Abstract
Background and Aims
Transgenic plants represent an excellent tool for experimental plant biology and are an important component of modern agriculture. Fully understanding the stability of transgene expression is critical in this regard. Most changes in transgene expression occur soon after transformation and thus unwanted lines can be discarded easily; however, transgenes can be silenced long after their integration.
Methods
To study the long-term changes in transgene expression in potato (Solanum tuberosum), the activity of two reporter genes, encoding green fluorescent protein (GFP) and neomycin phosphotransferase (NPTII), was monitored in a set of 17 transgenic lines over 5 years of vegetative propagation in vitro.
Key Results
A decrease in transgene expression was observed mainly in lines with higher initial GFP expression and a greater number of T-DNA insertions. Complete silencing of the reporter genes was observed in four lines (nearly 25 %), all of which successively silenced the two reporter genes, indicating an interconnection between their silencing. The loss of GFP fluorescence always preceded the loss of kanamycin resistance. Treatment with the demethylation drug 5-azacytidine indicated that silencing of the NPTII gene, but probably not of GFP, occurred directly at the transcriptional level. Successive silencing of the two reporter genes was also reproduced in lines with reactivated expression of previously silenced transgenes.
Conclusions
We suggest a hypothetical mechanism involving the successive silencing of the two reporter genes that involves the switch of GFP silencing from the post-transcriptional to transcriptional level and subsequent spreading of methylation to the NPTII gene.
Keywords: 5-Azacytidine, de novo regeneration, green fluorescent protein (GFP), kanamycin resistance test, DNA methylation, (P)TGS, reactivation, Solanum tuberosum, transgene silencing
INTRODUCTION
Over the past two decades, transgenic plants have become an indispensable tool in studies of plant physiology and functional analyses of new genes identified by genomic, transcriptomic and proteomic approaches. Balanced and stable expression of introduced genes is an important aspect regarding the utility of transgenic plants in both basic research and agriculture. Numerous studies have concentrated on this topic in different plant species, including both model plants and agriculturally important crops (e.g. Ottaviani et al., 1993; Vain et al., 2002; De Buck et al., 2004; Nocarova and Fischer, 2009).
The level and stability of transgene expression is influenced primarily by the composition of introduced rDNA. De Bolle et al. (2003) investigated the impact of different regulatory elements on the level and variability of transgene expression in Arabidopsis thaliana. Variability in expression was not affected by either terminators nor 5′ untranslated regions, in contrast to promoters which drastically influenced not only expression levels but also greatly affected expression variability (De Bolle et al., 2003).
The number of transgene insertions within the genome and the arrangement of individual copies in the insertion locus is another important factor affecting transgene expression level and stability (reviewed in Butaye et al., 2005). Hobbs et al. (1990) reported that transformants with high transgene expression had predominantly a single T-DNA insertion, whereas transformants with low expression had usually multiple insertions at the same or different loci. Transformed plants with multiple T-DNA copies also have greater tendency towards being silenced at the post-transcriptional level (post-transcriptional gene silencing, PTGS) compared with plants with a single transgene insertion (Sallaud et al., 2003; Tang et al., 2007; reviewed in Depicker et al., 2005). Multiple transgene insertions can be arranged as inverted repeats that may directly produce double-stranded RNA (dsRNA) via transcription (Muskens et al., 2000). Moreover, high expression levels of transgenes, potentially resulting from higher transgene copy number, are considered to be connected with accidental occurrence of aberrant transcripts, which are recognized by RNA-dependent RNA polymerase RDR6, which forms dsRNA (Luo and Chen, 2007). Dicer-like mediated cleavage of this dsRNA produces small interfering RNAs (siRNAs), which are central to numerous mechanisms of the RNA silencing machinery, including PTGS, mediated through either degradation of mRNA or interference with the process of translation (Dalmay et al., 2000; reviewed in Brodersen and Voinnet, 2006). In addition to acting on mRNA, siRNAs are also responsible for homology-dependent TGS. RNA polymerase V, involved in RNA-dependent DNA methylation, has been demonstrated to interact directly with the siRNA–Argonaute complex and to mediate de novo methylation of cytosines by DRM2 (Cao and Jacobsen, 2002; Kanno et al., 2005; Wierzbicki et al., 2008; Daxinger et al., 2009). Although the impact of methylation in the gene coding sequence remains elusive (Zilberman et al., 2007), cytosine methylation and formation of compactly arranged heterochromatin in the promoter region appear to be linked to reduced or silenced gene expression (Linn et al., 1990; Fojtová et al., 2003; Depicker et al., 2005; Zilberman et al., 2007).
Transgene expression can also be influenced by the chromosomal environment, indicating that heterochromatin features can spread to the neighbouring inserted DNA sequences, resulting in direct TGS (Pröls and Meyer, 1992; Kim et al., 2007; Gelvin and Kim, 2007). A previous study on transgenic BY-2 cell lines, however, documented that even identical transgene insertions could result in completely different patterns of expression; these patterns appeared to be randomly established and stabilized early after transgene insertion (Nocarova and Fischer, 2009). In contrast, De Buck et al. (2004) demonstrated that 19 of 21 single-copy T-DNA Arabidopsis transgenic lines showed comparable transgene expression levels, which were independent of the orientation of the T-DNA or its integration into an intergenic or gene region, or into an exon or an intron. This unexpected result could be partially biased (Gelvin and Kim, 2007; Kim et al., 2007) because only lines with active expression of the resistance gene were selected in the studies.
Most of the studies mentioned above focused on transgene expression variability early after transformation. To evaluate the nature and potential causes of gene silencing long after transformation, the expression of two reporter genes inserted together (within a T-DNA) was followed in potato plants (Solanum tuberosum). A set of 17 independent lines obtained from Agrobacterium-mediated transformation were characterized and monitored over 5 years of in vitro vegetative propagation. Expression of the two reporter genes encoding green fluorescent protein (GFP) and neomycin phosphotransferase (NPTII) was monitored based on the presence or biological activity of the reporter proteins and was confirmed at the RNA level. The unique phenomenon of successive/coordinated transgene silencing observed in several independent transgenic lines is discussed with regard to the known mechanisms of post-transcriptional and transcriptional gene silencing.
MATERIALS AND METHODS
Plant transformation
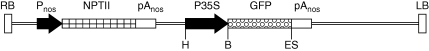
Potato plants (Solanum tuberosum L. ‘Désirée’) were cultured in vitro on LS medium (Linsmayer and Skoog, 1965) containing 3 % sucrose and were subcultured using apical or nodal cuttings every 4–6 weeks. Transformation of leaves was performed according to Dietze et al. (1995) using Agrobacterium tumefaciens (strain C58C1 with a plasmid pGV2260; Deblaere et al., 1985) carrying modified binary vector pCP60 (kindly provided by Dr P. Ratet, ISV-CNRS, France; Bolte et al., 2004) with a gene encoding red-shifted green fluorescent protein (RS-GFP, the excitation spectrum shifted to longer wavelengths; kindly provided by ABRC, Ohio State University, Columbus, OH, USA; Davis and Vierstra, 1998), inserted under the control of the CaMV 35S promoter with a single enhancer region. The T-DNA further contained the NPTII gene driven by a nopalinsynthase promoter (pNOS), which provided kanamycin resistance (Fig. 1). Regeneration of transgenic lines took about 2–3 months on the media containing kanamycin; subsequently, the regenerated lines were not selected for expression of the NPTII gene.
Fig. 1.
The scheme of T-DNA introduced to potato lines. Position of restriction sites used for Southern hybridization is indicated by letters; H, HindIII; B, BamHI; E, EcoRI; S, SacI.
Detection of the GFP gene by PCR and Southern hybridization
Total DNA was isolated from 150 mg (f. wt) of leaf according to Shure et al. (1983). The presence of the GFP gene was tested by PCR (in two replicates) with specific primers: gfp-F (3′AGTGGAAGTGGGAGAGGTGA5′) and gfp-R (3′CAGGTGTGTTAGACGGGAAA5′) using 200 ng of total DNA as a template.
Transgene copy number was determined by Southern hybridization (Sambrook et al., 1989) of genomic DNA, digested in separate reactions by EcoRI and HindIII. In unclear cases (potentially caused by star activity of the restrictases), additional digestions with SacI and BamHI were performed. DNA fragments were separated on agarose gels (0·8 %, 0·2 V cm−1; Sambrook et al., 1989) and blotted onto Nylon+ membrane (Roche, Mannheim, Germany). For probe preparation, the whole GFP gene was amplified by PCR with gfp-F and gfp-R primers in the presence of dUTP-DIG (Roche). The probe labelling, hybridization and detection procedure was performed according to the manufacturer's recommendations (Roche, 2010) with CDP-Star CL-AP chemiluminiscent substrate (Novagen-Merck, Darmstadt, Germany).
Semi-quantitative RT-PCR
Total RNA was isolated from leaves using the RNeasy Plant Mini Kit (Quiagen, Hilden, Germany). Total RNA (1 µg) was treated with DNase I, RNase-free (Fermentas, Burlington, ON, Canada), and used for reverse transcription with RevertAidTM M-MuLV Reverse Transcriptase (Fermentas) primed with anchored oligo-T23 primer. One microlitre of the reverse transcription reaction was used as a template for the subsequent PCR with specific primers for GFP: gfp-F and gfp-R (see above); NPTII: NPTF (3′GGTAGAACAAGTTAGTACGCTTT5′) and NPTR (3′GAACTGCTCAAGAAGACTCGC5′). The elongation factor EF1α (Nicot et al., 2005) was used as an internal standard with primers according to Dvorakova et al. (2007): EF1F (3′TACTGCACTGTGATTGATGCC5′) and EF1R (3′AGCAAATCATTTGCTTGACACC5′).
GFP fluorescence
GFP fluorescence was evaluated under an Olympus Provis AX70 fluorescence microscope equipped with a filter set for detection of fluorescein isothiocyanate (U-MWU). Images were grabbed with a digital camera Sony DXC-950P TV (Sony Corp., Tokyo, Japan). Relative intensity of the GFP fluorescence was determined by use of a macro in Lucia image analysis software (Laboratory Imaging, Prague, Czech Republic). Quantification was done in roots grown on apical cuttings of in vitro plants (3 weeks after planting). The roots were analysed at the beginning of the differentiation zone, i.e. about 5 mm from the root tip.
Immunodetection of GFP
Total soluble proteins were isolated from leaves (200 mg f. wt). The tissue was homogenized in liquid nitrogen and mixed with 200 µL phosphate-buffered saline (pH 7·4) supplemented with 1 mm phenylmethylsulfonyl fluoride (PMSF). The fraction of soluble proteins was obtained as a supernatant after centrifugation for 15 min, 20 000g, at 4 °C. The concentration of proteins was determined according to Popov et al. (1975). Proteins samples (50 µg per line) were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis as described by Sambrook et al. (1989) and electro-blotted onto the nitrocellulose membrane NC45 (Serva Electrophoresis GmbH, Heidelberg, Germany). Comparable amounts of proteins in individual lines and efficiency of blotting were confirmed by transient staining of the membranes with Ponceau S (Salinovich and Montelaro, 1986). Mouse monoclonal anti-GFP antibody (mixture of clone 7·1 and 13·1; Roche) was used at a dilution of 1 : 1000, and a secondary antibody against mouse IgG (from goat) was conjugated with alkaline phosphatase (Sigma-Aldrich, St Louis, MO, USA). The intensity of the colour precipitate was measured by a macro in Lucia image analysis software after incubation of the membranes in the BCIP/NBT-liquid substrate system (Sigma-Aldrich). In order to deal with minor differences between repetitions and differences between immunodetection and quantification of fluorescence, the signal intensities were classed into only three categories; high, low and undetectable.
Kanamycin resistance test
Resistance to kanamycin was tested based on callus formation. Pieces of leaves or stem internodal segments (0·5–1 cm long) from in vitro grown plants were cut with a razor blade and cultivated for 2–3 weeks on solidified LS medium supplemented with 5 mg L−1 NAA, 0·1 mg L−1 BAP and 100 L−1 kanamycin. The medium was mashed with forceps to provide maximal contact with the segments (leaf segments were inserted upside down).
AzaC treatment
Leaf explants from in vitro plants with silenced transgene expression were transiently treated with 10 µm 5-azacytidine (AzaC) and subsequently cultured on the medium supplemented with 50 mg L−1 kanamycin (E. Nocarova and L. Fischer, unpubl. res). Reactivation of NPTII expression was visible as formation of calli after 2–3 weeks. Reactivation of GFP expression was determined by fluorescence microscopy (see above) 3–7 d after AzaC treatment.
RESULTS
Generation and selection of transgenic lines; determination of T-DNA copy number
Thirty-one potato clones were regenerated after Agrobacterium-mediated transformation with a T-DNA containing two reporter genes encoding GFP and NPTII. For long-term monitoring, 17 lines that matched the following criteria were selected: normal growth characteristics and presence of the GFP gene, as confirmed by PCR (data not shown).
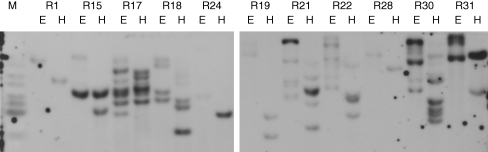
The number of T-DNA insertions per genome ranged from one to approx. six, as estimated roughly from Southern hybridization of genomic DNA cleaved by two or four restriction endonucleases (Fig. 2). One or two copies per genome were detected in about 60 % of the lines (Table 1).
Fig. 2.
Southern hybridization of total genomic DNA. A DIG-labelled probe of the GFP gene was hybridized with DNA cleaved by EcoRI (E) and HindIII (H). T-DNA copy number was estimated as the number of hybridizing bands.
Table 1.
Overview of the transgenic lines ordered according to T-DNA copy number
| Expression of the reporter genes |
|||||||
|---|---|---|---|---|---|---|---|
| 6 months |
1·5 years |
4·5 years |
|||||
| Line | T-DNA copy number | NPT II | GFP | NPT II | GFP | NPT II | GFP |
| R1 | 1 | R | ++ | R | ++ | R | ++ |
| R10 | 1 | R | ++ | R | ++ | R | ++ |
| R24 | 1 | R | + | R | + | R | + |
| R25 | 1 | R | ++ | R | ++ | R | ++ |
| R26 | 1 | R | ++ | R | ++ | R | ↓+ |
| R28 | 1 | R | ++ | R | ↓− | S | − |
| R15 | 2 | R | ++ | R | ++ | R | ++ |
| R16 | 2 | R | + | R | + | R | + |
| R19 | 2 | R | ++ | R | ++ | R | ++ |
| R31 | 2 | R | ++ | R | ++ | R | ++ |
| R18 | 4 | R | + | R | + | R | + |
| R22 | 4 | R | ++ | R | ↓+ | R | + |
| R23 | 4 | R | + | R | + | R | + |
| R5 | 5 | R | + | R | + | R | + |
| R21 | 5 | R | + | R | ↓− | S | − |
| R17 | 6 | R | + | R | ↑+ + | S | ↓− |
| R30 | 6 | R | ↓− | S | − | S | − |
Expression of the reporter genes (NPTII and GFP) was determined 0·5, 1·5 and 4·5 years from the beginning of transformation. NPTII expression was evaluated as resistance (R) or sensitivity (S) to kanamycin. Levels of GFP (determined on two parallel immunostained Western blots) were sorted into three categories indicating high, low and no expression, as shown with ++ , + and −, respectively. Arrows (↑ ↓ ) indicate a decrease or increase in expression as compared with the previous determination.
Determination of GFP levels
The intensity of green fluorescence was measured in different potato organs and tissues (apical meristems, leaves and roots) to estimate the level of GFP gene expression. However, the results were generally variable amongst repetitions. The most reproducible results were obtained using roots of in vitro plants grown from apical cuttings, where fluorescence was determined at the beginning of the differentiation zone, i.e. approximately 5 mm from the root tip (see Supplementary Information). Moderate fluorescence variability in these root samples was predominantly connected with slightly varying root thickness.
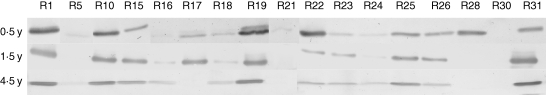
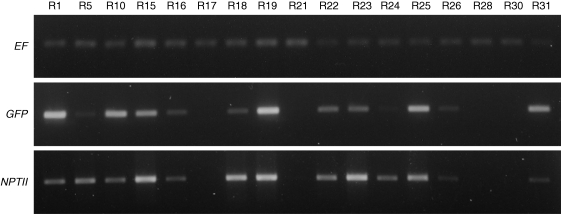
An alternative method of GFP quantification was based on immunodetection. This technique clearly differentiated between GFP levels in individual lines (see Supplementary Information) and therefore was used as a complementary method for monitoring of GFP expression in subsequent experiments (Fig. 3). Immunodetection results closely matched the GFP transcript levels detected by semi-quantitative RT-PCR (Fig. 4).
Fig. 3.
Immunodetection of GFP on western blots in the transgenic potato lines during 4·5 years of cultivation. The intensity of colour precipitate, a product of alkaline phosphatase activity (conjugated with the secondary antibody), corresponds to the GFP levels in the samples (representative blots of two replicates); y, year from the beginning of transformation.
Fig. 4.
Transcript levels of the GFP and NPTII genes in the transgenic lines 4·5 years after transformation. Shown are ethidium bromide-stained gels of semi-quantitative RT-PCR (representative gels of two replicates). Expression of the EF1α gene was used as the internal standard.
Determination of kanamycin resistance
To monitor the activity of the second reporter gene, NPTII, inhibition of root and shoot growth was evaluated on medium supplemented with kanamycin. The results were not reliable or reproducible, and therefore a simple test was introduced based on callus formation on either leaf pieces or stem internodal cuttings that were cultured on the callus-inducing medium supplemented with kanamycin (100 mg L−1). After 2–3 weeks of cultivation, massive callus formation was observed on explants from resistant plants, which remained dark green. In contrast, callus formation was completely inhibited on explants from non-resistant plants and the segments became yellowish (Fig. 5). The results obtained using this simple test correlated well with the detection of NPTII transcripts using RT-PCR (Fig. 4).
Fig. 5.
Kanamycin resistance test. (a) Leaf and (b) stem-internodal segments were cultivated for 20 d on the callus-inducing medium supplemented with 100 mg L−1 kanamycin. R, green segments of resistant plants forming massive calli; WT, yellowish segments of wild-type plants with no callus formation.
Long-term changes in reporter gene activities
GFP levels and kanamycin resistance were first determined approx. 6 months after co-cultivation with Agrobacterium when all regenerated plants were stably introduced in the culture (all regenerated plants were resistant to kanamycin, as kanamycin was applied for selection after co-cultivation). Thereafter, the determination was repeated after an additional 1 year and 4 years, i.e. 1·5 and 4·5 years from the beginning of transformation.
Based on the GFP levels that were determined by immunodetection and quantification of GFP fluorescence in roots, the lines were classed into three categories, with high, low and undetectable signal intensity (as indicated ++ , + and –, respectively, in Table 1). Counts of the lines in the three categories are shown in Fig. 6.
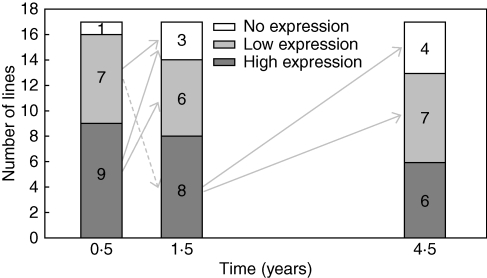
Fig. 6.
Changes in GFP levels in the transgenic potato lines during 4 years of cultivation. The lines were sorted into three categories with high, low and no expression, according to the levels of GFP. Numerals in the columns indicate numbers of lines in a given category. The arrows indicate changes in expression levels (transfers between the categories) in the respective periods; the dashed arrow shows an increase in GFP expression in line R17.
The decline in the GFP level or complete silencing of the GFP gene was observed in six lines. By contrast, in one line (R17), expression increased during the first year (Fig. 3). Six months after the transformation, expression of the GFP gene was silenced in a single line (R30). Three lines (R21, R22, R28) showed decreased or silenced GFP expression during the subsequent year, and similarly two other lines (R17, R26) during the last 3 years of cultivation (Table 1). The majority of these lines (four of six) contained four or more copies of T-DNA, representing more than 50 % of these ‘multicopy’ lines. In contrast, decreased or silenced GFP expression was observed in only 20 % of lines (two of ten) with a single or two T-DNA copies. Consequently, lines with low T-DNA copy number predominated within those expressing high levels of GFP at the end of the study. By contrast, high initial levels of GFP were characteristic for the majority of lines (four of five) that subsequently decreased or silenced GFP gene expression.
Six months after the transformation, all selected lines were resistant to kanamycin, as this was one of the selection criteria. During the subsequent year, loss of resistance was observed in a single line, R30, in which GFP expression had been silenced before the first determination (Table 1). All other lines remained kanamycin resistant, although lines R21 and R28 had already silenced expression of the GFP gene. Four and half years after the transformation, all lines with silenced GFP expression also lost kanamycin resistance (Table 1). The presence of the GFP and NPTII genes in these lines was revealed by PCR (data not shown) and AzaC treatment (see below).
Reactivation of transgene expression by AzaC
Treatment of leaf segments taken from the lines with silenced expression of the two transgenes (R17, R21, R28 and R30) with AzaC, an inhibitor of maintenance of methylation (Santi et al., 1984), resulted in partial reactivation of the two reporter genes. GFP reactivation was visible as GFP fluorescence in individual cells a few days after AzaC treatment. NPTII proved to be reactivated in some cells, as calli were forming on the explants even in the presence of kanamycin (E. Nocarova and L. Fischer, unpubl. res.). No GFP fluorescence or callus formation was observed on AzaC-untreated segments taken from the same plants and on AzaC-treated leaves from untransformed controls (data not shown).
Transient treatment of leaf pieces with AzaC, followed by kanamycin selection, was optimized to regenerate the whole plants with reactivated expression of silenced transgenes – either the NPTII gene alone or both NPTII and GFP (E. Nocarova and L. Fischer, unpubl. res.). The response to AzaC treatment and the stability of reactivated expression of the two reporter genes differed between the multicopy line R17 and the single-copy line R28. Two of six R17 regenerants showed reactivated expression of both transgenes. Although GFP reactivation was clearly visible after AzaC treatment in the R28 line as well, none of four regenerants from this line showed active GFP expression.
Activity of the two transgenes was monitored in these ten regenerated lines every 2 months. Lines with silenced expression of any of the two genes were also periodically treated with AzaC to test whether the genes were silenced at the transcriptional level and whether it was possible to reactivate their expression through DNA demethylation (Table 2); PTGS mediated by siRNAs is not substantially affected by DNA demethylation (Wang and Waterhouse, 2000). In R28 regenerants, reactivation of the silenced GFP gene was not possible during the first 4 months after regeneration. Two months later, GFP fluorescence was visible after AzaC treatment in all R28 lines. Thereafter, 2–4 months later, three lines (R28A1, A2/1, A2/2) silenced NPTII expression. The last line (R28A2/3) has remained resistant. Reactivation of the NPTII gene by AzaC was tested directly after the detection of silencing; it was successful in all three lines (Table 2).
Table 2.
Monitoring of GFP and NPTII activity in ten lines with reactivated expression of the reporter genes
| 0 months |
2 months |
4 months |
6 months |
8 months |
10 months |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Line | GFP | GFP-AzaC | NPTII | NPTII-AzaC | GFP | GFP-AzaC | NPTII | NPTII-AzaC | GFP | GFP-AzaC | NPTII | NPTII-AzaC | GFP | GFP-AzaC | NPTII | NPTII-AzaC | GFP | GFP-AzaC | NPTII | NPTII-AzaC | GFP | GFP-AzaC | NPTII | NPTII-AzaC |
| R28 (silenced) | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + |
| R28A1(A2/1) | − | − | + | n.d. | − | − | + | n.d. | − | − | + | n.d. | − | + | + | n.d. | − | + | + | n.d. | − | + | − | + |
| R28A2/2 | − | − | + | n.d. | − | − | + | n.d. | − | − | + | n.d. | − | + | + | n.d. | − | + | − | n.d. | − | + | − | + |
| R28A2/3 | − | − | + | n.d. | − | − | + | n.d. | − | − | + | n.d. | − | + | + | n.d. | − | + | + | n.d. | − | + | + | n.d. |
| R17 (silenced) | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + |
| R17A4 (A6) | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. | + | n.d. |
| R17A1 (A5) | − | − | + | n.d. | − | −* | −* | n.d. | − | + | −* | −* | − | + | − | + | − | + | − | + | n.d. | n.d. | n.d. | n.d. |
| R17A2 | − | − | + | n.d. | − | + | + | n.d. | − | + | + | n.d. | − | + | − | n.d. | − | + | − | + | n.d. | n.d. | n.d. | n.d. |
| R17A3 | − | − | + | n.d. | − | + | + | n.d. | − | + | − | + | − | + | − | n.d. | − | + | − | + | n.d. | n.d. | n.d. | n.d. |
The lines were obtained by reactivation of transgene expression in completely silenced lines R28 and R17 (included as controls) via AzaC treatment (E. Nocarova and L. Fischer, unpubl. res.). The ability to reactivate transgene expression by AzaC (GFP-AzaC; NPTII-AzaC) was usually determined as late as after detection of silencing (i.e. about 2–4 weeks later – the results are given in the column for the next assessment). +, Active gene or activated by AzaC; –, inactive gene or incapable of being activated by AzaC; n.d., not determined.
* Non-typical time behaviour of silencing.
Two R17 regenerants (R17A4 and A6) with reactivated GFP have shown continued active expression of both transgenes (more than 1 year; Table 2). The other R17 regenerants (R17A1, A2, A3, A5) had GFP silenced already after regeneration. The NPTII gene was silenced in all these lines during the subsequent 2–6 months. In lines R17A1 and A5, the GFP gene could be reactivated by AzaC as late as 2 months after silencing of the NPTII gene. Moreover, the resistance to kanamycin could not be directly reactivated by AzaC treatment in these two lines (Table 2). The progress of silencing in the lines R17A2 and A3 was similar to that of R28 regenerants, although faster (Table 2).
DISCUSSION
Mitotic instability of transgene expression
Two recent studies that focused on long-term monitoring of transgenic perennial Prunus subhirtella and vegetatively propagated Agapanthus demonstrated very stabile expression of the reporter gene (Maghuly et al., 2007; Mori et al., 2007). In contrast, the present results demonstrate relatively frequent silencing of the reporter genes in vegetatively propagated potato plants even quite time after the transformation. As the same promoter (CaMV 35S) controlled the expression of the reporter genes in all three studies, either the gene sequence itself or the acceptor species might be responsible for the differences in the frequency of silencing; for example, members of the family Solanaceae were reported to be very active in systemic spreading of siRNAs involved in post-transcriptional silencing (reviewed in Voinnet, 2005). The other possible cause could reside in the different culture conditions; in vitro culture is generally stressful and suboptimal conditions were reported to induce transgene silencing (Meyer et al., 1992; Walter et al., 1992).
Factors responsible for long-term transgene silencing
The initial expression level and the transgene copy number appeared to be the key factors influencing transgene silencing in our study. This finding corresponds well with the factors identified in short-term studies (e.g. Hobbs et al., 1990; Linn et al., 1990; Sallaud et al., 2003). As similar factors influenced both short- and long-term silencing, we assume that these factors act as a predisposition; whether the silencing really occurs or not is probably accidental and/or depends on multiple factors, as also indicated by different timing of silencing in genetically identical lines obtained after reactivation of silenced transgenes by AzaC (Table 2; E. Nocarova and L. Fischer, unpubl. res.).
Successive silencing of tandem transgenes
The site of insertion influences transgene expression, so the expression of two tandem genes transferred within T-DNA might be expected to correlate. Mlynárová et al. (2002), however, observed interconnections between the expressions of tandem transgenes only when the T-DNA contained matrix-associated regions (MARs). In the present study, we used T-DNA lacking MARs and observed a strong correlation between silencing of the two reporter genes. Silencing of one gene was always accompanied by silencing of the other. This observation contrasts with the results of Ottaviani et al. (1993), who studied the expression of GUS and NPTII genes in transgenic potato plants. They demonstrated silencing of only the GUS gene in three of seven lines. Given that our results indicate that there is a gap (several months) between the GFP and NPTII gene silencing, the absence of NPTII silencing in Ottaviani et al. (1993) probably resulted from too short an evaluation period.
In the present experiments, silencing of the GFP gene preceded that of the NPTII gene in all cases. In line R30, this succession could be connected with the initial selection of regenerants for resistance to kanamycin, whereas in the other lines preferential silencing of GFP resulted from the higher expression level – the GFP gene was controlled by a stronger promoter (CaMV 35S) than NPTII (nopalin synthase promoter). This assumption is also supported by the results of Ottaviani et al. (1993), who observed only silencing of the GUS gene driven by the CaMV 35S promoter, but no silencing of the NPTII gene controlled by nopalin synthase promoter. The high (supraliminal) levels of transcription from the strong 35S promoter can lead to formation of aberrant mRNAs, which are templates for RNA-dependent RNA polymerase (Luo and Chen, 2007). This enzyme forms dsRNA, an initial substrate for siRNA production involved in PTGS (Dalmay et al., 2000; reviewed in Brodersen and Voinnet, 2006).
Silencing of the GFP gene was demonstrably followed with silencing of NPTII in three genetically different (independent) transgenic lines. Moreover, in lines R17 and R28 this sequential silencing was repeated after reactivation of the previously silenced transgenes (E. Nocarova and L. Fischer, unpubl. res.). We therefore consider that the independent silencing events shared a similar mechanism. The following hypothetical four-step scenario is suggested: (1) the primary GFP silencing at the post-transcriptional level (PTGS) probably occurred accidentally in few cells of the plant; (2) siRNAs, produced primarily in these cells, spread subsequently throughout the plant (as reviewed in Voinnet, 2005); (3) GFP silencing at the post-transcriptional level switched in individual cells to transcriptional silencing accompanied by promoter methylation (TGS; Fojtová et al., 2003) – this step was indirectly confirmed by the effect of the DNA demethylation drug AzaC (Santi et al., 1984; Wang and Waterhouse, 2000); and finally (4) the methylation spread from the GFP/35S promoter to the NPTII gene and nopalin synthase promotor. This spreading could be mediated by RNA polymerase IV, which interacts with methylated DNA and generates templates for production of secondary siRNAs (Daxinger et al., 2009). These secondary siRNAs can cause, when complementary to the promoter region, direct silencing at the transcriptional level through the action of RNA polymerase V and DRM methyltransferases (Wierzbicki et al., 2008). Silencing of NPTII expression directly at the transcriptional level was indicated by the effect of AzaC in the majority of lines with reactivated expression of the NPTII gene (Table 2). Although the suggested model remains speculative, the corresponding time behaviour and the central switch from PTGS to TGS of the GFP gene was documented in the reactivated lines R28A1, A2/1, A2/2, R17A2 and A3. In these lines GFP had been presumably reactivated only transiently during regeneration and subsequently silenced at the post-transcriptional level, because AzaC was not effective in its reactivation.
The stepwise character of silencing of the two reporter genes indicates an interconnection between these processes. This kind of interaction between unrelated, physically connected transgenes is of general interest because it might occur also between transgenes and neighbouring plant genes. The mechanism of this connection and the reasons for the relatively long time lapse (several months) between the silencing of the GFP and NPTII genes require further study, including detailed analysis of transgene methylation and detection of specific siRNAs.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr S. J. Davis and Dr R. D. Vierstra (University of Wisconsin-Madison, USA) and the Arabidopsis Biological Research Center for providing us with the soluble-modified RS-GFP gene, and to Dr P. Ratet (ISV-CNRS, France) for providing the binary vector pCP60. This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (grant numbers LC06004, LC06034 and MSM0021620858).
LITERATURE CITED
- Bolte S, Brown S, Satiat-Jeunemaitre B. The N-myristoylated Rab-GTPase m-Rab(mc) is involved in post-Golgi trafficking events to the lytic vacuole in plant cells. Journal of Cell Science. 2004;117:943–954. doi: 10.1242/jcs.00920. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends in Genetics. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Butaye KMJ, Cammue BPA, Delaure SL, De Bolle MFC. Approaches to minimize variation of transgene expression in plants. Molecular Breeding. 2005;16:79–91. [Google Scholar]
- Cao XF, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Current Biology. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Molecular Biology. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Kanno T, Bucher E, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO Journal. 2009;28:48–57. doi: 10.1038/emboj.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle MFC, Butaye KMJ, Coucke WJW, et al. Analysis of the influence of promoter elements and a matrix attachment region on the inter-individual variation of transgene expression in populations of Arabidopsis thaliana. Plant Science. 2003;165:169–179. [Google Scholar]
- De Buck S, Windels P, De Loose M, Depicker A. Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable beta-glucuronidase accumulation levels. Cellular and Molecular Life Sciences. 2004;61:2632–2645. doi: 10.1007/s00018-004-4284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, Degreve H, et al. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene-transfer to plants. Nucleic Acids Research. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A, Sanders M, Meyer P. Transgene silencing. In: Meyer P, editor. Plant epigenetics. Vol. 19. Oxford: Blackwell Publishing Ltd; 2005. pp. 1–31. [Google Scholar]
- Dietze J, Blau A, Willmitzer L. Agrobacterium-mediated transformation of potato (Solanum tuberosum) In: Potrykus I, Spangenberg G, editors. Gene transfer to plants. Berlin: Springer-Verlag; 1995. pp. 24–29. [Google Scholar]
- Dvorakova L, Cvrckova F, Fischer L. Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genomics. 2007;8(412) doi: 10.1186/1471-2164-8-412. doi:10.1186/1471-2164-8-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojtová M, Van Houdt H, Depicker A, Kovarik A. Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiology. 2003;133:1240–1250. doi: 10.1104/pp.103.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB, Kim SI. Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biochimica et Biophysica Acta-Gene Structure and Expression. 2007;1769:410–421. doi: 10.1016/j.bbaexp.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Kpodar P, Delong CMO. The effect of T-DNA copy number, position and methylation on reporter gene-expression in tobacco transformants. Plant Molecular Biology. 1990;15:851–864. doi: 10.1007/BF00039425. [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature Genetics. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kim SI, Veena, Gelvin SB. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant Journal. 2007;51:779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- Linn F, Heidmann I, Saedler H, Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida – role of numbers of integrated gene copies and state of methylation. Molecular & General Genetics. 1990;222:329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- Linsmayer EM, Skoog F. Organic growth factor requirements of tobacco tissue cultures. Physiologia Plantarum. 1965;18:100–127. [Google Scholar]
- Luo ZH, Chen ZX. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghuly F, Machado AD, Leopold S, Khan MA, Katinger H, Laimer M. Long-term stability of marker gene expression in Prunus subhirtella: a model fruit tree species. Journal of Biotechnology. 2007;127:310–321. doi: 10.1016/j.jbiotec.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H. Endogenous and environmental-factors influence 35S promoter methylation of a maize A1 gene construct in transgenic Petunia and its color phenotype. Molecular & General Genetics. 1992;231:345–352. doi: 10.1007/BF00292701. [DOI] [PubMed] [Google Scholar]
- Mlynárová L, Loonen A, Mietkiewska E, Jansen RC, Nap JP. Assembly of two transgenes in an artificial chromatin domain gives highly coordinated expression in tobacco. Genetics. 2002;160:727–740. doi: 10.1093/genetics/160.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Oka E, Umehara H, et al. Somaclonal variation and stability of GUS gene expression in transgenic agapanthus (Agapanthus praecox ssp orientalis) plants at the flowering stage. In vitro Cellular & Developmental Biology-Plant. 2007;43:79–87. [Google Scholar]
- Muskens MWM, Vissers APA, Mol JNM, Kooter JM. Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Molecular Biology. 2000;43:243–260. doi: 10.1023/a:1006491613768. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Nocarova E, Fischer L. Cloning of transgenic tobacco BY-2 cells; an efficient method to analyse and reduce high natural heterogeneity of transgene expression. BMC Plant Biology. 2009;9(44) doi: 10.1186/1471-2229-9-44. doi:10.1186/1471-2229-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani MP, Smits T, Tencate CHH. Differential methylation and expression of the beta-glucuronidase and neomycin phosphotransferase genes in transgenic plants of potato cv bintje. Plant Science. 1993;88:73–81. [Google Scholar]
- Popov N, Schmitt S, Schulzeck S, Matthies H. Eine störungsfreie Mikromethode zur Bestimmung des Proteingehaltes in Gewebshomogenaten. Acta biologica et medica Germanica. 1975;34:1441–1446. [PubMed] [Google Scholar]
- Pröls F, Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant Journal. 1992;2:465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Roche. The DIG system user's guide for filter hybridization. Mannheim, Germany: F: Hoffmann–La Roche Ltd; 2010. [Google Scholar]
- Salinovich O, Montelaro RC. Reversible staining and peptide-mapping of proteins transferred to nitrocellulose after separation by sodium dodecyl-sulfate polyacrylamide-gel electrophoresis. Analytical Biochemistry. 1986;156:341–347. doi: 10.1016/0003-2697(86)90263-0. [DOI] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, et al. Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theoretical and Applied Genetics. 2003;106:1396–1408. doi: 10.1007/s00122-002-1184-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. Molecular-identification and isolation of the waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Tang W, Newton RJ, Weidner DA. Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. Journal of Experimental Botany. 2007;58:545–554. doi: 10.1093/jxb/erl228. [DOI] [PubMed] [Google Scholar]
- Vain P, James VA, Worland B, Snape JW. Transgene behaviour across two generations in a large random population of transgenic rice plants produced by particle bombardment. Theoretical and Applied Genetics. 2002;105:878–889. doi: 10.1007/s00122-002-1039-5. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Non-cell autonomous RNA silencing. FEBS Letters. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Walter C, Broer I, Hillemann D, Puhler A. High-frequency, heat treatment-induced inactivation of the phosphinothricin resistance gene in transgenic single cell-suspension cultures of Medicago sativa. Molecular & General Genetics. 1992;235:189–196. doi: 10.1007/BF00279360. [DOI] [PubMed] [Google Scholar]
- Wang MB, Waterhouse PM. High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Molecular Biology. 2000;43:67–82. doi: 10.1023/a:1006490331303. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genetics. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.