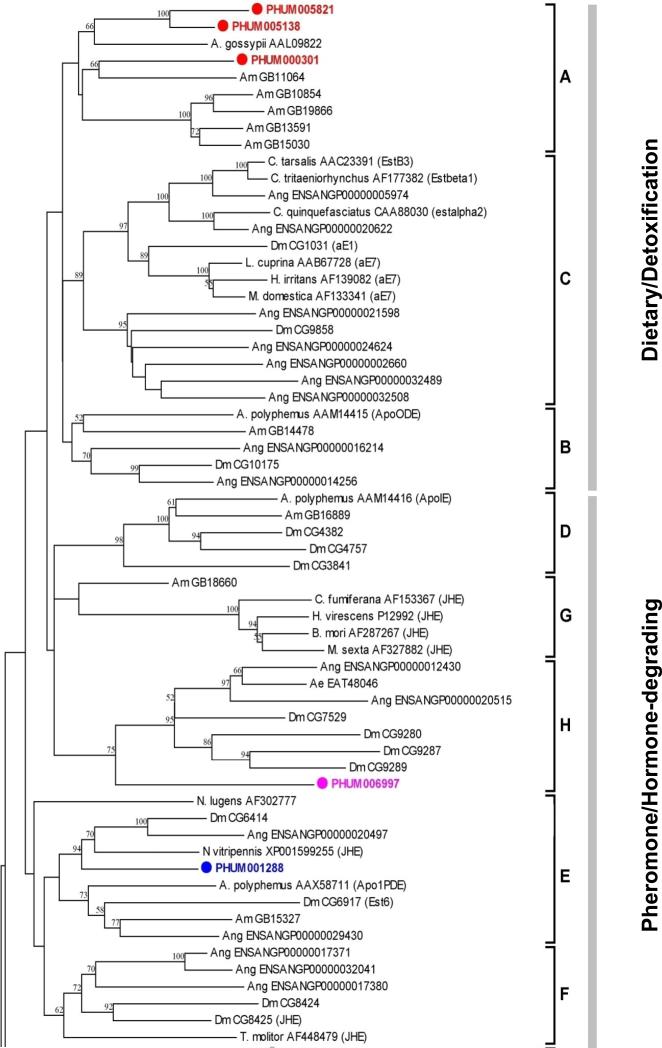
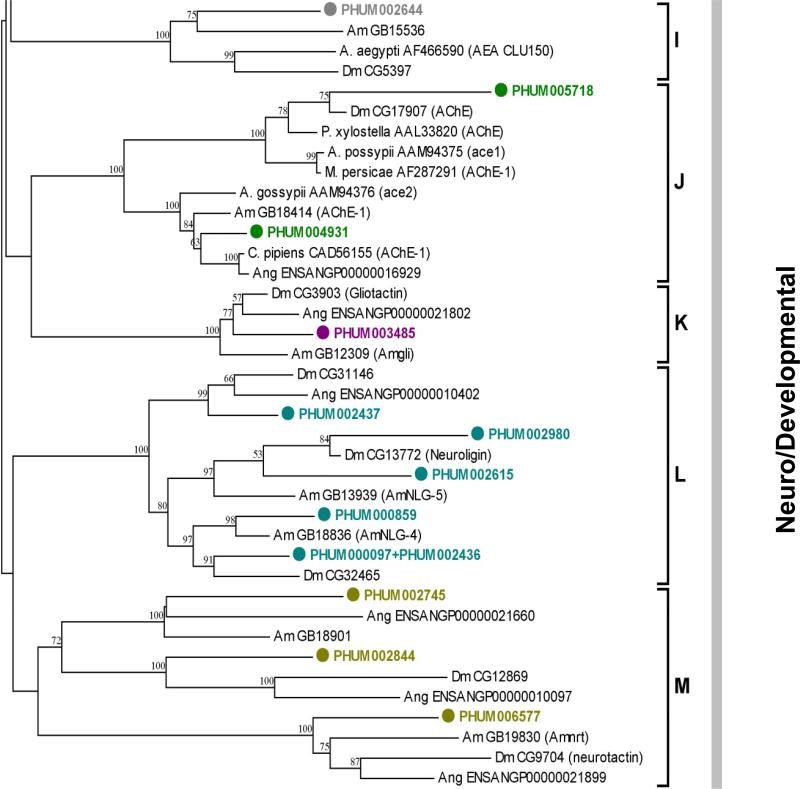
Figure 3.
Unrooted phylogenetic tree of the predicted Est proteins of P. h. humanus, D. melanogaster (Dm), An. gambiae (Ang), A. mellifera (Am), T. castaneum (Tc) and other several insect species. Sequences from other insects except P. h. humanus are given with NCBI accession number. Deduced amino acid sequences were aligned using ClustalW (Thompson et al., 1994). The highly variable amino and carboxy terminal extensions of all the neuro/developmental esterase class were excluded for better alignment (Claudianos et al., 2006). The resulting sequences, spanning the core region (amino acid residues 65–558) of D. melanogaster AChE (CG17907-PA), was used for the generation of the tree. The trimmed alignment was used for the generation of tree by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). The D. melanogaster serine protease (CG1505-PA) was used as an outgroup. Three esterase classes containing a total of 13 major clades are indicated on the right of the tree. Nodes with >50% bootstrap support were only marked with percentage values.
The unique expansion of Clade B dietary Ests in An. gambiae may also be due, in part, to the marked differences in life history and ecology compared to P. h. humanus. The absence of any Clade A Est in P. h. humanus, as is also the case in D. melanogaster and An. gambiae, supports the notion that this group of Ests specifically diversified in hymenopterans, including A. mellifera (Claudianos et al., 2006). One P. h. capitis Est, which is orthologous to the P. h. humanus PHUM005138 belonging to this class, is associated with malathion resistance in the head louse (Lee and Clark, unpublished data), suggesting that the dietary class of Ests is also exploited in the evolution of insecticide resistance in Pediculus lice. Three dietary Ests (PHUM005821, PHUM005138, PHUM00031) are categorized as serine-hydrolases by the presence of the catalytic triad (serine, glutamic acid and histidine) and an additional conserved serine residue near the catalytic triad (Thomas et al., 1999).
Catalytic triad consensus motifs (-GESAG-, -EGL-, and –C/SHG/AD-) and other essential active site oxyanion hole motifs (-GGG/A- and –GESAG-) are well conserved in these Ests (Fig. 4). These Ests are clearly distinguished from putative P. h. humanus acetylcholinesterases in that they lack the conserved choline binding tryptophan and tyrosine residues in the N-terminal region but have the three highly conserved basic contact residues in the amphipathic helix that are specific to most Ests (Fig. 4). Some of the acyl binding pocket residues (W251, Val307, Phe309, and Phe421 in the Lucilia cuprina E3 carboxylesterase; Heidari et al., 2004) are not conserved, suggesting they may have different substrate specificities. Signal peptide prediction for the three serine-hydrolases suggests that only PHUM000301 is a secretory enzyme. The other two closely related Ests (PHUM005821 and PHUM005138) do not have signal peptide sequences, suggesting a cytosolic origin.
Figure 4.
Amino acid sequence alignment of tentative catalytic ESTs from P. h. humanus. Conserved catalytic triad residues and one additional serine residue are marked with yellow bars. Conserved GE/QSAG motif region is marked by a dotted blue box. Highly conserved oxyanion hole residues are indicated with red boxes whereas less conserved acyl binding pocket residues are marked with green boxes. Consensus choline binding residues that are specific to acetylcholinesterase are indicated by purple boxes. The locations of three basic amino acid residues forming the amphipathic helix, which are conserved in Lepidopteran JHEs, are marked with blue lines above sequences. Parts of N-terminal and C-terminal regions of the genes were not included for better alignment. Due to unresolved sequences in parts of PHUM004931 in the PhumU1.1 version, previously reported P. h. humanus ace2 gene sequences (BAF46104) were adapted for alignment.
P. h. humanus appears to possess only two Ests in the pheromone/hormone processing class, in marked contrast with other insect groups, which usually retain 5~12 such Ests. One of the Ests (PHUM001288), categorized as a Clade E (pheromone esterase) Est, is closely related to the juvenile hormone esterase (JHE) from Nasonia vitripennis according to a BlastP search analysis. Additionally, an amino acid sequence alignment revealed that it has three basic amino acid residues (lysine) forming the amphipathic helix located in the active site, which are conserved in lepidopteran JHEs (Thomas et al., 1999; Campbell et al., 2001) (Fig. 4). PHUM001288 lacks, however, the JHE-specific consensus GQSAG nucleophilic elbow motif in its predicted active site, which is highly conserved in most insect JHEs. Instead, it has the GESAG consensus motif that is universal in most other general Ests. The glutamine residue in the GQSAG motif is predicted to shorten the hydrogen bond between the histidine and glutamic acid residues in the catalytic triad, thereby increasing the affinity for the juvenile hormone substrate (Thomas et al., 1999). The other Est (PHUM006997) in the pheromone/hormone processing class, however, does have the GQSAG motif. Although it is the only Est possessing the GQSAG consensus motif out of the 18 Ests of P. h. humanus, PHUM006997 does not have the three basic contact residues in the amphipathic helix that are critical for its function as a JHE. In addition, it shows a higher sequence identity (36.2%) to the N-terminal Est domain of Drosophila glutactin (CG9280) than to any known insect JHEs. Taken together, PHUM001288 appears to be a better candidate for the functional P. h. humanus JHE but this aspect remains to be confirmed.
Glutactin is a basement membrane-related glycoprotein, which is particularly abundant in the embryonic envelope of the central nervous system, muscle apodemes and dorsal median cell processes, and is involved in intercellular ordering and adhesion (Olson et al., 1990; Darboux et al., 1996). The N-terminal domain of glutactin resembles serine hydrolases but is catalytically inactive due to the lack of the serine residue in the catalytic triad. Although PHUM006997 is localized to Clade H together with Drosophila glutactin ortholog, this is due principally to its high sequence similarity to the N-terminal domain of glutactin. Unlike glutactin, however, it lacks the glutamine and glutamic acid-rich acidic C-terminal domain, has a signal peptide at the N-terminus and retains the catalytic triad residues, suggesting its catalytic function as a secretory Est.
The lack of most pheromone-degrading Ests associated with Clade D in the P. h. humanus genome may be related to its simple life history and ecology on the highly specialized habitat of its human host, where sophisticated chemical communication via pheromones may not be required.
In general, Est genes that appear catalytically active share common structural properties in terms of their exon-intron arrangements (7~9 exons) and in the conservation of relatively short introns (average 114 bp).
In contrast with the first and second classes of Ests, which show a dramatic reduction in gene number, there are 12 Ests localized in the Clade I-M neuro/developmental class, including the acetylcholinesterase (Clade J). This number is very similar to the corresponding gene numbers in other insects (10, 12, and 11 in D. melanogaster, An. gambiae, and A. mellifera, respectively), indicating that the neuro/developmental class Ests have evolutionarily conserved roles in insects. Except for cyclorraphan Diptera including Drosophila spp., P. h. humanus, like most other insect species, has two putative acetylcholinesterase (AChE) genes, ace1 and ace2. Both ace1-type (PHUM004931) and ace2-type (PHUM005718) genes in P. h. humanus have all the conserved amino acid residues forming the catalytic triad, oxyanion hole, acyl binding pocket, and choline binding site, suggesting that both express catalytically sufficient AChEs. The ace1-type PHUM004931 has been reported to be more predominantly expressed than the ace2-type PHUM005718 (Lee at al., 2007), indicating that P. h. humanus expresses the major form of AChE actively involved in the modulation of synaptic cholinergic communication.
There is one gliotactin-orthologous gene (PHUM0034850) in P. h. humanus that is located in Clade K, a finding that matched that reported for A. mellifera, D. melanogaster and An. gambiae. Gliotactin is a catalytically inactive cholinesterase-like protein that is expressed in glial cells and has the properties of adhesion proteins in D. melanogaster (Auld, 1995). It plays a critical role in the formation of the glial-based blood-nerve barrier, thus ensuring proper insulation between nerves and hemolymph in insects (Auld, 1995). The presence of a single gliotactin ortholog across a variety of insect species implies that it has been evolutionary well conserved in insect taxa. Although one gene (PHUM002745) is located in a branch near gliotactin (Fig. 3), it, along with PHUM002644, forms a group with some other genes previously categorized as Clade I (uncharacterized neuro/developmental group).
Like gliotactin, both neuroligin and neurotactin also posses a large cholinesterase-like extracellular domain, a transmembrane region and a protruding cytoplasmic domain and are involved in cell-cell adhesion and signaling (Soreq and Sedman, 2001). P. h. humanus appears to have five neuroligins that are localized to Clade L. Among these, two genes (217.m01901 and 217.m001826) are adjacent to each other and share a similar feature of having long introns, indicative of gene duplication. Other neuroligins are found on different genomic scaffolds, which indicate that they are dispersed in different locations in the genome. Two neurotactin orthologs that are located in Clade M are present in P. h. humanus genome. They are found on separate genomic scaffolds but share the attribute of having 10-12 short introns dispersed evenly over a 3.3-3.6 kb gene segment. Their low sequence identity (14.9%), however, indicates that they diverged a relatively long time ago.
Taken together, the neuro/developmental class Ests in P. h. humanus genome is similar in composition to those in other insect species, suggesting that these Ests play essential and housekeeping roles in insects across different taxa and have been conserved.
ABC transporter superfamily genes
ABC-transporters are members of a very large and ancient superfamily, which are preserved in all forms of life, including insects (Dassa and Bouige, 2001). ABC transporters are involved in many cellular processes, including the export or import of a wide variety of substances across biological membranes, such as metabolic products, lipids and xenobiotics. They are responsible for the widely known phenotype of multi-drug resistance (MDR) in cancer cells and bacteria. Typical ABC transporters contain two transmembrane domains (TMs), each of which consists of 6-11 α-helix transmembrane segments, and two ATP-binding domains (ABDs) (Dassa and Bouige, 2001). Classification of ABC transporters is based on the sequence and organization of their ABDs.
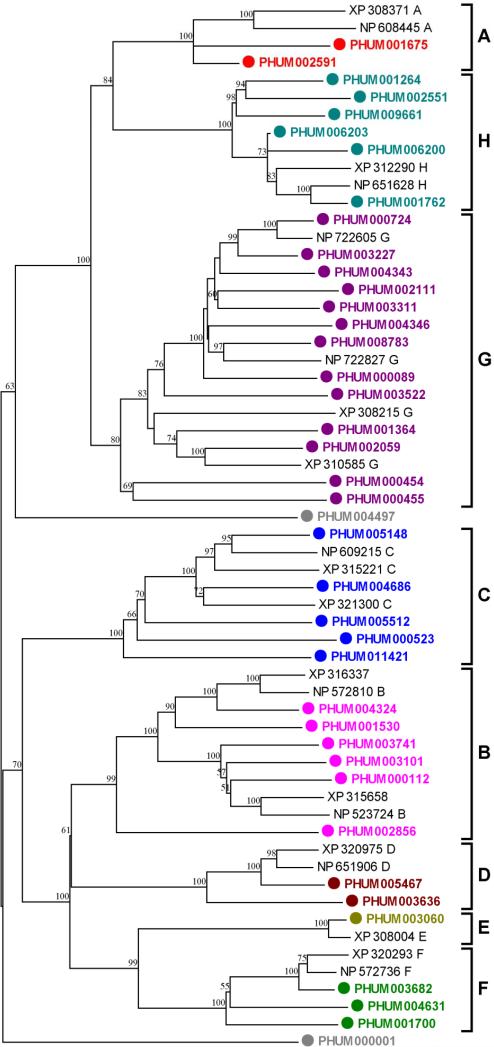
We found a total of 40 ABC transporters in the annotated protein set derived from the P. h. humanus genome. Among these, four appear to be nonfunctional in that they lack the critical ATP-binding sequence motif in the ABD domain. If they are in fact nonfunctional, the number of functional ABC transporters in P. h. humanus is the same as that reported for A. mellifera (36 ABC transporters) but fewer than that reported for An. gambiae (44 ABC transporters) and substantially fewer than that in D. melanogaster (65 ABC transporters) (Roth et al., 2003) (Table 1). Phylogenetic tree construction with some of the known ABC transporters from D. melanogaster and An. gambiae demonstrates that P. h. humanus ABC transporters are localized to the standard ABC subfamilies (A-H) based on the nomenclature convention for human ABC genes (Nomenclature for Human ABC-Transporter Genes; http://www.gene.ucl.ac.uk/nomenclature/genefamily/abc.html) (Table 1 and Fig. 5). Comparing the number of genes in each subfamily reveals that the underrepresentation of ABC transporters in P. h. humanus is due primarily to the significant reduction of those in the subfamilies A and C (ABCA and ABCC). The average pairwise sequence identity between ABC transporters is substantially low (14.9%), indicating that they are ancient in origin and have been preserved over time. Multiple gene duplication events are evident in three different lineages (1 in ABCA, 2 in ABCG, and 2 in ABCH lineages), indicating an ongoing process of gene evolution.
Figure 5.
Phylogenetic relationships of P. h. humanus ABC transporters with some representative homologues from D. melanogaster and An. gambiae. Deduced amino acid sequences of ABC transporters were aligned using ClustalW (Thompson et al., 1994) and the variable amino and carboxy terminal extensions (amino acid residues XX-YY in ZZZ) were removed. The trimmed alignment was used for the generation of an unrooted tree by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). The ABC transporters of D. melanogaster and An. gambiae are indicated with NP and XP prefix accession numbers, respectively. Seven ABC transporter subfamilies are indicated on the right of tree. Nodes with >50% bootstrap support were only marked with percentage values.
ABC transporters conferring the MDR phenotype are of particular interest because they provide a fundamental cellular defense system, which has often been associated with insecticide resistance in insects. At least three subfamilies of ABC transporters confer the MDR phenotype (Cole and Deeley, 1998). P-glycoproteins (P-gps) that belong to the B subfamily (ABCB) efflux hydrophobic compounds from the cell, thereby playing a critical role in the intrinsic defense mechanism against hydrophobic xenobiotics, such as insecticides (Sheps et al., 2004; Sarkadi et al., 2006). Macrocyclic lactones that function as anthelmintics, including ivermectin, are high affinity substrates for P-gps and their overexpression has been associated with ivermectin resistance in parasitic nematodes (Prichard and Roulet, 2007). P-gps are also associated with insecticide resistance in the tobacco budworm (Heliothis virescens) by reducing cuticular penetration of thiodicarb, a carbamate insecticide (Lanning et al., 1996). Overexpression of P-gps is also involved in insecticide resistance of Helicoverpa armigera larvae (Srinivas et al., 2004). The number of functional ABCB type transporters in P. h. humanus (6) is comparable to those reported for D. melanogaster (10) and An. gambiae (5), suggesting that P. h. humanus has maintained a sufficient number of P-gp type ABC transporters to provide defense and/or drug resistance against xenobiotics. All of the P. h. humanus ABCB transporters have the typical topology of either 2(TM + ABD) or TM + ABD. All P. h. humanus ABCB transporters have both the consensus ATP-binding sequence motifs (-GXSGS/T/CGKS/T-) in the ABD domains and ABC-2 type signature motifs (-SGGQ/EKQRIAIARAL/V-), suggesting that they are functional transporters.
The second group of ABC transporters that are similarly responsible for the MDR phenotype belongs to the subfamily C (ABCC type). The MDR mediated by the ABCC proteins has a slightly different mechanism in that they co-transport toxic xenobiotics as organic anion conjugates (e.g., glutathione conjugates) (Cole and Deeley, 1998; Ballatori et al., 2005). P. h. humanus has a substantially reduced number of ABCC type transporters (5) compared to the two dipterans (12 in D. melanogaster and 14 in An. gambiae, Table 1). Phylogenetic analysis reveals that the P. h. humanus ABCC transporters form a separate clade from the B, D, E and F subfamilies, suggesting that they are relatively more ancient and have not been expanded or lost over the course of evolution, perhaps due to the simplified life history of P. h. humanus. The typical topology of 2(TM + ABD) is preserved in all five ABCC transporters in P. h. humanus. Because ABCC transporters are largely responsible for glutathione conjugate export that is associated with the detoxification of harmful electrophilic compounds, the loss of ABCC proteins may similarly be linked to the same coevolutionary processes that resulted in the loss of a substantial number of GSTs in P. h. humanus.
Finally, some G subfamily ABC transporters (ABCG type) also exhibit broad substrate specificity for xenobiotic compounds and are associated with MDR in humans (Sarkadi et al., 2006; Kusuhara and Sugiyama, 2007). Other ABCG transporters play a crucial role in efflux of membrane lipids such as cholesterol (Kusuhara and Sugiyama, 2007). Unlike typical human ABCG transporters with the topology of ABD-TM, all 13 ABCG transporters in P. h. humanus contain only a single ABD domain without the TM domain, suggesting that they are not involved in MDR but function in some other cellular processes and sequestration. The number of P. h. humanus ABCG transporters (13) is similar to the number found in D. melanogaster (15) and An. gambiae (12), suggesting that they are conserved across species. Four ABCG genes (PHUM004343 and PHUM004346; PHUM000454 and PHUM000455) are found in two separate clusters. Among these, PHUM000454 and PHUM000455 are located in a tail-to-tail arrangement and both appear nonfunctional, suggesting that the parental gene lost its function before the duplication event.
The number of ABCA transporters is dramatically reduced in P. h. humanus (2) compared to D. melanogaster (19) and is one third of that found in An. gambiae (6). ABCA transporters are involved primarily in lipid trafficking in a wide range of eukaryotic cells and tissues (Dassa and Bouige, 2001), suggesting that they play essential roles in lipid homeostasis. It remains to be elucidated why this subfamily is greatly underrepresented in P. h. humanus and perhaps in An. gambiae.
The remaining ABC transporter subfamilies, including ABCD, ABCE and ABCF, show high degrees of conservation across different insect species, indicative of their housekeeping functions. A slightly increased number of ABCH transporters are present in P. h. humanus (6-7) when compared to the two dipteran species (3 in D. melanogaster and 2 in An. gambiae) (Table 1).
In summary, the overall decrease in the number of ABC transporters is consistent with the general tendency toward detoxification gene reduction in P. h. humanus. This reduction is not as dramatic, however, as that seen in the other detoxification gene superfamilies, such as P450s, GSTs and ESTs. Interestingly, P. h. humanus still retains a similar repertoire of ABCB transporters that are most responsible for MDR phenotype, suggesting that it has a comparable potential for the evolution of toxicokinetic resistance against pediculicides.
Neuronal channel superfamily genes
Genes encoding neuronal components, such as VDSC, TipE homologue and nAChR, have been identified in the P. h. humanus genome. Two VDSC genes (PHUM001860 and PHUM001207), which are orthologous to para and NCP60E (CG9071) sodium channel α-subunits from D. melanogaster, respectively, were found in the P. h. humanus genome (Supplementary Fig. 1A). This inventory is identical to that of other sequenced insect genomes, including An. gambiae, A. mellifera, and T. castaneum, in which single orthologs for each VDSC are present (data not shown). P. h. humanus also possess five homologues to the Drosophila tipE gene, also known as the insect sodium channel auxiliary subunit gene (Supplementary Fig. 1B). Each gene of the tipE family is represented by a single orthologous gene in all of the insect genomes examined, demonstrating its high degree of conservation across insects. All five P. h. humanus tipE homologues are present in a cluster, indicating ancient gene duplication events. Similar genomic clustering of tipE homologous genes is also apparent in other insects, including D. melanogaster, An. gambiae, and A. mellifera (Derst et al., 2006), and suggests that gene duplication events occurred long before the diversification of insect taxa.
A total of nine genes homologous to nAChRs were found in P. h. humanus. The putative nAChR genes were further categorized into eight groups (a single gene in each of the groups Dα1, Dα2, Dα3, Dα4, Dβ1, Dβ2, and more distantly related Dβ3 versus two genes in groups Dα5-7) (Jones et al., 2007) (Supplementary Fig. 1C). Other insects, including D. melanogaster, An. gambiae and A. mellifera, possess ten nAChRs and their distribution is very similar to that of P. h. humanus (Jones et al., 2007). The difference in gene number between P. h. humanus and other insects is due to the makeup of groups Dα5-7 (two in P. h. humanus versus three in other insects). The remarkable similarity in the gene number and composition of nAChRs suggests that they are highly conserved across insect taxa even with remarkably different life history and ecology, reflecting their evolutionarily retained function. In summary, unlike most of the detoxification genes, all of the examined neuronal component genes are highly conserved across different insect taxa and do not reflect the great differences in their life history, physiology, ecology, and environmental settings. These findings are consistent, however, with the unique and essential functions that they perform in the insect nervous system.
Transcription factor binding motifs
In order to provide future researchers with core information on potential regulation of P450s, GSTs, and Ests in P. h. humanus, we analyzed their putative transcription factor binding motifs. There were four potential transcription factor binding motifs (TFBMs) (Hand1:E47-like, SOX-9-like, MAZR-like, and NF-kappaB-like, Supplementary Fig. 2) associated with the P450, three motifs (GATA-1-like, PPAR-like, and SF-1-like, Supplementary Fig. 3) associated with the GST, and five potential motifs (FOXJ2-like, Sp3-like, STAT1-like, FOXO3-like, and ISRE-like, Supplementary Fig. 4) observed with the Est genes in P. h. humanus. Because we had a group of potential TFBMs with similarities to those observed in mammals, we named these potential TFBMs in P. h. humanus using the term from the similar mammalian motifs followed by “-like”. Some of the potential TFBMs that we observed are similar to known TFBMs in mammals associated with regulation of P450s and GSTs. For example, in mammals: (i) NF-kappa B is known to be involved in P450 regulation (Gu et al., 2006) and (ii) PPAR influences GST expression (Park et al., 2004). It remains to be determined if any of these potential TFBMs are functional, and if so, what is their specific involvement in regulation of these P450s, GSTs, and Ests.
Conclusion
The body louse genome has the smallest number of P450s and Ests identified in an insect species to date, and as well this genome contains a small number of GSTs. This small number of detoxification genes has the potential to make the louse an excellent model system to understand the role of these aforementioned genes, and their resultant enzymes, in (i) basic insect detoxification/defense physiology and (ii) metabolic resistance to pesticides. Due the reduced number of detoxification genes in P. h. humanus, the elucidation of their function will be more feasible than in any other insect species with large numbers of detoxification genes in their genomes.
Methods
The PhumU1.1 peptide database (http://phumanus.vectorbase.org/Tools/BLAST/) was searched by Blastp (Altschul et al., 1997) using complete peptide sequences of well characterized representative reference genes (mostly from D. melanogaster, An. gambiae, A. mellifera, T. castaneum, etc.) as queries. Members of a target gene family showing highly significant matches (mostly >40%) were first retrieved, and then, using the P. h. humanus sequences as queries in turn, the PhumU1.1 peptide database Blastp search was repeated until no new target genes were found. Once putative target gene sets were identified from the human body louse genome, they were subsequently used as queries for the NCBI Blastp search to verify their identity and phylogenetic relationships with other known genes. Exon-intron structure of each gene was obtained from the TIGR Pediculus humanus genome browser (http://www.tigr.org/tigr-scripts/gbrowse/gbrowse/louse/) or through comparison with intron-exon structure of reference genes. Conserved protein domains and functional sites were identified by using Prosite (Hofmann et al., 1999; http://ca.expasy.org/prosite/). Signal peptides were predicted using SignalP 3.0 (Bendtsen et al., 2004; http://www.cbs.dtu.dk/services/signalP/). Deduced amino acid sequences of target genes were aligned with homologous genes from other insects using ClustalW method (Thompson et al., 1994). In the cases of Ests and ABC transporters, the highly variable amino and carboxy terminal extensions were trimmed from the alignment. Aligned sequences were used to generate matrices of mean distances among proteins using Pam250, and these matrices were used to generate phylogenetic trees by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). If available, apparent outgroups were incorporated for the creation of rooted trees. If any outgroup is not available, unrooted trees were generated.
The statistical analysis to identify possible transcription factor binding motifs followed the same procedure as Li et al., (2008). More specifically, promoter sequences were analyzed by variety of de novo motif discovery algorithms and the motif results were combined by clustering analysis.
Supplementary Material
Supplementary Figure 1. Phylogenetic relationships of three P. h. humanus neuronal component protein families, including (A) voltage-dependent sodium channel α-subunit (VDSC), (B) sodium channel auxiliary subunit and (C) nicotinic acetylcholine receptor subunit (nAChR), with respective homologues from D. melanogaster (Dm). Deduced amino acid sequences were aligned using ClustalW (Thompson et al., 1994), and rooted trees were generated from the alignments by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). The D. melanogaster voltage-dependent calcium channel (Dm VDCC, CG4894-PA), Homo sapiens calcium-activated potassium channel beta 2 subunit (Hs KCNMB2, NP_005828), and D. melanogaster Rdl GABA receptor subunit (CG10537-PA) were used as outgroups for the tree generation of VDSC, sodium channel auxiliary subunit, and nAChR, respectively. Nodes with >50% bootstrap support were only marked with percentage values.
Supplementary Figure 2. Transcription factor binding motifs observed in cytochrome P450s of P. h. humans. The sequences include 800 base pairs upstream of all P450 genes’ transcription start sites, and 200 base pairs after the transcription start sites.
Supplementary Figure 3. Transcription factor binding motifs observed in GSTs of P. h. humans. The sequences include 800 base pairs upstream of the genes’ transcription start site, and 200 base pairs after the transcription start site.
Supplementary Figure 4. Transcription factor binding motifs observed in Esterases of P. h. humans. The sequences include 800 base pairs upstream of the gene's transcription start site, and 200 base pairs after the transcription start site.
Acknowledgements
This work was supported by a grant from NIH (5 R01 AI045082-05). Mr. JS Kang and Ms. JS Min were supported in part by Brain Korea 21 program. We thank the Body Louse Genome Sequencing Consortium for their efforts on the Body Louse Genome Sequencing Consortium paper (Kirkness et al., 2010) and for their permission to work on and publish this companion publication.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld VJ, Fetter RD, Broadie K, Goodman CS. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in Drosophila. Cell. 1995;81:757–767. doi: 10.1016/0092-8674(95)90537-5. [DOI] [PubMed] [Google Scholar]
- Ballatori N, Hammond CL, Cunningham JB, Krance SM, Marchan R. Molecular mechanisms of reduced glutathione transport: role of the MRP/CFTR/ABCC and OATP/SLC21A families of membrane proteins. Toxicol. Appl. Pharmacol. 2005;204:238–255. doi: 10.1016/j.taap.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. ( http://www.cbs.dtu.dk /services/SignalP/) [DOI] [PubMed]
- Brandt A, Scharf M, Pedra JH, Holmes G, Dean A, Kreitman M, Pittendrigh BR. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst(2) DDT locus. Insect Mol Biol. 2002;11(4):337–341. doi: 10.1046/j.1365-2583.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- Clayton JD, Cripps RM, Sparrow JC, Bullard B. Interaction of troponin-H and glutathione S-transferase-2 in the indirect flight muscles of Drosophila melanogaster. J. Muscle Res. Cell Motil. 1998;19:117–127. doi: 10.1023/a:1005304527563. [DOI] [PubMed] [Google Scholar]
- Campbell PM, Harcourt RL, Crone EJ, Claudianos C, Hammock BD, Russell RJ, Oakeshott JG. Identification of a juvenile hormone esterase gene by matching its peptide mass fingerprint with a sequence from the Drosophila genome project. Insect Biochem. Mol. Biol. 2001;31:513–520. doi: 10.1016/s0965-1748(01)00035-2. [DOI] [PubMed] [Google Scholar]
- Carino FA, Koener JF, Plapp FW, Feyereisen R. Constitutive overexpression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect. Biochem. Mol. Biol. 1994;24:411–418. doi: 10.1016/0965-1748(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Chiu TL, Wen Z, Rupasinghe SG, Schuler MA. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proc Natl Acad Sci U S A. 2008;105(26):8855–60. doi: 10.1073/pnas.0709249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, Feyereisen R, Oakeshott JG. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SPC, Deeley R. Multidrug resistance mediated by the ATP-binding cassette transporter protein MRP. BioEssays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Danielson PB, MacIntyre RJ, Fogleman JC. Molecular cloning of a family of xenobiotic-inducible drosophilid cytochrome p450s: Evidence for involvement in host-plant allelochemical resistance. Proc. Natl. Acad. Sc.i U S A. 1997;94:10797–10802. doi: 10.1073/pnas.94.20.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darboux I, Barthalay Y, Piovant M, Hipeau-Jacquotte R. The structure-function relationships in Drosophila neurotactin show that cholinesterasic domains may have adhesive properties. EMBO J. 1996;15:4835–4843. [PMC free article] [PubMed] [Google Scholar]
- Dassa E, Bouige P. The ABC of ABCs: A phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- Derst C, Walther C, Veh RW, Wicher D, Heinemann SH. Four novel sequences in Drosophila melanogaster homologous to the auxiliary para sodium channel subunit TipE. Biochem. Biophys. Res. Commun. 2006;339:939–948. doi: 10.1016/j.bbrc.2005.11.096. [DOI] [PubMed] [Google Scholar]
- Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Insect cytochrome P450s. In: Gilbert LI, Iatron K, Gill SS, editors. Comprehensive Molecular Insect Science. Elsevier Pergamon; Oxford: 2005. pp. 1–77. [Google Scholar]
- Feyereisen R. Evolution of insect P450. Biochem. Soc. Trans. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, Gallo MA, Xie W, Tian Y. Role of NF-kappa B in regulation of PXR-mediated gene expression: A mechanism for the suppression of cytochrome P450 3A4 by proinflammatory agents. J. Biol. Chem. 2006;281:17882–17889. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- Guo IC, Shih MC, Lan HC, Hsu NC, Hu MC, Chung BC. Transcriptional regulation of human CYP11A1 in gonads and adrenals. J Biomed Sci. 2007;14:509–15. doi: 10.1007/s11373-007-9177-z. [DOI] [PubMed] [Google Scholar]
- Guzov VM, Unnithan GC, Chernogolov AA, Feyereisen R. CYP12A1, a mitochondrial cytochrome P450 from the house fly. Arch. Biochem. Biophys. 1998;359:231–240. doi: 10.1006/abbi.1998.0901. [DOI] [PubMed] [Google Scholar]
- Heidari R, Devonshire AL, Campbell BE, Bell KL, Dorrian SJ, Oakeshott JG, Russell RJ. Hydrolysis of organophosphorus insecticides by in vitro modified carboxylesterase E3 from Lucilia cuprina. Insect Biochem. Mol. Biol. 2004;34:353–363. doi: 10.1016/j.ibmb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Helvig C, Tijet N, Feyereisen R, Walker FA, Restifo LL. Drosophila melanogaster CYP6A8, an insect P450 that catalyzes lauric acid (omega-1)-hydroxylation. Biochem. Biophys. Res. Commun. 2004;325:1495–1502. doi: 10.1016/j.bbrc.2004.10.194. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Mille r J., Mumcuoglu KY. Pyrethroid resistance mechanisms in the head louse Pediculus capitis from Israel: implications for control. Med. Vet. Entomol. 1999;13:89–96. doi: 10.1046/j.1365-2915.1999.00141.x. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Flaquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Holvoet P. Relations between metabolic syndrome, oxidative stress and inflammation and cardiovascular disease. Verh K Acad Geneeskd Belg. 2008;70:193–219. [PubMed] [Google Scholar]
- Honey Bee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Brown LA, Sattelle DB. Insect nicotinic acetylcholine receptor gene families: from genetic model organism to vector, pest and beneficial species. Invert. Neurosci. 2007;7:67–73. doi: 10.1007/s10158-006-0039-6. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome Sequences of the Human Body Louse and its Primary Endosymbiont Provides Insights into the Permanent Parasitic Lifestyle. PNAS. 2010 doi: 10.1073/pnas.1003379107. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuhara H, Sugiyama Y. ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch. 2007;453:735–744. doi: 10.1007/s00424-006-0134-x. [DOI] [PubMed] [Google Scholar]
- Lanning CL, Ayad HM, Abou-Donia MB. P-glycoprotein involvement in cuticular penetration of [14C]thiodicarb in resistant tobacco budworms. Toxicol. Lett. 1996;85:127–133. doi: 10.1016/0378-4274(96)03654-5. [DOI] [PubMed] [Google Scholar]
- Lee SW, Kasai S, Komagata O, Kobayashi M, Agui N, Kono Y, Tomita T. Molecular characterization of two acetylcholinesterase cDNAs in Pediculus human lice. J. Med. Entomol. 2007;44:72–79. doi: 10.1603/0022-2585(2007)44[72:mcotac]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Li HM, Buczkowski G, Mittapalli O, Xie J, Wu J, Westerman R, Schemerhorn BJ, Murdock LL, Pittendrigh BR. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol Biol. 2008;17:325–39. doi: 10.1111/j.1365-2583.2008.00808.x. [DOI] [PubMed] [Google Scholar]
- Li W, Schuler MA, Berenbaum MR. Diversification of furanocoumarinmetabolizing CYP6B cytochrome P450s in papilionids: specificity and substrate encounter rate. Proc. Nat. Acad. Sci. USA 100 Suppl. 2003;2:14593–8. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- Liu N, Scott JG. Genetic analysis of factors controlling high-level expression of cytochrome P450, CYP6D1, cytochrome b5, P450 reductase, and monooxygenase activities in LPR house flies, Musca domestica. Biochem. Genet. 1996;34:133–148. doi: 10.1007/BF02396246. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hao H, Liu C, Wang G, Xie H. Drugs as CYP3A probes, inducers, and inhibitors. Drug Metab. Rev. 2007;39:699–721. doi: 10.1080/03602530701690374. [DOI] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Monti-Dedieu L, Aragon S, Dauphin-Villemant C. A new cytochrome P450 from Drosophila melanogaster, CYP4G15, expressed in the nervous system. Biochem. Biophys. Res. Commun. 2000;273:1132–1137. doi: 10.1006/bbrc.2000.3058. [DOI] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Jacquin-Joly E, François MC, Nagnan-Le Meillour P. cDNA cloning of biotransformation enzymes belonging to the cytochrome P450 family in the antennae of the noctuid moth Mamestra brassicae. Insect Mol. Biol. 2002;11:273–281. doi: 10.1046/j.1365-2583.2002.00335.x. [DOI] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Niconov A, Ishida Y, Jacquin-Joly E, Leal WS. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. U S A. 2004;101:11459–11464. doi: 10.1073/pnas.0403537101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Schuler MA, Berenbaum MR. Cytochrome P450s in Papilio multicaudatus and the transition from oligophagy to polyphagy in the Papilionidae. Insect Mol Biol. 2007;16(4):481–90. doi: 10.1111/j.1365-2583.2007.00741.x. [DOI] [PubMed] [Google Scholar]
- Mao W, Rupasinghe S, Zangerl AR, Schuler MA, Berenbaum MR. Remarkable substrate-specificity of CYP6AB3 in Depressaria pastinacella, a highly specialized caterpillar. Insect Mol. Biol. 2006;15:169–179. doi: 10.1111/j.1365-2583.2006.00623.x. [DOI] [PubMed] [Google Scholar]
- Mittapalli O, Neal JJ, Shukle RH. Differential expression of two cytochrome P450 genes in compatible and incompatible Hessian fly/wheat interactions. Insect Biochem. Mol. Biol. 2005;35:981–989. doi: 10.1016/j.ibmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Mittapalli O, Neal JJ, Shukle RH. Tissue and life stage specificity of glutathione S-transferase expression in the Hessian fly, Mayetiola destructor: Implications for resistance to host allelochemicals. J. Insect Sci. 2007;7:1–13. doi: 10.1673/031.007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics. 2007;8:36. doi: 10.1186/1471-2164-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson PF, Fessler LI, Nelson RE, Sterne RE, Campbell AG, Fessler JH. Glutactin, a novel Drosophila basement membrane-related glycoprotein with sequence similarity to serine esterases. EMBO J. 1990;9:1219–1227. doi: 10.1002/j.1460-2075.1990.tb08229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EY, Cho IJ, Kim SG. Transactivation of the PPAR-responsive enhancer module in chemopreventive glutathione S-transferase gene by the peroxisome proliferator-activated receptor-gamma and retinoid × receptor heterodimer. Cancer Research. 2004;64:3701–3713. doi: 10.1158/0008-5472.CAN-03-3924. [DOI] [PubMed] [Google Scholar]
- Pittendrigh BR, Clark JM, Johnston JS, Lee SH, Romero-Severson J, Dasch GA. Sequencing of a new target genome: the Pediculus humanus humanus (Phthiraptera: Pediculidae) genome project. J. Med. Entomol. 2006;43:1103–1111. doi: 10.1603/0022-2585(2006)43[1103:soantg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Prichard RK, Roulet A. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134:1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2(11):e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, O'Connor MB, Gilbert LI. Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation. Ins. Biochem. Mol. Biol. 2007;37:741–753. doi: 10.1016/j.ibmb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Raoult D, Roux V. The body louse as a vector of reemerging human disease. Clin. Infect. Dis. 1999;29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- Roth CW, Holm I, Graille M, Dehoux P, Rzhetsky A, Wincker P, Weissenbach J, Brey PT. Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol. Cells. 2003;30:15:150–158. [PubMed] [Google Scholar]
- Rupasinghe SG, Duan H, Schmidt HLF, Berthold DA, Rienstra CM, Schuler MA. High-yield expression and purification of isotopically labeled cytochrome P450 monooxygenases for solid-state NMR spectroscopy. Biochim. Biophys. Acta. 2007;1768:3061–3070. doi: 10.1016/j.bbamem.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B, Homolya L, Szakács G, Váradi A. Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol. Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- Sheps JA, Ralph S, Zhao Z, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Coronella JA, Benes H, Cochrane BJ, Zimniak P. Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1–1 (GST-2) in conjugation of lipid peroxidation end products. Eur. J. Biochem. 2001;268:2912–2923. doi: 10.1046/j.1432-1327.2001.02179.x. [DOI] [PubMed] [Google Scholar]
- Soreq H, Sedman S. Acetylcholinesterase - new roles for an old actor. Nat. Rev. Neurosci. 2001;2:294–302. doi: 10.1038/35067589. [DOI] [PubMed] [Google Scholar]
- Srinivas R, Udikeri SS, Jayalakshmi SK, Sreeramulu K. Identification of factors responsible for insecticide resistance in Helicoverpa armigera. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;137:261–269. doi: 10.1016/j.cca.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Sutherland TD, Unnithan GC, Andersen JF, Evans PH, Muralaliev MB, Szabo LZ, Mash EA, Bowers WS, Feyereisen R. A cytochrome P450 terpenoid hydroxylase linked to the suppression of insect juvenile hormone synthesis. Proc. Natl. Acad. Sci. U S A. 1998;95:12884–12889. doi: 10.1073/pnas.95.22.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. and Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomas BA, Church WB, Lane TR, Hammock BD. Homology model of juvenile hormone esterase from the crop pest, Heliothis virescens. Proteins. 1999;34:184–196. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Baudry J, Berenbaum MR, Schuler MA. Ile115Leu mutation in the SRS1 region of an insect cytochrome P450 (CYP6B1) compromises substrate turnover via changes in a predicted product release channel. Protein Eng. Des. Sel. 2005;18:191–199. doi: 10.1093/protein/gzi023. [DOI] [PubMed] [Google Scholar]
- Wen Z, Rupasinghe S, Niu G, Berenbaum MR, Schuler MA. CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): adaptive evolution through subfunctionalization. Mol Biol Evol. 2006;23(12):2434–43. doi: 10.1093/molbev/msl118. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Fujii H, Aso Y, Banno Y, Koga K. Expression and characterization of a Sigma-class glutathione S-transferase of the fall webworm, Hyphantria cunea. Biosci. Biotechnol. Biochem. 2007;71:553–560. doi: 10.1271/bbb.60592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Phylogenetic relationships of three P. h. humanus neuronal component protein families, including (A) voltage-dependent sodium channel α-subunit (VDSC), (B) sodium channel auxiliary subunit and (C) nicotinic acetylcholine receptor subunit (nAChR), with respective homologues from D. melanogaster (Dm). Deduced amino acid sequences were aligned using ClustalW (Thompson et al., 1994), and rooted trees were generated from the alignments by the neighbor-joining method with 500 bootstrap replicates using MEGA4.1 package (Tamura et al., 2007). The D. melanogaster voltage-dependent calcium channel (Dm VDCC, CG4894-PA), Homo sapiens calcium-activated potassium channel beta 2 subunit (Hs KCNMB2, NP_005828), and D. melanogaster Rdl GABA receptor subunit (CG10537-PA) were used as outgroups for the tree generation of VDSC, sodium channel auxiliary subunit, and nAChR, respectively. Nodes with >50% bootstrap support were only marked with percentage values.
Supplementary Figure 2. Transcription factor binding motifs observed in cytochrome P450s of P. h. humans. The sequences include 800 base pairs upstream of all P450 genes’ transcription start sites, and 200 base pairs after the transcription start sites.
Supplementary Figure 3. Transcription factor binding motifs observed in GSTs of P. h. humans. The sequences include 800 base pairs upstream of the genes’ transcription start site, and 200 base pairs after the transcription start site.
Supplementary Figure 4. Transcription factor binding motifs observed in Esterases of P. h. humans. The sequences include 800 base pairs upstream of the gene's transcription start site, and 200 base pairs after the transcription start site.