Abstract
Background and Aims
Species delimitation can be problematic, and recently diverged taxa are sometimes viewed as the extremes of a species' continuum in response to environmental conditions. Using population genetic approaches, this study assessed the relationship between two Casearia sylvestris (Salicaceae) varieties, which occur sympatrically and allopatrically in the landscape of south-east Brazil, where intermediate types are also found.
Methods
In total, 376 individuals from nine populations in four different ecosystems were sampled, and nine microsatellite markers were used to assess the relative effects of the ecosystems and varieties on the distribution of genetic diversity among populations of this species.
Key Results
As a by-product of this study, several PCR products with more than two alleles were observed. The possibility that extra bands represent non-specific amplification or PCR artefacts was discarded by sequencing a sample of these bands. We suggest that (partial) genome duplication in C. sylvestris most probably explains this phenomenon, which may be a key factor in the differentiation of the two taxa, as it was markedly more frequent in one of the varieties. AMOVA indicated that approx. 22 % of the total genetic diversity was found between the two varieties. Bayesian analysis identified varieties and ecosystems as evolutionary units, rather than the individual populations sampled.
Conclusions
The results are in agreement with field observations and support the recognition of two varieties, as well as documenting the occurrence of hybridization between them.
Keywords: Atlantic Forest; Casearia sylvestris; Cerrado; ecotones; hybrid zone; microsatellites; population genetic structure; SSR, sympatry
INTRODUCTION
Ecology is of fundamental importance in speciation (Orr and Smith, 1998; Schluter, 2001; Wiens, 2004; Gegear and Burns, 2007), as incipient species occur in different ecological locations, and local adaptation leads to evolutionary divergence (Wiens, 2004). Such divergence may arise through the evolution of ecotypes (Abbott and Comes, 2007), which are distinct forms of a species adapted to different environmental conditions or habitats (Turesson, 1922). However, reproductive isolation between recently diverged taxa may be incomplete, hindering the delimitation of the boundaries among them (Rieseberg and Willis, 2007). In plants, phenotypic variation does not necessarily assort into discrete categories, and, for this reason, and due to interspecific hybridization, the definition of a species has been a major impediment to botanical studies of speciation (Rieseberg and Willis, 2007).
Casearia sylvestris Sw. (Salicaceae – APG II, 2003; Flacourtiaceae – Cronquist, 1981) is a shrub-to-tree species that is widespread in Central and South America, from Mexico to Argentina and Uruguay (Sleumer, 1980). It presents hermaphrodite flowers that are attractive to insects, mainly diptera, while its fruits are utilized by birds (Torres and Ramos, 2007). Two varieties, proposed by Sleumer (1980), are traditionally recognized: C. sylvestris var. sylvestris, which inhabits humid, dense forests, and C. sylvestris var. lingua, which is restricted to open, xeric habitats (Sleumer, 1980). The latter variety has also been recognized at the level of species [Casearia lingua (Cambess.) Eichler, Saint-Hilaire, 1829], and presents several traits related to drought and fire stress, including smaller, coriaceous leaves and shrubby habit (Silva, 1996). Despite differences in morphology and habitat preference, a gradation of intermediate forms can be found, making it difficult to delimit the taxa (Sleumer, 1980; Torres and Yamamoto, 1986; Silva et al., 2006). For this reason, in a recent review of Flacourtiaceae in São Paulo State, Brazil, Torres and Ramos (2007) did not recognize infraspecific taxa in C. sylvestris. Due to a lack of studies on the genetic structure of this species, whether the two taxa are independent biological units or represent the genetic continuum of a single species in response to different environmental conditions remains unresolved.
Molecular methods for assaying genetic diversity provide numerous tools to estimate relationships among natural populations of closely related organisms, including detecting species at shallow levels of evolutionary divergence (Drummond and Hamilton, 2007). In this context, microsatellite markers have proved to be powerful tools to solve biological problems, and are largely used to track the biological history of populations (Chambers and MacAvoy, 2000), providing a better understanding of species boundaries and of interspecific hybridization (e.g. Lexer et al., 2005; Drummond and Hamilton, 2007).
The present study characterizes the genetic relationships among populations of two closely related taxa, C. sylvestris var. sylvestris and C. sylvestris var. lingua, which occur sympatrically and allopatrically in different ecosystems in south-east Brazil. Microsatellite markers are used to assess the relative effects of the ecosystems and varieties in the distribution of genetic diversity in this species, thus helping to clarify the genetic relationships between its varieties. We hypothesized that if the varieties are in fact divergent lineages rather than the continuum of a single taxon, distinct populations of the same variety would tend to be more closely related than sympatric populations of different varieties. In other words, geographical distribution of sampling sites would not be the major factor determining the distribution of genetic diversity.
MATERIALS AND METHODS
Sampling
Although Casearia sylvestris var. sylvestris is widespread throughout the neotropics, C. sylvestris var. lingua has a more restricted distribution and occurs in open areas in eastern and south-eastern South America, more specifically in the Brazilian Cerrado and Caatinga, usually in poor soils and at low altitudes (Sleumer, 1980). This study focuses on south-east Brazil, more specifically the State of São Paulo, which is a mosaic of ecosystems where the two varieties occur sympatrically and allopatrically.
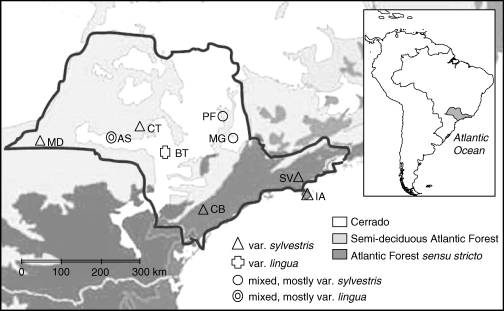
We sampled nine locations (Table 1, Fig. 1) representing four different ecosystems [Atlantic Forest sensu stricto (s.s.), semi-deciduous Atlantic forest, Cerrado and ecotones). The Atlantic Forest s.s. is a dense, humid, evergreen forest that occurs in the mountains along the coast, while the semi-deciduous Atlantic Forest occurs in more continental areas and has a well-characterized dry season. The Cerrado is characterized by shrub savannas under fire regime occurring on acid, aluminium-rich soils, with a severe dry season (Ratter et al., 1997). Contact zones (ecotones) between the semi-deciduous Atlantic Forest and the Cerrado are found throughout the landscape.
Table 1.
Sampling of Casearia sylvestris: ecosystems sampled, name of sampling sites and their characteristics, and number of individuals of each variety and of dubious morphology sampled in each location and ecosystem
| Sampling site and ecosystem | Coordinates | Altitude (m) | Area (ha) | Dry season | Site characteristics | var. sylvestris | var. lingua | Dubious | Total |
|---|---|---|---|---|---|---|---|---|---|
| Atlantic Forests.s. | 122 | – | – | 122 | |||||
| Ilha Anchieta | 23°32′S, 45°03′W | 100 | 828 | Absent | Oceanic island/moderate disturb | 32 | – | – | 32 |
| Santa Virginia | 23°20′S, 45°08′W | 900 | 17 000 | Absent | Part of a larger continuous forest/low disturbance | 39 | – | – | 39 |
| Carlos Botelho | 24°02′S, 47°57′W | 800 | 37 000 | Absent | Part of a larger continuous forest/low disturbance | 51 | – | – | 51 |
| Semi-deciduous Atlantic Forest | 87 | – | – | 87 | |||||
| Morro do Diabo | 22°30′S, 52°19′W | 450 | 34 000 | Moderate | Large fragment isolated/low disturbance | 54 | – | – | 54 |
| Caetetus | 22°23′S, 49°42′W | 600 | 2178 | Moderate | Medium fragment isolated/medium disturbance | 33 | – | – | 33 |
| Ecotones | 48 | 10 | 34 | 92 | |||||
| Mogi-Guaçu | 22°15′S, 47°11′W | 650 | 980 | Moderate | Isolated/disturbed | 16 | 6 | 25 | 47 |
| Porto Ferreira | 21°51′S, 47°25′W | 560 | 612 | Moderate | Isolated/disturbed | 32 | 4 | 9 | 45 |
| Cerrado | 4 | 68 | 3 | 72 | |||||
| Assis | 22°34′S, 50°24′W | 550 | 1170 | Severe | Isolated/disturbed | 4 | 39 | 3 | 43 |
| Botucatu | 22°56′S, 48°27′W | 850 | 33 | Severe | Very small fragment isolated and disturbed | – | 29 | – | 29 |
| Total | 261 | 78 | 37 | 376 | |||||
Fig. 1.
Map showing (approximately) the original ecosystem distribution in Sao Paulo state, south-east Brazil, and populations of Casearia sylvestris sampled. AS = Assis, BT = Botucatu, CB = Carlos Botelho, CT = Caetetus, IA = Ilha Anchieta, MD = Morro do Diabo, MG= Mogi-Guaçu, PF = Porto Ferreira, SV = Santa Virginia.
Sampling sites differed in size, isolation from other natural areas and conservation status (Table 1), with Carlos Botelho (Atlantic Forest s.s.) and Morro do Diabo (semi-deciduous Atlantic Forest) being the largest and least disturbed localities. Ilha Anchieta (Atlantic Forest s.s.) is an oceanic island separated from the mainland by about 500 m. The population in Botucatu (Cerrado) is particularly small and disturbed: until 1970 it was a cattle pasture, and since then natural regeneration of Cerrado has been taking place. Although the site covers 33 ha, C. sylvestris is only found in an area of approx. 1 ha.
The average distance between sampling sites is 320 km, varying from 25 km (Ilha Anchieta – Santa Virginia) to 745 km (Ilha Anchieta – Morro do Diabo). The number of individuals sampled in each site is given in Table 1. Leaf samples were dried in sealed plastic bags containing silica gel and stored at –20 °C prior to DNA extraction.
Classifying individuals at the level of variety
For each individual sampled, a voucher was deposited in the IAC (Instituto Agronômico de Campinas) Herbarium, Campinas, Brazil. Morphological traits were used to assign the individuals to a variety according to Sleumer (1980): var. lingua presents more coriaceous, oval leaves, with prominent reticulation and a slender petiole. We also considered cymbiform leaves to be characteristic of var. lingua, based on the field and taxonomic experience of R.B.T. on the genus Casearia. Individuals were classified as lingua, sylvestris or dubious.
DNA isolation
DNA was isolated from leaf samples according to the acid protocol of Csaikl et al. (1998). We added 2 % (w/v) sodium bisulfite to the extraction buffer to avoid oxidation, particularly observed in var. sylvestris samples.
Markers, PCR and electrophoresis conditions, and allele scoring
Ten microsatellite markers specifically designed for C. sylvestris (Cavallari et al., 2008) were tested. Locus Csy08 presented null alleles according to MICRO-CHECKER software (Van Oosterhout et al., 2004) and was not retained for subsequent analysis. The remaining nine loci were polymorphic and were thus used in this study. PCR conditions followed Cavallari et al. (2008). The amplified fragments were analysed at 700 and 800 nm by electrophoresis on an IR-DNA analyser (LI-COR 4200 sequencer, www.licor.com) at the Montpellier Languedoc-Roussillon Genopole genotyping platform. Allele scoring was performed with the SAGA Generation 2 v.3·2 (LI-COR) software.
Verifying the microsatellite identity of the bands
We observed several PCR products with more than two alleles (see Results), which is not expected for microsatellite markers in diploid species (the ploidy of C. sylvestris is not known). A new set of PCR reactions was performed with 90 randomly chosen individuals (for all primer pairs), and the same results were obtained. To follow up, we randomly chose three individuals and two primer pairs (Csy04 and Csy09) to perform a more detailed study. For this study, PCR products were run in a 3·8 % agarose gel and detected bands were excised from the gel under UV light. Gel slices containing the bands were passed through a purification column (Wisard SV Gel and PCR Clean-Up System, Promega) and the purified products were ligated into pGEM-T Easy vector (Promega, www.promega.com) and used to transform DH5α competent cells. Five positive colonies (using blue/white β-galactosidase selection) from each Petri dish (i.e. representing each of the bands detected in the gel) were sequenced using the SP6 primer and BigDye terminator version 3·1 Cycle Sequencing Kit (Applied Biosystems, www.appliedbiosystems.com) and run on a 3100 DNA Analyser (Applied Biosystems). The sequences obtained were examined for the presence of the microsatellite repeats and their flanking regions.
Statistical analysis
Genetic diversity
Due to the detection of several PCR products with more than two alleles (see Results), correct allele frequencies could not be calculated, and most of the analyses that are usually performed with co-dominant markers could not be performed (HE, HO, FIS, etc.). Descriptive analyses of genetic diversity were thus restricted to those that were completely independent of allele frequencies, i.e. by estimating the number of alleles per locus and allele richness (El Mousadik and Petit, 1996). This enabled us to describe and compare the genetic diversity of sampling sites, ecosystems and varieties without making assumptions on the ploidy level of the species. Allele richness in particular has been considered to be one of the most relevant criteria for measuring diversity, especially in the context of genetic conservation (reviewed by Petit et al., 1998). To perform these analyses, PCR products with more than two alleles (which correspond to only 3·4 % of the total PCR products – see Results) were removed from the data set; this did not distort the results as all the alleles that were excluded were present in other individuals of the same population (data not shown). Analyses were performed using FSTAT software (Goudet, 1995). Sampling sites, ecosystems and varieties were compared to get an idea of their relative genetic diversity. For each variety, we also determined the size range of alleles, the number of private alleles and the frequency of the most common allele. To this end, all morphologically dubious individuals were excluded.
In addition, we calculated Shannon's information index to estimate the degree of genetic diversity within each sampling site and ecosystem using POPGENE software v.1·31 (Yeh et al., 1997). This was done by coding alleles as dominant marker bands (binary coding), considering all PCR products, including those with more than two alleles.
Population genetic structure
The usual F-statistics performed with co-dominant markers could not be carried out due to the lack of allele frequencies. Based on the presence/absence of bands (binary coding, considering all PCR products), population genetic structure was inferred by analysis of molecular variance (AMOVA) with ARLEQUIN v.2·0 software (Schneider et al., 2000), estimating Φ-statistics (Excoffier et al., 1992), which are analogous to Wright's hierarchical fixation indices under the island model of gene flow (Wright, 1951). However, it should be borne in mind that the alleles from a given microsatellite locus are not independent samples and, for this reason, P values obtained in the AMOVAs may present some bias. Two independent AMOVAs were performed, which hierarchically partitioned genetic diversity by (1) ecosystems/sampling sites/individuals and (2) varieties/sampling sites/individuals.
Population genetic structure was also revealed using the Bayesian approach of Pritchard et al. (2000), implemented in the software STRUCTURE. The latest versions (2·2·3 and 2·3·2) of the software include the algorithm of Falush et al. (2007), which can handle genotypic ambiguity (where it is not possible to identify the exact genotypes in heterozigotes, e.g. tetraploid individuals displaying two or three alleles). As we observed individuals with up to four bands in some loci, STRUCTURE was run considering all the individuals as tetraploids. Following the user's guide for the software, the four rows of each individual at each locus (representing the four alleles) were filled up with the alleles observed (one, two, three or four alleles). The integer for missing data was used to complete the infile, and the option RECESSIVEALLELES was set to 1. The program thus considers that the four alleles are co-dominant to each other, and that there is genotypic ambiguity. In this way, we incorporated all alleles observed in the sample in the study of population genetic structure.
STRUCTURE 2·3·2 was used to check the ancestry of individuals within populations, ecosystems and varieties. The optimal number of clusters (K) was determined by varying the value of K from 1 to 10, and five runs of each K value were performed without any prior information using the admixture model and correlated alleles frequencies, with 200 000 generations sampled as burn-in and 500 000 generations sampled in the Monte Carlo Markov chain (MCMC). The ad hoc statistics ΔK (Evanno et al., 2005) and Pr(X|K), given by lnP(D) (Pritchard et al., 2000), were used as predictors of the real number of clusters. At K = 2, the clusters that formed agreed closely with the morphological classification of the individuals (see Results), and, for this reason, it was possible to use the admixture proportions (Q) of each individual to reveal admixed individuals between the two varieties and to give an idea of the extent of hybridization in each population. This was possible because the admixture model estimates the proportion of each individual's genome that originated from each of the K inferred clusters (Pritchard et al., 2000). Although the threshold adopted here is somewhat arbitrary, Vähä and Primmer (2006) and Lepais et al. (2009) demonstrated through simulation studies that a threshold of 10 % is probably the best choice to correctly classify purebreds and hybrids in such situations. Following these authors, we considered as admixed between the two varieties all individuals presenting a Q value of between 0·1 and 0·9. The STRUCTURE results were displayed graphically by the software DISTRUCT (Rosenberg, 2004).
RESULTS
Classification of individuals into varieties and their distribution among ecosystems
For the majority of individuals sampled, classification at the variety level was not problematic. Key morphological features were generally clear, leading to immediate recognition of the variety. As a general rule, all individuals sampled in the Cerrado were easily recognized as var. lingua, and all individuals sampled in the Atlantic Forest (both s.s. and semi-deciduous) were undoubtedly var. sylvestris. We found the two varieties occurring sympatrically in the ecotones, where individuals of dubious morphology were also observed. The population of Assis (Cerrado) also included the two varieties and intermediate morphs, with var. lingua restricted to Cerrado s.s. (shrub savannas) areas with var. sylvestris and the intermediate morphs restricted to ‘Cerradão’ areas (dense arrangements of Cerrado trees). The number of individuals of each variety sampled in each ecosystem/sampling site is given in Table 1.
Detection of PCR products with more than two alleles and study of band identity
Several PCR products with more than two alleles were detected (see below). Sequencing of some of these PCR products (three individuals each for primer pair Csy04 and Csy09) revealed that all bands contained the expected microsatellite repeats (CTn for Csy04 and AGn for Csy09; Cavallari et al., 2008) located between the same flanking regions. This result shows that the additional bands observed are true microsatellite alleles, and not PCR artefacts.
PCR products with more than two alleles were observed in all but one locus (Csy18), with locus Csy04 being the most affected (12 % of PCR products). Considering all loci, multiple alleles were observed in 3·4 % of the PCR products, and detected in 83 (22 %) of the 376 individuals analysed. In most of them (64 individuals), the phenomenon was restricted to only one locus, although some individuals had more than two alleles at two, three or four loci. We detected many more cases of PCR products with three alleles (106) than with four alleles (only four). Of the 78 var. lingua individuals genotyped, 53 (68 %) presented more than two alleles in at least one locus. PCR products with more than two alleles were observed in only 7·7 % (20 of 261 individuals) of var. sylvestris samples and in 27 % (ten of 37 individuals) of the morphologically dubious individuals. Possible explanations for the existence of more than two alleles per locus are provided in the Discussion.
Genetic diversity
The number of alleles sampled and the allele richness of each sampling site, ecosystem and variety are presented in Table 2. The mean number of alleles per locus per sampling site was 8·87, ranging from 3·1 (Botucatu) to 12·2 (Porto Ferreira). Botucatu displayed a very particular genetic composition: in addition to the extremely low number of alleles per locus (ranging from one to six alleles, whereas in other sampling sites the number ranged from three to 21) and low total allele richness (19·89, whereas the mean total allele richness was 37·89), in six of the nine loci no homozygotes were observed. In the remaining sampling sites, homozygotes were observed at all loci.
Table 2.
Mean number of alleles sampled per locus, total number of alleles sampled, mean allele richness per locus and total allele richness of each ecosystem, sampling site and variety studied
| Number of alleles sampled |
Allele richness* |
|||
|---|---|---|---|---|
| Ecosystems/sampling sites/varieties | Mean | Total | Mean | Total |
| Atlantic Forests.s. | 12·56 | 113 | 9·97 | 89·77 |
| Ilha Anchieta | 8·89 | 80 | 4·33 | 38·94 |
| Santa Virginia | 9·33 | 84 | 4·12 | 37·05 |
| Carlos Botelho | 9·00 | 81 | 4·19 | 37·71 |
| Semi-deciduous Atlantic Forest | 11·67 | 105 | 9·95 | 89·52 |
| Morro do Diabo | 10·00 | 90 | 4·59 | 41·34 |
| Caetetus | 9·44 | 85 | 4·67 | 42·01 |
| Cerrado (combined) | 9·56 | 86 | 8·06 | 72·56 |
| Assis (combined) | 9·00 | 81 | 4·55 | 40·97 |
| Botucatu | 3·11 | 28 | 2·21 | 19·89 |
| Cerrado (only major variety) | 8·67 | 78 | 7·53 | 67·80 |
| Assis (only major variety) | 8·11 | 73 | 4·26 | 38·31 |
| Botucatu | 3·11 | 28 | 2·21 | 19·89 |
| Ecotones (combined) | 13·22 | 119 | 10·48 | 94·32 |
| Mogi-Guaçu (combined) | 8·89 | 80 | 4·08 | 36·76 |
| Porto Ferreira (combined) | 12·22 | 110 | 5·15 | 46·35 |
| Ecotones (only major variety) | 10·67 | 96 | 9·84 | 88·53 |
| Mogi-Guaçu (only major variety) | 5·00 | 45 | 3·51 | 31·55 |
| Porto Ferreira (only major variety) | 9·78 | 88 | 4·83 | 43·46 |
| var. sylvestris | 15·56 | 140 | 11·36 | 102·21 |
| var. lingua | 9·44 | 85 | 7·76 | 69·82 |
| unknown var. | 10·11 | 91 | 9·73 | 87·60 |
* Allele richness for sampling sites based on minimum sample size of five diploid individuals; for ecosystems based on minimum sample size of 33 diploid individuals; and for varieties based on minimum sample size of 30 diploid individuals.
The remaining sampling sites displayed very similar patterns of diversity (Table 2), with the mean number of alleles per locus per sampling site ranging from 8·88 (Mogi-Guaçu, Ilhan Anchieta) to 12·2 (Porto Ferreira), and mean allele richness per locus ranging from 4·12 (Santa Virginia) to 5·15 (Porto Ferreira).
As a whole, the ecotones presented the highest number of alleles per locus and the highest allele richness. When the minor variety (var. lingua) in these sites was excluded from the data set, these numbers decreased and Atlantic Forest s.s. and semi-deciduous Atlantic Forest appear as the most genetically diverse ecosystems (Table 2).
The allele richness observed in each variety differed significantly (Table 2). Of the total number of alleles detected, 64 (42·1 %) were unique to var. sylvestris and eight (5·25 %) were unique to var. lingua (Table 3), implying that only 52·6 % of the bands are shared by the two varieties (dubious individuals were not taken into consideration). The size range of the alleles, the most common allele and its frequency, and the number of private alleles for each locus and each variety are shown in Table 3. The varieties differed significantly in the presence and frequency of alleles, especially at loci Csy04, Csy06, Csy11, Csy14 and Csy18 (Table 3).
Table 3.
Comparison between varieties of C. sylvestris: size range of alleles, most common allele and number of private alleles detected in each locus studied
| Range size of alleles |
Most common allele* |
Number of private alleles |
|||||
|---|---|---|---|---|---|---|---|
| Locus | var. sylvestris | var. lingua | Total | var. sylvestris | var. lingua | var. sylvestris | var. lingua |
| Csy_04 | 115–167 | 113–159 | 113–167 | 145 (0·49) | 115 (0·28) | 9 | 5 |
| Csy_06 | 276–322 | 276–318 | 276–326 | 280 (0·13) | 280 (0·60) | 14 | 0 |
| Csy_07 | 246–276 | 246–276 | 244–276 | 246 (0·45) | 246 (0·60) | 6 | 0 |
| Csy_09 | 185–209 | 191–202 | 185–209 | 191 (0·60) | 199 (0·47) | 5 | 0 |
| Csy_11 | 140–182 | 142–182 | 140–186 | 162 (0·23) | 172 (0·31) | 3 | 2 |
| Csy_14 | 218–244 | 218–242 | 218–244 | 230 (0·66) | 226 (0·37) | 6 | 0 |
| Csy_15 | 251–285 | 251–287 | 251–287 | 271 (0·24) | 263 (0·22) | 7 | 1 |
| Csy_16 | 272–286 | 276–282 | 270–286 | 278 (0·67) | 278 (0·55) | 3 | 0 |
| Csy_18 | 271–313 | 273–297 | 271–313 | 287 (0·26) | 273 (0·90) | 11 | 0 |
| Total | 64 | 8 | |||||
*Allele frequency in parentheses.
Values obtained for Shannon's information index (data not shown) were similar for all sampling sites, ranging from 0·16 (Santa Virginia) to 0·21 (Porto Ferreira). The only discrepancy was observed for Botucatu (0·05). The total diversity observed in all populations was 0·23. Ecosystems and varieties showed similar Shannon's information index values ranging from 0·17 (Cerrado) to 0·20 (ecotones). Varieties presented similar levels of genetic diversity (Shannon's information index approx. 0·21 in each variety).
Population genetic structure
The first hierarchical AMOVA (Table 4) revealed that approx. 13 % of the total molecular genetic diversity was attributable to differences among ecosystems. The second AMOVA revealed that differences between the varieties accounted for approx. 22 % of the total genetic diversity sampled. Estimated Φ-statistics were shown to be significant at the 1 % level (P < 0·01) in both AMOVAs, but this result should be interpreted with caution due to the lack of independence between alleles of the same locus. In both AMOVAs, the greater part of the genetic diversity sampled was within populations, but diversity was greater when individuals of the same variety were grouped together in the same sampling sites (first AMOVA, 76 %) than when they were separated into different populations (second AMOVA, 64 %).
Table 4.
Results of AMOVA
| Source of variation | d.f. | SSD | Percentage total variance | Φ-statistics |
|---|---|---|---|---|
| AMOVA ecosystems | ||||
| Ecosystems | 3 | 492·505 | 0·1281 | ΦCT = 0·13 |
| Sampling sites within ecosystems | 5 | 251·099 | 0·1146 | ΦSC = 0·11 |
| Individuals within sampling sites | 367 | 2587·465 | 0·7573 | 1 − ΦST = 0·76 |
| Total | 375 | 3331·069 | ||
| AMOVA varieties | ||||
| Varieties | 1 | 331·316 | 0·2199 | ΦCT = 0·22 |
| Sampling sites within varieties | 10 | 455·916 | 0·1385 | ΦSC = 0·14 |
| Individuals within sampling sites | 327 | 2197·718 | 0·6416 | 1 − ΦST = 0·64 |
| Total | 338 | 2984·95 | ||
d.f., degrees of freedom; SSD, sum of squared deviations.
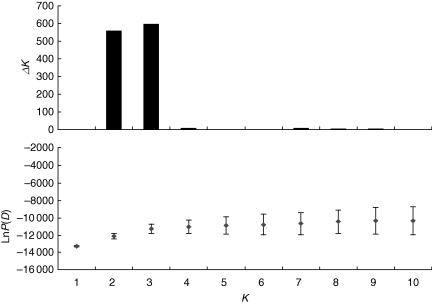
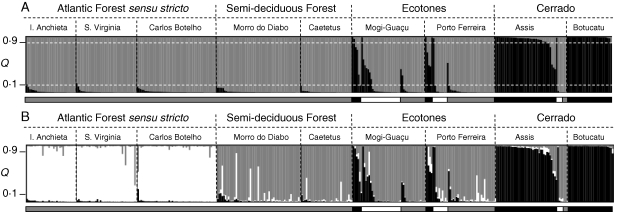
Bayesian analysis performed using STRUCTURE with K from 1 to 10 provided either two or three clusters (K = 2 and K = 3; Fig. 2). Figure 3 shows clustering results for K = 2 and K = 3. At K = 2 (Fig. 3A), clustering closely matched our morphological classification. When the threshold of 10 % was used, individuals with an admixture coefficient (Q) below 0·1 or above 0·9 can be considered as not admixed (or as ‘pure grey’ and ‘pure black’, respectively, in Fig. 3A). Of the 69 ‘pure black’ individuals, 65 (94·2 %) were morphologically classified as var. lingua (underlined by a black bar in Fig. 3). In the same way, of the 287 ‘pure grey’ individuals, 261 (91 %) were morphologically classified as var. sylvestris (underlined by a grey bar in Fig. 3). There were no individuals with var. lingua morphology in the ‘grey’ cluster, and vice versa. Individuals of intermediate morphology (underlined by a white bar in Fig. 3) were revealed to have different degrees of admixture, ranging from ‘pure black’ to ‘pure grey’.
Fig. 2.
Plots of lnP(D) (Pritchard et al., 2000) and ΔK (Evanno et al., 2005) for each K obtained from the STRUCTURE analysis.
Fig. 3.
Graphical representation of results of STRUCTURE obtained for two (A) and three (B) clusters (K). Each individual is represented by a vertical line divided into K coloured fragments proportional to its membership in the corresponding genetic cluster. Q denotes the proportion of admixture for each individual and the threshold of 10 % is represented by a dashed white line in (A). Dashed black lines separate individuals from different populations and ecosystems, which are indicated at the top. The thin bar under the figures indicates the morphological classification of individuals (black = var. lingua morphology, grey = var. sylvestris morphology, white = individuals with dubious morphology).
Once we confirmed that this clustering agreed closely with our morphological classification, the admixture coefficient for each individual (Q) was used to reveal putatively admixed individuals between the two varieties. Thirty of 376 individuals (7·9 %) presented a value of Q of between 0·1 and 0·9, and can be considered as admixed between the two varieties. Only seven putatively admixed individuals out of 238 (i.e. 2·9 %) were found in the populations of the Atlantic Forest, semi-deciduous Forest and Cerrado (only Botucatu). In the mixed populations (Assis, Mogi-Guaçu and Porto Ferreira), 23 of 138 individuals (16·6 %) can be considered as putatively admixed between the two varieties, and this proportion increased to 19·5 % (18 of 92) when only the ecotones (Mogi-Guaçu and Porto Ferreira) were taken in account.
At K = 3, clustering agreed closely with the ecosystems sampled (Fig. 3B), showing the ability of the analysis to detect biologically meaningful clusters.
DISCUSSION
Genome duplication in C. sylvestris
The unusual microsatellite banding pattern obtained suggests the occurrence of genome duplication in C. sylvestris, although it was not possible to distinguish between chromosome duplication (polyploidy, aneuploidy) and small-scale duplication (only some genes duplicated) with the tools used.
We observed individuals with up to four alleles in three loci, and with up to three alleles in eight of the nine loci studied. Moreover, in Botucatu, which had a low number of alleles in all loci, six of the nine loci showed fixed heterozygous profiles. This pattern is similar to that obtained by Segarra-Moragues et al. (2004) in Borderea pyrinaica (Dioscoreaceae): they observed individuals with up to four bands in 11 of 17 microsatellite loci they screened, whereas two loci showed fixed heterozygous profiles. They interpreted the fixed heterozygous profile of these loci as the amplification of two duplicate loci that are fixed for one allele each, and based on overall patterns observed they suggest that this species is allotetraploid.
One of the obvious approaches to solve this question is cytogenetics, which will be done in the near future. Cytogenetic studies on Casearia are very rare in the literature. The Index to Plant Chromosome Numbers (available at http://mobot.mobot.org/W3T/Search/ipcn.html), which contains data from published indices from 1979 onward, lists only one publication on Casearia chromosome numbers (n = 21 for C. elliptica Tul.; Bir et al., 1980). However, in Salicaceae, aneuploids have been reported in Populus (Bradshaw and Stettler, 1993; Cervera et al., 2001; Yin et al., 2004), while Salix species present various ploidy levels (Leskinen and Alström-Rapaport, 1999).
Molecular and genetic studies in plants have shown that autopolyploidy is much more common than traditionally assumed (Soltis et al., 2003). A large number of angiosperm families, including Salicaceae, which were traditionally considered to be diploids, are probably the products of ancient polyploid events (Soltis and Soltis, 2000). These ancient polyploids exhibit extra copies of some of their genes above the level that one would expect for diploid plants (Gottlieb, 1982). For example, Populus trichocarpa has 8000 duplicated genes (Tuskan et al., 2006). The complete sequencing of the P. trichocarpa genome and the comparison of Populus and Salix orthologous genes revealed that the common ancestor of these two genera was polyploid (Tuskan et al., 2006). Thus, we may find that C. sylvestris, as a member of Salicaceae, shares this history and also presents duplicated genes.
Duplicated genes and genomes can undergo divergent evolution and evolve new functions (Soltis and Soltis, 2000; Cui et al., 2006), facilitating the colonization of unstable habitats (Lawton-Rauh, 2003). It is important to note that in the present study, the great majority of individuals with gene (or chromosome) duplication were found in the Cerrado (var. lingua), an unstable, harsh environment under fire regime.
Genetic diversity
The primer pairs utilized in this work were developed from C. sylvestris var. sylvestris and tested for transferability for other Casearia species, with no positive results (Cavallari et al., 2008). In the present work, these markers successfully amplified the DNA of the two varieties of C. sylvestris, but a high percentage of private alleles (47·35 %) was observed, and the most common allele and its frequency differed markedly between the two varieties at most loci. When studying the boundaries between two sympatric varieties of Lupinus microcarpus (Leguminosae), Drummond and Hamilton (2007) observed a similar pattern of allele frequency distribution, revealing a mix of shared and private alleles. According to these authors, this result is consistent with shared ancestral polymorphism and recent divergence between species.
The low genetic diversity observed in Botucatu is congruent with the recent (after 1970) foundation of the population. The distribution of individuals (restricted to 1 ha) also suggests that few founders arrived in the locality (which has a total area of 33 ha). Although fragmentation and a reduction in population size are expected to decrease gene diversity (Young et al., 1996), apart from Botucatu, observed gene diversity was not positively correlated with fragment size or conservation status. Sampling sites harbouring the two varieties, although relatively isolated and disturbed, presented high values of Shannon's information index and allele richness. The ecotones as a whole had the highest values of Shannon's index and allele richness, higher than the main values observed for ecosystems. Our sampling strategy (two varieties occurring sympatrically and allopatrically) and the high percentage of bands restricted to one or the other variety may explain these trends. Indeed, excluding from the data set the minor variety occurring in these sampling sites/ecosystem resulted in decreased diversity (although this was not the case of Porto Ferreira, which even without considering the minor variety still displayed the highest genetic diversity). These results are in accordance with recent research on a wide range of taxa, which suggests that environmental gradients are important in diversification and speciation, and may deserve special attention (Smith et al., 2001).
Population genetic structure: main components
The population structure of C. sylvestris in this region can be described in terms of ‘varieties’ and ‘ecosystems’, which were shown to be significant factors in the distribution of genetic diversity. This was predicted from field observations and corroborated by statistical analyses, especially by the Bayesian analysis.
Clustering by STRUCTURE at K = 2 (Fig. 3A) and at K = 3 (Fig. 3B) grouped the individuals in a way that is highly concordant with varieties and ecosystems, respectively. The agreement between clusters and varieties indicates a non-random distribution of alleles and allele frequencies between them, the same applying to the ecosystems. As ΔK (Fig. 2) provided poor statistical support for K > 3, the subdivision of the species' genetic diversity into varieties and ecosystems was very strong, with each variety or ecosystem recognized as a discrete unit, although this was not the case for individual sampling sites.
According to Minder and Widmer (2008), ‘good’ species are expected to be genetically distinct from each other, with geographically distant populations of the same species being genetically more closely related to each other than to geographically proximate populations of the other species. This pattern was clearly supported by our Bayesian analysis. In the Bayesian analysis, the clusters obtained agreed closely with our morphological classification. Thus, in addition to being morphologically recognizable, the two varieties also possess significant differences at neutral (microsatellite) loci, suggesting little connectivity between the two taxa. Other results that also support divergence between them are the marked differences in the proportion of individuals with evidence of partial genome duplication in each variety, the high proportion of private alleles in each variety (47·4 %) and results of the AMOVA showing that approx. 22 % of the total genetic diversity sampled is attributable to genetic differences between varieties. The results of the AMOVA are similar to those observed between Croton alabamensis varieties separated by more than 1000 km (Van Ee et al., 2006) and between two species of Iliamna (Malvaceae) (Slotta and Porter, 2006), and, although there is no absolute measure of genetic differentiation for determining species separation (Morgan-Richards and Wolff, 1999), our results indicate strong differentiation between the two taxa.
The partitioning of the genetic diversity of the species according to the ecosystem was supported by the clustering at K = 3 (Fig. 3B). Individuals of var. sylvestris were separated into two clusters: one composed of individuals from the Atlantic Forest s.s., and the other of individuals from the remaining ecosystems. This is reasonable as the semi-deciduous Atlantic Forest and the Cerrado are connected by ecotones, allowing more connectivity between its individuals than between individuals from the Atlantic Forest s.s., which is more isolated. Genetic divergence between Atlantic Forest s.s. and the semi-deciduous Atlantic Forest (both harbouring exclusively var. sylvestris) may be explained by genetic drift, as they are all isolated fragments.
Putative hybrid zones
The literature on C. sylvestris refers to the existence of morphologically intermediate individuals (Sleumer, 1980; Torres and Ramos, 2007) without referring to their habitat preference or co-occurrence with ‘pure forms’. We observed that intermediate forms occur exclusively in the presence of both of the ‘pure’ varieties. The sympatric occurrence of the three forms (var. sylvestris, var. lingua and intermediate forms) suggests that morphologically intermediate individuals are the product of hybridization, rather than of phenotypic responses to environmental intermediacy. For instance, we visited several open Cerrado-like areas within the Atlantic Forest, where although the trees had a somewhat different morphology due to full exposure to the sun, they could undoubtedly be classified as pure var. sylvestris individuals. Of course, to disentangle phenotypic plasticity and genetic divergence leading to divergent morphology, other approaches should also be used (e.g. reciprocal transplants, quantitative genetic analyses of morphological and adaptive traits to separate genetic effects from environmental effects), and in this sense, our results are preliminary.
In addition, the literature on C. sylvestris refers to the existence of two varieties inhabiting different environments (var. sylvestris in dense forests and var. lingua in open areas) (Sleumer, 1980; Silva et al., 2006), and, to our knowledge, sympatry of these forms has never been directly reported. Our fieldwork allowed us to observe sympatry of the two varieties: in the ecotones, individuals of each variety were found side by side.
As expected, populations harbouring the two varieties contained a greater number of admixed individuals displaying a wide range of admixture proportions (revealed by Q values), suggesting the coexistence of many hybrid generations and backcrosses. According to these observations, these regions can be considered putative hybrid zones, a pattern extensively reported in the literature at the species level (e.g. Fritsche and Kaltz, 2000; Lexer et al., 2005; Valbuena-Carabaña et al., 2005; Remington and Robichaux, 2007).
The study of plant species hybrid zones in the Cerrado/Atlantic Forest geographical region is in its infancy. A putative hybrid zone was studied by Lacerda et al. (2002) and Goulart et al. (2005), in ecotonal regions where the Fabaceae species Plathymenia reticulata (from Cerrado) and P. foliolosa (from the Atlantic Forest) meet. These authors evoked the ‘refugium theory’ (Haffer, 1969) to explain the origin of the two species: changes in vegetation cover and in the distribution of plant species during climatic changes in the Pleistocene would have restricted some populations to dry areas, which may have evolved into P. reticulata, whereas populations of warmer and humid refuges evolved into P. foliolosa. This hypothesis may be applicable to C. sylvestris populations in the same region, leading to the differentiation between populations from humid refuges (which may have evolved into var. sylvestris) and xeric areas (which may have evolved into var. lingua). It is reasonable to propose that the ecological differences between these habitats (e.g. different pollinators, habitat preference of pollinators and seed dispersers, different pluviometric regimes leading to flowering asynchrony) today represent strong barriers to gene exchange between varieties, thus helping to maintain separation of the taxa.
It would be worth checking if there are preferential crossing patterns or partial reproductive barriers between the two taxa in the ecotones. In plant species, many closely related taxa differ in flowering time to minimize niche competition (Friberg et al., 2008). During sampling, we observed that pure parental forms persist sympatrically with intermediate forms. Our field observations indicate the absence of flowering synchrony between the two varieties (in Assis, Mogi-Guaçu and Porto Ferreira, we noted that only individuals of var. lingua were flowering), but there is no statistical robustness in this observation, and more detailed studies are necessary.
In summary, our field observations and experimental results suggest that the two C. sylvestris varieties are relatively independent biological units, rather than representing the genetic continuum of a single species in response to different environmental conditions. We also documented the existence of putative hybrid zones between these two varieties. To corroborate these findings, additional studies with chloroplast DNA microsatellites, cpDNA sequences and rDNA sequences will be performed in the near future. It is important to stress that our sampling was restricted to the geographical region of south-east Brazil, which harbours a particular assortment of ecosystems. It is possible that the genetic relationships between the two varieties of C. sylvestris differ in central northern Brazil, where they also may occur sympatrically.
ACKNOWLEDGEMENTS
We thank the staff of the Conservation Units visited, especially Dr Giselda Durigan (Assis) and Dr Léo Zimback (Botucatu), Instituto Florestal de São Paulo, for the information they provided on site history and distribution of C. sylvestris, Eduardo Moretti for help with fieldwork, Dr Mateus Mondin (ESALQ/USP) for critical review of part of the manuscript and Dr Aluana G. Abreu (IAC) for insightful discussion. This work was partially supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). M.M.C. received a PhD fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Part of this work was carried out using the resources of the CBSU from Cornell University, which is partially funded by Microsoft Corporation.
LITERATURE CITED
- Abbott RJ, Comes HP. Blowin' in the wind – the transition from ecotype to species. New Phytologist. 2007;175:197–200. doi: 10.1111/j.1469-8137.2007.02127.x. [DOI] [PubMed] [Google Scholar]
- APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Bir SS, Gill BS, Bedi YS, Singhal VK. Evolutionary status of the woody taxa of Garhwal Himalaya. In: Khosla PK, editor. Improvement of forest biomass. Solan: Society of Tree Scientists; 1980. pp. 81–96. [Google Scholar]
- Bradshaw HD, Stettler RF. Molecular genetics of growth and development in Populus. I. Triploidy in hybrid poplars. Theoretical and Applied Genetics. 1993;86:301–307. doi: 10.1007/BF00222092. [DOI] [PubMed] [Google Scholar]
- Cavallari MM, Billot C, Bouvet J-M, et al. Isolation and characterization of microsatellite markers for Casearia sylvestris Sw. (Salicaceae), a neotropical medicinal tree. Molecular Ecology Resources. 2008;8:802–804. doi: 10.1111/j.1755-0998.2007.02069.x. [DOI] [PubMed] [Google Scholar]
- Cervera M-T, Storme V, Ivens B, et al. Dense genetic linkage maps of three Populus species. Populus deltoides, P. nigra and P. trichocarpa based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. doi: 10.1093/genetics/158.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers GK, MacAvoy ES. Microsatellites: consensus and controversy. Comparative Biochemistry and Physiology. 2000;126:455–476. doi: 10.1016/s0305-0491(00)00233-9. [DOI] [PubMed] [Google Scholar]
- Cronquist A. An integrated system of classification of flowering plants. New York: Columbia University Press; 1981. [Google Scholar]
- Csaikl UM, Bastian H, Brettschneider R, et al. Comparative analysis of different DNA extraction protocols: a fast, universal maxi-preparation of high quality plant DNA for genetic evaluation and phylogentic studies. Plant Molecular Biology Reporter. 1998;16:69–86. [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond CS, Hamilton MB. Hierarchical components of genetic variation at a species boundary: population structure in two sympatric varieties of Lupinus microcarpus. (Leguminosae) Molecular Ecology. 2007;16:753–769. doi: 10.1111/j.1365-294X.2006.03186.x. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial – DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg M, Bergman M, Kullberg J, Wahlberg N, Wiklund C. Niche separation in space and time between two sympatric sister species—a case of ecological pleiotropy. Evolutionary Ecology. 2008;22:1–18. [Google Scholar]
- Fritsche F, Kaltz O. Is the Prunella (Lamiaceae) hybrid zone structured by an environmental gradient? Evidence from a reciprocal transplant experiment. American Journal of Botany. 2000;87:995–1003. [PubMed] [Google Scholar]
- Gegear RJ, Burns JG. The birds, the bees, and the virtual flowers: can pollinator behavior drive ecological speciation in flowering plants? American Naturalist. 2007;170:551–556. doi: 10.1086/521230. [DOI] [PubMed] [Google Scholar]
- Gottlieb LD. Conservation and duplication of isozymes in plants. Science. 1982;216:373–380. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT version 1·2: a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Goulart MF, Lemos Filho JP, Lovato MB. Phenological variation within and among populations of Plathymenia reticulata in Brazilian Cerrado, the Atlantic Forest and transitional sites. Annals of Botany. 2005;96:445–455. doi: 10.1093/aob/mci193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffer J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Lacerda DR, Lemos Filho JP, Acedo MDP, Lovato MB. Molecular differentiation of two vicariant neotropical tree species, Plathymenia foliolosa and P. reticulata. (Mimosoideae), inferred using RAPD markers. Plant Systematics and Evolution. 2002;235:67–77. [Google Scholar]
- Lawton-Rauh A. Evolutionary dynamics of duplicated genes in plants. Molecular Phylogenetics and Evolution. 2003;29:396–409. doi: 10.1016/j.ympev.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, et al. Species relative abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Leskinen E, Alström-Rapaport C. Molecular phylogeny of Salicaceae and closely related Flacourtiaceae: evidence from 5·8S, ITS1, and ITS2 of the rDNA. Plant Systematics and Evolution. 1999;215:209–227. [Google Scholar]
- Lexer C, Fay MF, Joseph JA, Nica MS, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): the role of ecology and life history in gene introgression. Molecular Ecology. 2005;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. [DOI] [PubMed] [Google Scholar]
- Minder AM, Widmer A. A population genomic analysis of species boundaries: neutral processes, adaptive divergence and introgression between two hybridizing plant species. Molecular Ecology. 2008;17:1552–1563. doi: 10.1111/j.1365-294X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- Morgan-Richards M, Wolff K. Genetic structure and differentiation of Plantago major reveals a pair of sympatric sister species. Molecular Ecology. 1999;8:1027–1036. [Google Scholar]
- Orr MR, Smith TB. Ecology and speciation. Trends in Ecology and Evolution. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratter JA, Riveiro JF, Bridgewater S. The Brazilian cerrado vegetation and threats to its biodiversity. Annals of Botany. 1997;80:223–230. [Google Scholar]
- Remington DL, Robichaux RH. Influences of gene flow on adaptive speciation in the Dubautia arborea–D. ciliolata complex. Molecular Ecology. 2007;16:4014–4027. doi: 10.1111/j.1365-294X.2007.03447.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Saint-Hilaire A. Casearia lingua. Cambess. (Eichler) Flora brasiliae meridionalis. 1829;2:236–236. [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology and Evolution. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. ARLEQUIN: a software for population data analysis. Geneva: University of Geneva; 2000. version 2·0. [Google Scholar]
- Segarra-Moragues JG, Palop-Esteban M, Gonzalez-Candelas F, Catalan P. Characterization of seven (CTT)n microsatellite loci in the Pyrenean endemic Borderea pyrenaica (Dioscoreaceae): remarks on ploidy level and hybrid origin assessed through allozymes and microsatellite analyses. Journal of Heredity. 2004;95:177–183. doi: 10.1093/jhered/esh028. [DOI] [PubMed] [Google Scholar]
- Silva JF. Biodiversity and stability in tropical savannas. In: Solbrig OT, Medina E, Silva JF, editors. Biodiversity and savanna ecosystem processes: a global perspective. Berlin: Springer-Verlag; 1996. pp. 161–171. [Google Scholar]
- Silva MAS, Ming LC, Pereira AMS, Bertoni BW, Batistini AP, Pereira PS. Phytochemical and genetic variability of Casearia sylvestris Sw. from São Paulo State Atlantic Forest and Cerrado populations. Revista Brasileira de Plantas Medicinais. 2006;8:159–166. [Google Scholar]
- Sleumer HO. Flora Neotropica Monograph n.22. FLACOURTIACEAE. New York: The New York Botanical Garden; 1980. [Google Scholar]
- Slotta TAB, Porter DM. Genetic variation within and between Iliamna corei and I. remota (Malvaceae): implications for species delimitation. Botanical Journal of the Linnean Society. 2006;151:345–354. [Google Scholar]
- Smith TB, Kark S, Schneider CJ, Wayne RK, Moritz C. Biodiversity hotspots and beyond: the need for preserving environmental transitions. Trends in Ecology and Evolution. 2001;16:431. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2003;161:173–191. [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences. 2000;97:7051–7057. doi: 10.1073/pnas.97.13.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres RB, Ramos R. Flacourtiaceae. In: Wanderley MGL, Shepherd GJ, Melhem TSA, Giulietti AM, editors. Flora Fanerogâmica do Estado de São Paulo. São Paulo: Instituto de Botânica; 2007. pp. 201–223. [Google Scholar]
- Torres RB, Yamamoto K. Taxonomia das espécies de Casearia Jacq. (Flacourtiaceae) do estado de São Paulo. Revista Brasileira de Botânica. 1986;9:239–258. [Google Scholar]
- Turesson G. The genotypical response of the plant species to the habitat. Hereditas. 1922;3:211–350. [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa. Torr. and Gray. Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Vähä J-P, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Valbuena-Carabaña M, González-Martínez SC, Sork VL, et al. Gene flow and hybridization in a mixed oak forest [Quercus pyrenaica Willd. and Q. petraea (Matts). Liebl.] in central Spain. Heredity. 2005;95:457–465. doi: 10.1038/sj.hdy.6800752. [DOI] [PubMed] [Google Scholar]
- Van Ee BW, Jelinski N, Berry PE, Hipp AL. Phylogeny and biogeography of Croton alabamensis (Euphorbiaceae), a rare shrub from Texas and Alabama, using DNA sequence and AFLP data. Molecular Ecology. 2006;15:2735–2751. doi: 10.1111/j.1365-294X.2006.02970.x. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Wiens J. Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution. 2004;58:193–197. doi: 10.1111/j.0014-3820.2004.tb01586.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:395–420. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX. POPGENE, the user-friendly shareware for population genetic analysis. Edmonton: Molecular Biology and Biotechnology Centre, University of Alberta; 1997. [Google Scholar]
- Yin TM, DiFazio SP, Gunter LE, Riemenschneider D, Tuskan GA. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theoretical and Applied Genetics. 2004;109:451–463. doi: 10.1007/s00122-004-1653-5. [DOI] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]