Abstract
The plastid is an organelle vital to all photosynthetic and some non-photosynthetic eukaryotes. In the model plant Arabidopsis thaliana, a number of nuclear genes encoding plastid proteins have been found to be necessary for embryo development. However, the exact roles of plastids in this process remain largely unknown. Here we use publicly available datasets to obtain insights into the relevance of plastid activities to A. thaliana embryogenesis. By searching the SeedGenes database (http://www.seedgenes.org) and recent literature, we found that, of the 339 non-redundant genes required for proper embryo formation, 108 genes likely encode plastid-targeted proteins. Nineteen of these genes are necessary for development of preglobular embryos and/or their conversion to globular embryos, of which 13 genes encode proteins involved in non-photosynthetic metabolism. By contrast, among 38 genes which are dispensable for globular embryo formation but necessary for further development, only one codes for a protein involved in metabolism. Products of 21 of the 38 genes play roles in plastid gene expression and maintenance. Examination of RNA profiles of embryos at distinct growth stages obtained in laser-capture microdissection coupled with DNA microarray experiments revealed that most of the identified genes are expressed throughout embryo morphogenesis and maturation. These findings suggest that metabolic activities are required at preglobular and throughout all stages of embryo development, whereas plastid gene expression becomes necessary during and/or after the globular stage to sustain various activities of the organelle including photosynthetic electron transport.
Keywords: Arabidopsis thaliana, embryogenesis, globular embryo, microarray, plastid, preglobular embryo, SeedGenes.
INTRODUCTION
Plastids are organelles derived from an ancient form of cyanobacteria by endosymbiosis [1] and are vital for all photosynthetic and some nonphotosynthetic eukaryotes. In higher plants, plastids are present in all cell types except male gametophytes of certain species [2, 3]. Plastids exist in several distinct forms, such as chloroplasts in photosynthetic tissues, chromoplasts in yellow, orange, and some red fruits and flower petals, amyloplasts in non-colored storage tissues, and undifferentiated proplastids in meristematic cells. Most of these plastids are inter-convertible, and their development is closely associated with plant growth and development [4]. In addition to the oxygenic photosynthetic activity of chloroplasts, numerous metabolic processes such as the biosynthesis and accumulation of starch, lipids, amino acids, and various isoprenoids, including carotenoids and precursors to gibberellins, take place in plastids [5-8]. Hence, properly-functioning plastids are essential for the viability of plants, although this idea has not been systematically addressed. During the evolution of plastids, most of the genes in the cyanobacterial endosymbiont have been transferred to the host nuclear genome [1]. The resultant plastid still contains its own genome, which encodes about 100 proteins including major components of the photosynthetic electron transport machineries and the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase [9]. However, most plastid proteins are encoded in the nuclear genome, and the majority of these proteins are synthesized on cytoplasmic ribosomes as a precursor with an N-terminal extension called the transit peptide. Transit peptide-dependent protein import across the double-membrane envelope of plastids is catalyzed by two distinct protein complexes in the outer and inner membranes called TOC and TIC (Translocon at the Outer and Inner-envelope membrane of Chloroplasts), respectively [10]. Based on extensive evaluation of several prediction programs that identify proteins with a transit peptide, a total of 2,100 nuclear genes were predicted to encode plastid proteins in the model plant Arabidopsis thaliana [11]. Furthermore, no more than 100 plastid proteins encoded by nuclear genes are synthesized without a transit peptide; they include most outer envelope proteins [12], a few inner envelope proteins [13, 14] and α-carbonic anhydrase that is sorted through a secretory pathway [15].
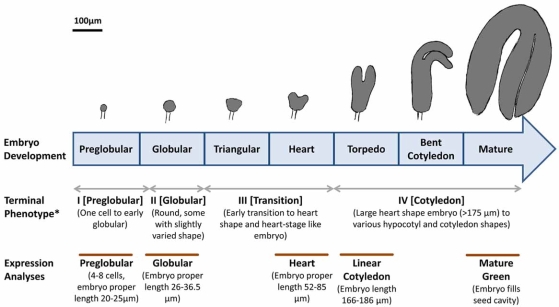
In the life cycle of flowering plants, embryogenesis is a crucial developmental period, which can be divided into two distinct phases [16]. The first phase is morphogenesis during which the basic body plan of the plant is established. The second is the maturation phase that involves cell growth and expansion, and accumulation of macromolecules that promote tolerance to the desiccation period and seedling growth. Embryo morphogenesis begins with the single-celled zygote which, in A. thaliana, undergoes a stereotypical cell division pattern giving rise to preglobular, globular, heart, torpedo, linear cotyledon, bent-cotyledon, and mature green stage embryos. Undifferentiated plastids begin to develop into chloroplasts and increase their numbers at the torpedo stage before embryos enter into the maturation phase (Fig. 1) [17]. At the maturation phase, storage products such as starch, lipid and proteins accumulate in the embryo in preparation for a period of metabolic quiescence and developmental arrest. Embryos resume development as seedlings when the appropriate environmental conditions are met, and seeds germinate. Molecular genetic studies have identified genes encoding proteins involved in controlling nuclear gene expression and auxin transport as key embryonic regulators in A. thaliana [18]. However, our understanding of the molecular mechanisms underlying seed development of this model plant is not complete. Functional genomics provides information that can be used to better understand the molecular basis for embryo development. Several projects with data publicly available are underway, such as the “Gene Networks in Seed Development project” (http://seedgene-network.net), which utilizes laser capture microdissection, microarray and high-throughput sequencing technologies to profile the mRNA sets present in different seed regions and compartments throughout development (John J. Harada, unpublished). Another example is the “SeedGenes project” (http://www.seedgenes.org), which presents comprehensive information about A. thaliana genes that are essential for seed development [19, 20].
Fig. (1). Overview of terminal phenotype classification of SeedGenes and microarray analyses on embryo development.
A series of embryo development stages are listed in different boxes in the arrow (from left to right: early to late stages) and corresponding embryos (approximately to scale) are shown above the arrow. The stages at which embyos were taken for laser capture microdissection and microarray analyses (http://seedgenenetwork.net) are listed below the arrow and indicated by brown lines. Gene Expression Omnibus Accession numbers of the data are: GSE11262, 12403, 12404, 15160, and 15165. The terminal phenotypes of embryo-defective mutants were defined by SeedGenes (http://www.seedgenes.org).
*According to SeedGenes database, mutant embryos were removed from seeds prior to desiccation and examined under a dissecting microscope. Seeds classified as I [preglobular] often contain an early globular embryo too small to be seen upon seed dissection. These early globular embryos can be seen using Nomarski optics.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper).
A cytological study showed that plastids in A. thaliana embryonic cells remain as undifferentiated non-photosynthetic forms without detectable starch accumulation until the late globular stage when grana become visible [17]. Although the exact roles of these plastids remain unclear, a number of nuclear genes encoding plastid proteins have been found to be required for embryogenesis (see below). We are interested in elucidating roles of plastids vital for various stages of plant development. In this article, we make use of publicly available datasets to shed light on the relevance of plastid activity to plant embryogenesis.
IDENTIFICATION OF NUCLEAR GENES ENCODING PLASTID PROTEINS NECESSARY FOR EMBRYOGENESIS IN ARABIDOPSIS THALIANA
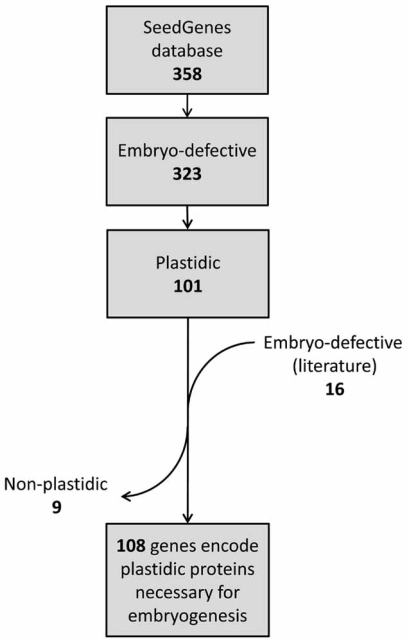
The SeedGenes database (Release 7, December, 2007) [20] lists 358 genes that give a mutant seed phenotype when disrupted by mutation. Knockout mutations of 323 genes cause arrests at various stages of embryo development. Seeds of some mutants showing an arrest phenotype at the late stage of embryo morphogenesis (cotyledon stage) can germinate and sometimes develop into mature plants (e.g., [21]). The SeedGenes database includes corresponding genes because they are needed for normal growth and development of seeds [22]. Since the latest release of SeedGenes, an additional 16 genes have been reported to be necessary for embryo development in A. thaliana [23-36], making the total number of genes known to be required for embryogenesis 339. This number corresponds to about 30-60% of all the genes necessary for proper embryo development in this model species based on previous estimates [22, 37].
Null-mutants of most of these genes are arrested at a single stage. However, in some cases, a single null mutation causes embryos to arrest at a wide range of developmental stages (e.g., [38]). It has also been shown that different null mutant alleles of a single gene can result in different terminal phenotypes (e.g. [39]; SeedGenes Database). These findings may indicate that a gene is required at the beginning of a certain embryonic stage but the mutation does not immediately cause an arrest of development. Alternatively, the mutation may only indirectly affect embryogenesis, having a primarily effect in a seed compartments other than embryos [38].
By a thorough search of the available literatures and the Plant Proteome DataBase [40], and also by using a computer prediction program to detect transit peptides [41], we estimated that 101 out of 323 genes in the SeedGenes database and seven of the 16 recently reported genes most likely encode proteins targeted to plastids (Fig. 2; Table 1). Hence, 108 out of 339, or about one third of non-redundant genes necessary for A. thaliana embryogenesis encode plastid proteins. This fraction is about three times larger than the proportion in A. thaliana nuclear genes encoding plastid-targeted proteins, which include proteins with a transit peptide (8%; [11]) and those without (less than 1%: including most outer envelope proteins [12], two inner envelope proteins [13, 14] and α-carbonic anhydrase [15]). This apparent overrepresentation of genes encoding plastid proteins may suggest that functional plastids are required for normal embryo development [20]. However, we cannot completely exclude a possibility that availability of embryo-defective mutants may be skewed toward genes encoding plastid proteins for some unknown reasons. Genome-wide bioinformatics analyses are necessary to address these possibilities. Recently, 122 independent lines with mutations in nuclear genes encoding plastid proteins were reported from A. thaliana as potential embryo-lethal mutants based on the lack of viable homozygous mutants [42]. Interestingly, among the 91 genes represented by these lines, only 16 genes are found in our list (Table 1). It remains to be determined whether the inability to obtain viable homozygous mutants corresponding to the other 76 genes is due to embryo-lethality.
Fig. (2). Flow chart indicating the identification of Arabidopsis thaliana genes encode plastid proteins indispensable for embryogenesis.
The SeedGenesdatabase (http://www.seedgenes.org; last updated December, 2007) contains 358 A. thaliana genes that give a seed phenotype when disrupted by mutation. Among these genes, 323 of them are necessary for embryogenesis and their disruption results in arrests in development. To determine the localization of encoded proteins, three approaches were used: literature search, Plant Proteome Database (PPDB) search, and compurter algorithm prediction (TargetP). Literature search also revealed that 16 additional genes are necessary for embryogenesis and 7 of them encode plastid proteins, resulting in a total of 108 non-redudant genes necessary for embryogenesis.
Table 1.
Nuclear Genes Encoding Plastid Proteins Required for Arabidopsis thaliana Embryogenesis
| Genea | IDb | Function (Cat)c | Referencesd | |||
|---|---|---|---|---|---|---|
| EMB | F | L | ||||
| I. Genes whose disruption causes embryo abortion at preglobular stage (19) | ||||||
| At1g34430 | NC | dihydrolipoamide S-acetyltransferase# | (M) | S | [45] | P |
| At4g33680 | C | aminotransferase class I and II family protein# | (M) | [46] | [46] | [46] |
| At1g74960 | C | ketoacyl-acyl carrier protein synthase# | (M) | [47] | – | P |
| At4g26900 | C | imidazole glycerol phosphate synthase# (His biosynthesis) | (M) | [48] | – | P |
| At2g36230 | C | N'-5'-phosphoribosyl-formimino-5-aminoimidazole-4-carboxamide ribonucleotide isomerase# (His biosynthesis) | (M) | [48] | [49] | P |
| At1g31860 | C2’ | phosphoribosyl-ATP pyrophosphohydrolase/phosphoribosyl-AMP cyclohydrolase (His biosynthesis) | (M) | [48] | – | P |
| At5g10330+ | C | histidinol phosphate aminotransferase# (His biosynthesis) | (M) | [48] | – | P |
| At5g14760 | C | L-asp oxidase (NAD biosynthesis) | (M) | [50] | [50] | [50] |
| At5g50210+ | C | ouinolinate synthase (NAD biosynthesis) | (M) | [50] | [50] | [50] |
| At2g01350 | C | ouinolinic acid phosphoribosyl transferase (NAD biosynthesis) | (M) | [50] | – | [50] |
| At2g28880 | NC | 4-amino-4-deoxychorismate synthase (folate biosynthesis) | (M) | S | [51] | [51] |
| At3g54660 | C2 | glutathione reductase# | (M) | S | [52] | [52] |
| At4g26500 | C | activator of plastidic and mitochondrial desulfurases (AtSufE) | (M) | [53] | [53] | [53] |
| At1g08840 | C2 | DNA replication helicase# | (PGME) | S | – | T |
| At5g24120 | C | RNA polymerase sigma subunit SigE# | (PGME) | [54] | – | [55] |
| At3g02660 | C | tyrosyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At3g18290 | C2’ | zinc finger protein-related# | (PGME) | S | – | T |
| At5g17710 | C | co-chaperone GrpE family protein# | (PH) | S | – | P |
| At3g46740+ | C | precursor protein import channel (Toc75) | (PT) | [58, 59] | [58, 59] | [58] |
| II. Genes whose disruption causes embryo abortion at globular stage (38) | ||||||
| At4g39120* | C | histidinol-phosphate phosphatase (His biosynthesis) | (M) | [36] | [36, 60] | [36] |
| At2g01860 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At5g03800 | C | similar to pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At1g30610+ | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At1g79490 | NC | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At4g39620 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At5g50280 | NC | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At3g49240 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | P |
| At2g38770 | C | U5 associated protein | (PGME) | S | – | P |
| At5g26742 | NC | DEAD/DEAH box RNA helicase# | (PGME) | S | – | P |
| At3g18390+ | NC | chloroplast splicing factor (CRS1) | (PGME) | S | – | P |
| At4g26300 | NC | arginyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At1g05190+ | NC | ribosomal protein L6 family protein# | (PGME) | S | – | P |
| At1g78630 | NC | ribosomal protein L13 family protein# | (PGME) | S | – | P |
| At4g04350 | C | leucyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At1g62750+ | C | elongation factor Tu family protein# | (PGME) | [62] | – | [63] |
| At2g04842 | NC | threoninyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At5g16715 | C | valyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At4g29060 | C | elongation factor Ts family protein# | (PGME) | S | – | P |
| At3g48110+ | C2 | glycyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At2g04530* | NC | RNase Z# | (PGME) | [27] | – | [27] |
| At5g18570*+ | C | GTP1/OBG family protein# | (PGME) | [32] | – | [32] |
| At3g10670 | C | plastidic SufC-like protein (Fe-S cluster biogenesis) | (PH) | [64] | [64] | [64] |
| At3g04340+ | NC | FtsH protease family protein# | (PH) | S | – | P |
| At5g18820 | NC | RuBisCO subunit binding-protein alpha subunit (Cpn60a)# | (PH) | S | – | T |
| At1g02560* | C | ATP-dependent Clp protease proteolytic subunit (ClpP5)# | (PH) | [28] | [28] | [65] |
| At1g06950+ | C | chloroplast protein import (Tic110) | (PT) | [66] | [66] | [67] |
| At2g31530 | C | secY family protein# | (PT) | S | – | T |
| At4g32400* | C | nucleotide export | (T) | [33] | [68] | [68] |
| At5g19620* | C | OEP80 | (U) | [35] | – | [69] |
| At5g66055+ | C | ankyrin repeat protein (AKRP) # | (U) | [70] | – | [70] |
| At1g10510 | C | leucine-rich repeat family protein, similar to ribonuclease inhibitor# | (U) | S | – | P |
| At3g12080 | C | GTP-binding family protein# | (U) | S | – | P |
| At5g63420 | C | metallo-beta-lactamase family protein# | (U) | S | – | P |
| At3g24560 | C | ATP binding | (U) | [71] | – | T |
| At5g40160 | C | ankyrin repeat protein# | (U) | [72] | [73] | [70] |
| At2g25660 | C | unknown | (U) | S | – | T |
| At5g57930 | C | Fe-S cluster related | (U) | S | – | T |
| III. Genes whose disruption causes embryo abortion at the transition of globular to cotyledon stage (15) | ||||||
| At3g25860 | C | dihydrolipoamide S-acetyltransferase# | (M) | S | [74] | [74] |
| At5g16390 | C | biotin carboxyl carrier protein of acetyl-CoA carboxylase# | (M) | S | [75, 76] | [76] |
| At3g20440 | C2 | 1,4-alpha-glucan branching enzyme# (starch biosynthesis) | (M) | S | – | P |
| At5g67570 | C4 | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | [77] |
| At3g29290 | NC | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At1g75350 | NC | ribosomal protein L31 family protein# | (PGME) | S | – | P |
| At5g22800 | C2 | aminoacyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At1g23400 | C2 | chloroplast intron splicing factor | (PGME) | [78] | [78] | P |
| At2g04030+ | C | heat shock protein (Hsp90) | (PH) | S | [79, 80] | [79] |
| At1g79560 | C2’ | AAA and metalloprotease (FtsH12) | (PH) | S | – | [81] |
| At3g16290 | C2 | FtsH protease (AAA ATPase) | (PH) | S | – | P |
| At4g23430 | C | subunit of Tic complex (Tic32), short chain dehydrogenase | (PT) | [82] | – | [82] |
| At5g62990 | C | unknown | (U) | S | – | T |
| At3g61780 | C2 | unknown | (U) | S | – | T |
| At2g37920 | NC | copper transporter related | (U) | S | [83] | T |
| IV. Genes whose disruption causes embryo abortion at cotyledon stage (35) | ||||||
| At4g30580 | C | 2-acylglycerophosphoethanolamine acyltransferase | (M) | [84] | [85] | [84] |
| At1g48850 | NC | chorismate synthase/5-enolpyruvylshikimate-3-phosphate phospholyase# | (M) | S | – | P |
| At3g55610 | C | delta 1-pyrroline-5-carboxylate synthetase B# (Pro synthesis) | (M) | [86] | – | [86] |
| At5g61410 | C | ribulose-5-phosphate-3-epimerase# | (M) | S | – | P |
| At3g06350 | C | dehydroquinate dehydratase; shikimate dehydrogenase# | (M) | S | [87] | P |
| At1g08510 | C4 | acyl-acyl carrier protein thioesterase# | (M) | [21] | – | T |
| At4g23100 | C | gamma-glutamylcysteine synthetase# | (M) | [88] | [89] | [90] |
| At5g24400 | C | 6-phosphogluconolactonase | (M) | [91] | – | [91] |
| At5g52920 | C4 | similar to pyruvate kinase isozyme G# | (M) | [92] | – | [92] |
| At2g19450 | C4 | diacylglycerol O-acyltransferase / acyl CoA:diacylglycerol acyltransferase# | (M) | [93, 94] | [93, 94] | [95] |
| At1g78580 | C | trehalose-6-phosphate synthase 1# | (M) | [96] | [97] | T |
| At3g10690+ | C3 | DNA gyrase subunit A family protein# | (PGME) | [98] | – | [98] |
| At3g06430+ | NC | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At3g18110 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At3g49170 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At4g20090 | C | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | [61] | – | T |
| At5g27270 | NC | pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At1g06145 | UC | similar to pentatricopeptide (PPR) repeat-containing protein# | (PGME) | S | – | T |
| At1g79350 | NC | DNA-binding protein | (PGME) | S | – | T |
| At1g70070+ | C3 | DEAD/DEAH box helicase# | (PGME) | [99, 100] | – | P |
| At1g74970 | NC | Ribosomal protein S9# | (PGME) | S | – | [101] |
| At1g14610 | C | valyl-tRNA synthetase# | (PGME) | [56] | – | [57] |
| At5g02250 | C4 | ribonuclease II family protein# | (PGME) | [102] | [102] | [103] |
| At2g28000 | C3 | RuBisCO subunit binding-protein alpha subunit (Cpn60a) # | (PH) | [104] | [105] | P |
| At1g19800 | C4 | Permease (TGD1, trigalactosyldiacylglycerol 1) | (T) | [106] | [107] | [106] |
| At4g33460 | NC | ABC-type transport protein# | (T) | S | – | P |
| At2g01735 | C | zinc finger (C3HC4-type RING finger) family protein# | (U) | S | – | T |
| At5g22640 | C | MORN (Membrane Occupation and Recognition Nexus) repeat-containing protein# | (U) | S | – | P |
| At3g07430 | C | YGGT family protein# | (U) | S | – | P |
| At1g58210 | C | kinase interacting family protein, similar to kinase interacting protein 1# | (U) | S | – | T |
| At4g28210 | C | Unknown | (U) | S | – | T |
| At1g21390 | NC | Unknown | (U) | S | – | T |
| At1g56200 | C3 | Unknown | (U) | [108] | – | [108] |
| At5g53860 | C | Unknown | (U) | S | – | P |
| At1g49510 | C2 | Unknown | (U) | S | – | T |
| Genes whose disruption causes embryo lethality, but its terminal phenotype unknown (1) | ||||||
| At2g03050* | C | similar to the mitochondrial transcription termination factor | (PGME) | [34] | – | [34] |
Genes not listed in the SeedGenes database but reported in individual literatures are indicated with an asterisk (*), and those that give mutants with no viable homozygotes as reported by Myouga et al. [42] with a plus symbol (+).
ID indicates identity confidence as defined in SeedGenes database. C, confirmed by the presence of multiple alleles causing an embryo arrest or by the genetic complementation assay; C2, having multiple null-lines with insertions in different portions of exons showing different terminal phenotypes; C2’, having multiple alleles including the ones with 5’UTR insertion causing a phenotype different from those with coding region insertions; C3, having null-mutant seeds that can germinate and develop into seedlings but not beyond; C4, having null-mutant seeds that can germinate and develop into mature plants; NC, not confirmed (only a single mutant allele with sequence information is available); UC, uncertain (insertion or mutation site not within coding region or 5' UTR and either downstream of stop codon or more than 250 bp upstream of start codon. The information of identity confidence extracted from SeedGenes database has been further updated with recent reports.
Function is assigned based on annotation in public database (GreenPhylDB http://greenphyl.cirad.fr/cgi-bin/greenphyl.cgi as indicated with a number sign #) or individual publications. Cat, functional categories: M, metabolism; PGME, plastid gene maintenance and expression; PH, protein homeostasis; PT, protein trafficking; T, transport; U, unknown.
References are listed for EMB (embryo deficiency), F (function), and L (localization). S, embryo-defective mutants were reported only by SeedGenes database; P, subcellular localization was confirmed only by proteomic research (compliled by PPDB [40]) but not other means; T, subcellular localization was predicted by TargetP [41] but has not been confirmed by experiments.
FUNCTIONAL DISTRIBUTION OF PLASTID PROTEINS ENCODED BY GENES REQUIRED FOR VARIOUS STAGES OF EMBRYO DEVELOPMENT
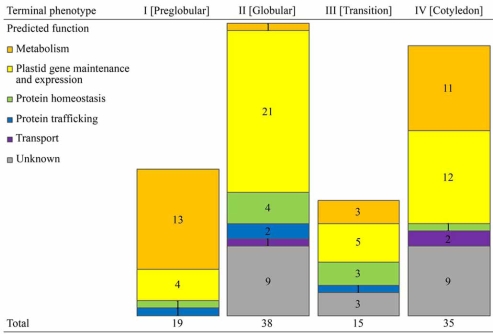
We next put each of the identified genes into one of the four groups based on the reported terminal phenotype of the null mutants (arrested at preglobular (I), globular (II), transition of globular to heart (III), and cotyledon stages (IV), respectively; Fig. 1) and also into one of six categories (metabolism, gene maintenance and expression, protein trafficking, protein homeostasis, membrane transport, and unknown) based on functions of their products as demonstrated by published studies and/or annotated in the publicly available databases (Table 1). For a gene with a single mutant allele showing heterogeneous seed phenotypes, or the one with multiple alleles showing different phenotypes, the earliest stage was used for grouping because we consider that it should be the stage when the gene is first required.
As shown in Fig. (3), our analysis revealed a clear shift in functionalities necessary at two early stages of embryo development. Group I consists of 19 genes, which are required for proper development of preglobular embryos and/or their conversion to globular embryos. Among them, 13 genes encode enzymes, including those responsible for the biosynthesis of acetyl-CoA, histidine, nicotinamide adenine dinucleotide, and folate, four code for proteins related to plastid DNA replication, transcription, and translation, and two others encode a precursor protein import channel (Toc75) and a molecular chaperone (GrpE) (Table 1). Group II consists of 38 genes which are dispensable for globular embryo formation but become necessary for further embryo morphogenesis and maturation. By contrast to Group I, Group II is enriched with genes encoding proteins involved in organellar gene expression and maintenance, such as pentatricopeptide repeat-containing proteins, tRNA synthetases, and ribosomal proteins (Table 1). Only one gene in Group II encodes a protein in the metabolism category, and its product is responsible for a later step of histidine biosynthesis [36]. Groups III and IV, which includes a total of 15 and 35 genes, respectively, are more diverse than the former two in functional categories (Fig. 3; Table 1). The clear functional differences between the first two groups of genes may be due to the necessities of operating basic metabolic pathways from a very early stage of embryogenesis, and/or producing a massive amount of proteins encoded by the plastid genome at the globular stage.
Fig. (3). Functional grouping of genes encoding plastid proteins essential for A. thaliana embryogenesis.
TGenes essential for A. thaliana embryogenesis and encoding plastidic proteins are grouped by their mutant phenotypes and predicted functions. Predicted functions are based on sequence comparison and/or experimental data, and divided into six categories: metabolism (orange blocks), plastid gene maintence and expession (yellow blocks), protein homeostasis (green blocks), protein trafficking (blue blocks), transport (purple blocks), and unknown (gray blocks).
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper).
EXPRESSION PATTERN OF GENES ENCODING PLASTID PROTEINS REQUIRED FOR EMBRYOGENESIS AT DIFFERENT STAGES OF SEEDS
To examine whether the plastid-protein-encoding genes we identified are expressed in embryos, we analyzed seed RNA profiles from DNA microarray experiments. The samples used for RNA extraction were captured from seven distinct seed compartments at five developmental stages obtained by laser capture microdissection, assuring precise sampling without contamination from adjacent compartments (Fig. 1). Among the 108 genes corresponding to embryo lethal mutations that encode plastid proteins (Table 1), unambiguous data for 95 genes were available (Table 2). Data for the rest of 13 genes were unavailable or ambiguous although one gene in this group was reported to be expressed in embryo (Table 3). Of the group of 95 genes, expression of eighty-one genes was confirmed, whereas expression of eight genes was under the detection limit of microarrays in any of the seed compartments. Interestingly, six other genes were not detectably expressed in embryos, but they were expressed in at least one of the other seed compartments (Table 4). It is possible that their functions in compartments other than embryos are required for proper embryo development, similar to previously reported cases [43, 44].
Table 2.
Unambiguous Gene Expression Data Available in GeneChip for Essential Plastid-Targeted Protein-Encoding Genes
| Terminal phenotype | I [Preglobular] | II [Globular] | III [Transition] | IV [Cotyledon] | Unknown |
|---|---|---|---|---|---|
| Total | 19 | 38 | 15 | 35 | 1 |
| Expression analyses available | 18 | 33 | 13 | 30 | 1 |
| Expressed at all stages of embryoa | 11 | 21 | 7 | 17 | 0 |
| Not detected in any stage of embryoa | 2 | 2 | 3 | 6 | 1 |
| Not detected in preglobular stage | 4 | 6 | 3 | 8 | 1 |
| Not detected in globular stage | 6 | 5 | 3 | 9 | 1 |
| Not detected in heart stage | 3 | 2 | 3 | 8 | 1 |
| Not detected in linear cotyledon stage | 4 | 7 | 3 | 7 | 1 |
| Not detected in mature green stage | 5 | 11 | 6 | 11 | 1 |
Different compartments of Arabidopsis seeds were collected at different developmental stages and gene expression profile of these compartments were analyzed. For the 108 embryogenesis-essential, plastidic protein-encoding genes, 95 of them have unambiguous probe sets on Arabidopsis whole genome ATH1 GeneChip.
The five stages at which embryo samples were taken for analyses (Fig. 1).
Table 3.
Genes Encoding Plastid Proteins Required for Embryogenesis whose Expression Data are Unavailable or Ambiguous on the Arabidopsis whole Genome ATH1 GeneChip
| Not covered in the Chip |
| At2g04842 |
| At3g06430 |
| At5g22800 |
| At5g26742 |
| A probe is available but shared with another gene |
| At2g31530 |
| At3g55610a |
| At4g23430 |
| At5g10330 |
| Two probes are available and results are not consistent |
| At5g63420 |
| Defined as distinct genes by AGI and locus identifier |
| At1g06145 |
| At1g21390 |
| At3g49170 |
| At5g03800 |
Expression in embryo was reported in the reference [86].
Table 4.
Genes Encoding Plastid Proteins Required for Embryogenesis that are Not Expressed in Embryos but in Other Seed Compartments
| Gene | Terminal Phenotype | Expression in non-embryo compartment(s) |
|---|---|---|
| At1g19800 | IV | Peripheral endosperm; Chalazal seed coat; Seed coat |
| At2g28880 | I | Micropylar endosperm; Peripheral endosperm; Chalazal endosperm; Chalazal seed coat |
| At2g37920 | III | Micropylar endosperm; Peripheral endosperm; Chalazal endosperm |
| At3g20440 | III | Peripheral endosperm |
| At4g30580 | IV | Peripheral endosperm |
| At4g33460 | IV | Peripheral endosperm; Seed coat |
Expression data for individual seed compartments is available at http://seedgenenetwork.net.
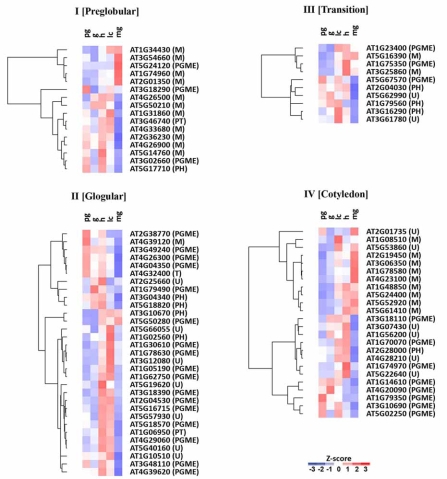
Within the group of 81 genes expressed in embryos, 56 genes are expressed at five distinct embryonic stages. We wondered if a gene is most highly expressed at the stage at which the corresponding mutant is arrested. However, no obvious correlation between expression level and terminal phenotype was observed with the possible exception of genes in Group IV. Approximately one-half of the genes that are necessary at the cotyledon stage are highly expressed in linear cotyledon-stage embryos (Fig. 4).
Fig. (4). Expression pattern of gene encoding plastid proteins necessary for A. thaliana embryogenesis.
Heat map showing the variation in levels (Z-score) of the indicated mRNAs encoding plastid proteins in embryos at different stages of development (columns: pg, preglobular; g, globular; h, heart; lc, linear cotyledon; mg, matrue green). Predicted functions of gene products are indicated in parentheses (M, metabolism; PGME, plastid gene maintenance and expression; PH, protein homeostasis; PT, protein trafficking; T, transport; U, unknown). Genes with an expression under the detection limit at all five stages were not included in the analysis.
CONCLUSIONS
As an endosymbiotic organelle, the plastid shares various properties with its prokaryotic relatives, the cyanobacteria. The plastids of higher plants have also gained the ability to develop into a variety of non-photosynthetic types and play vital roles in the growth and development of the organisms. However, plastid functions that are essential at each developmental stage are not known, except for chloroplasts in photosynthetic tissues. The current work takes an in silico approach to shed light on the functions of plastids during embryogenesis, the earliest stage of plant development following zygote formation. Although the analysis was limited to a set of non-redundant genes, our findings suggest that the non-photosynthetic metabolic activities of plastids is a prerequisite for the transition of preglobular to globular embryos and that the requirement for proteins involved in plastid gene expression becomes significant at or after the globular stage. Furthermore, analysis of the microarray data confirmed expression of most of these genes in the embryos. Based on these results, we hypothesize that i) the early stage of embryogenesis (from preglobular to globular) requires metabolic activities of plastids which are critical for various cellular processes possibly including those known to be essential for embryo development, i.e., cell division, nuclear gene expression, and auxin transport, and ii) activation of plastid gene expression that establishes various organelle activities including photosynthetic electron transport becomes necessary for the later stage of morphogenesis (from globular to heart), prior to when embryos start preparing for maturation. Furthermore, the current work poses several interesting questions. What are the effects of embryo-lethal mutations on the morphology of plastids? Apparently, some components of plastid gene expression and maintenance are not required for the formation of globular embryos. Does this mean that the organelle gene expression is not necessary at all until this stage of embryo morphogenesis? Development of strategies that allow visualization and morphological examination of aborted seed plastids, and also additional genetic and biochemical studies are needed to test these hypotheses and questions.
ACKNOWLEDGEMENTS
We greatly acknowledge support from the Plant Genome Program of the National Science Foundation (DBI-0501720 to J.J.H), and Energy Biosciences Program at the US Department of Energy DE-FE02-08ER15963 (to K.I.).
REFERENCES
- 1.Keeling P . The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corriveau J L, Coleman A W. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988;75:1443–1458. [Google Scholar]
- 3.Zhang Q, Liu Y. Sodmergen. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cyto-plasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- 4.Waters M, Pyke K. Plastid development and differentiation. In: Møller S G, editor. Annual Plant Reviews, Plastids. Vol. 13. Oxford: Blackwell; 2005. pp. 30–59. [Google Scholar]
- 5.Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 6.DellaPenna D, Pogson B J. Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- 7.Neuhaus H E, Emes M J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- 9.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- 11.Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–16. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Inoue K. The chloroplast outer envelope membrane: the edge of light and excitement. J. Integr. Plant Biol. 2007;49:1100–1111. [Google Scholar]
- 13.Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N. Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 2002;277:47770–47778. doi: 10.1074/jbc.M207477200. [DOI] [PubMed] [Google Scholar]
- 14.Nada A, Soll J. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci. 2004;117:3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- 15.Villarejo A, Buren S, Larsson S, Dejardin A, Monne M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, von Heijne G, Grebe M, Bako L, Samuelsson G. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg R B, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- 17.Mansfield S, Briarty L. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 1991;69:461–476. [Google Scholar]
- 18.Jenik P D, Gillmor C S, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- 19.Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. The Arabidopsis SeedGenes Project. Nucleic Acids Res. 2003;31:90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Bonaventure G, Salas J J, Pollard M R, Ohlrogge J B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell. 2003;15:1020–1033. doi: 10.1105/tpc.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman M A, Tossberg J, Nickle T, Levin J Z, Law M, Meinke D, Patton D. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olmo I D, López-González L, Martín-Trillo M M, Martínez-Zapater J M, Piñeiro M, Jarillo J A. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010;61:623–636. doi: 10.1111/j.1365-313X.2009.04093.x. [DOI] [PubMed] [Google Scholar]
- 24.Haussuehl K, Huesgen P F, Meier M, Dessi P, Glaser E B, Adamski J, Adamska I. Eukaryotic GCP1 is a conserved mitochondrial protein required for progression of embryo development beyond the globular stage in Arabidopsis thaliana. Biochem. J. 2009;423:333–341. doi: 10.1042/BJ20091023. [DOI] [PubMed] [Google Scholar]
- 25.Ni D A, Sozzani R, Blanchet S, Domenichini S, Reuzeau C, Cella R, Bergounioux C, Raynaud C. The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol. 2009;184:311–322. doi: 10.1111/j.1469-8137.2009.02961.x. [DOI] [PubMed] [Google Scholar]
- 26.Jurkuta R J, Kaplinsky N J, Spindel J E, Barton M K. Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell. 2009;21:1957–1971. doi: 10.1105/tpc.108.065284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canino G, Bocian E, Barbezier N, Echeverria M, Forner J, Binder S, Marchfelder A. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 2009;150:1494–1502. doi: 10.1104/pp.109.137950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Oli-nares P D B, van Wijk K J. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell. 2009;21:1669–1692. doi: 10.1105/tpc.108.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takemiya A, Ariyoshi C, Shimazaki K-i. Identification and functional characterization of inhibitor-3, a regulatory subunit of protein phosphatase 1 in plants. Plant Physiol. 2009;150:144–156. doi: 10.1104/pp.109.135335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronceret A, Gadea-Vacas J, Guilleminot J, Lincker F, Delorme V, Lahmy S, Pelletier G, Chabouté M-E, Devic M. The first zygotic division in Arabidopsis requires de novo transcription of thymidylate kinase. Plant J. 2008;53:776–789. doi: 10.1111/j.1365-313X.2007.03372.x. [DOI] [PubMed] [Google Scholar]
- 31.Jonczyk R, Ronconi S, Rychlik M, Genschel U. Pantothenate synthetase is essential but not limiting for pantothenate biosynthesis in Arabidopsis. Plant Mol. Biol. 2008;66:1–14. doi: 10.1007/s11103-007-9248-6. [DOI] [PubMed] [Google Scholar]
- 32.Bang W Y, Hata A, Jeong I S, Umeda T, Masuda T, Chen J, Yoko I, Suwastika I N, Kim D W, Im C H, Lee B H, Lee Y, Lee K W, Shiina T, Bahk J D. AtObgC, a plant ortholog of bacterial Obg, is a chloroplast-targeting GTPase essential for early embryogenesis. Plant Mol. Biol. 2009;71:379–390. doi: 10.1007/s11103-009-9529-3. [DOI] [PubMed] [Google Scholar]
- 33.Meinke D, Sweeney C, Muralla R. Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE. 2009;4:e7386. doi: 10.1371/journal.pone.0007386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meskauskiene R, Würsch M, Laloi C, Vidi P-A, Coll N S, Kessler F, Baruah A, Kim C, Apel K. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J. 2009;60:399–410. doi: 10.1111/j.1365-313X.2009.03965.x. [DOI] [PubMed] [Google Scholar]
- 35.Patel R, Hsu S-C, Bédard J, Inoue K, Jarvis P. The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 2008;148:235–245. doi: 10.1104/pp.108.122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen L N, Marineo S, Mandala S, Davids F, Sewell B T, Ingle R A. The missing link in plant histidine biosynthesis: Arabidopsis myoinositol monophosphatase-like2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol. 2010;152:1186–1196. doi: 10.1104/pp.109.150805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franzmann L H, Yoon E S, Meinke D W. Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 1995;7:341–350. [Google Scholar]
- 38.Meinke DW. Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor. Appl. Genet. 1985;69:543–552. doi: 10.1007/BF00251102. [DOI] [PubMed] [Google Scholar]
- 39.Patton D A, Franzmann L H, Meinke D W. Mapping genes essential for embryo development in Arabidopsis thaliana. Mol. Gen. Genet. 1991;227:337–347. doi: 10.1007/BF00273921. [DOI] [PubMed] [Google Scholar]
- 40.Sun Q, Zybailov B, Majeran W, Friso G, Olinares P D B, van Wijk K J. PPDB, the Plant Proteomics Database at Cornell. Nucl. Acids Res. 2009;37:D969–974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 42.Myouga F, Akiyama K, Motohashi R, Kuromori T, Ito T, Iizumi H, Ryusui R, Sakurai T, Shinozaki K. The Chloroplast Function Database: a large-scale collection of Arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J. 2010;61:529–42. doi: 10.1111/j.1365-313X.2009.04074.x. [DOI] [PubMed] [Google Scholar]
- 43.Garcia D, Saingery V, Chambrier P, Mayer U, Jurgens G, Berger F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003;131:1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo M, Dennis E S, Berger F, Peacock W J, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mentzen W, Peng J, Ransom N, Nikolau B, Wurtele E. Articulation of three core metabolic processes in Arabidopsis: Fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol. 2008;8:76. doi: 10.1186/1471-2229-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song J T, Lu H, Greenberg J T. Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell. 2004;16:353–366. doi: 10.1105/tpc.019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pidkowich M S, Nguyen H T, Heilmann I, Ischebeck T, Shanklin J. Modulating seed β-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc. Natl. Acad. Sci. USA. 2007;104:4742–4747. doi: 10.1073/pnas.0611141104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol. 2007;144:890–903. doi: 10.1104/pp.107.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujimori K, Tada S, Kanai S, Ohta D. Molecular cloning and characterization of the gene encoding N'-[(5'-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase from Arabidopsis thaliana. Mol. Gen. Genet. 1998;259:216–223. doi: 10.1007/s004380050807. [DOI] [PubMed] [Google Scholar]
- 50.Katoh A, Uenohara K, Akita M, Hashimoto T. Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 2006;141:851–857. doi: 10.1104/pp.106.081091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basset G J C, Quinlivan E P, Ravanel S, Rébeillé F, Nichols B P, Shinozaki K, Seki M, Adams-Phillips L C, Giovannoni J J, Gregory J F, Hanson A D. Folate synthesis in plants: The p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids. Proc. Natl. Acad. Sci. USA. 2004;101:1496–1501. doi: 10.1073/pnas.0308331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chew O, Whelan J, Millar A H. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003;278:46869–46877. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- 53.Xu X M, Moller S G. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 2006;25:900–909. doi: 10.1038/sj.emboj.7600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao J, Roy-Chowdhury S, Allison L A. AtSig5 is an essential nucleus-encoded Arabidopsis σ-like factor. Plant Physiol. 2003;132:739–747. doi: 10.1104/pp.102.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl. Acad. Sci. USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berg M, Rogers R, Muralla R, Meinke D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 2005;44:866–878. doi: 10.1111/j.1365-313X.2005.02580.x. [DOI] [PubMed] [Google Scholar]
- 57.Duchêne A-M, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters N M, Zaepfel M, Maréchal-Drouard L, Small I D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baldwin A, Wardle A, Patel R, Dudley P, Park S K, Twell D, Inoue K, Jarvis P. A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 2005;138:715–733. doi: 10.1104/pp.105.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hust B, Gutensohn M. Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol. (Stuttg) 2006;8:18–30. doi: 10.1055/s-2005-873044. [DOI] [PubMed] [Google Scholar]
- 60.Torabinejad J, Donahue J L, Gunesekera B N, Allen-Daniels M J, Gillaspy G E. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009;150:951–961. doi: 10.1104/pp.108.135129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cushing D A, Forsthoefel N R, Gestaut D R, Vernon D M. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta. 2005;221:424–436. doi: 10.1007/s00425-004-1452-x. [DOI] [PubMed] [Google Scholar]
- 62.Ruppel N, Hangarter R. Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 2007;7:37. doi: 10.1186/1471-2229-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albrecht V, Ingenfeld A, Apel K. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 2006;60:507–518. doi: 10.1007/s11103-005-4921-0. [DOI] [PubMed] [Google Scholar]
- 64.Xu X M, Møller S G. AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:9143–9148. doi: 10.1073/pnas.0400799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peltier J-B, Ytterberg J, Liberles D A, Roepstorff P, van Wijk K J. Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2001;276:16318–16327. doi: 10.1074/jbc.M010503200. [DOI] [PubMed] [Google Scholar]
- 66.Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 2005;41:412–428. doi: 10.1111/j.1365-313X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 67.Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell D J. Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon K, Joachim T, Neuhaus H E. Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 2008;56:51–63. doi: 10.1111/j.1365-313X.2008.03583.x. [DOI] [PubMed] [Google Scholar]
- 69.Inoue K, Potter D. The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J. 2004;39:354–365. doi: 10.1111/j.1365-313X.2004.02135.x. [DOI] [PubMed] [Google Scholar]
- 70.Garcion C, Guilleminot J, Kroj T, Parcy F, Giraudat J, Devic M. AKRP and EMB506 are two ankyrin repeat proteins essential for plastid differentiation and plant development in Arabidopsis. Plant J. 2006;48:895–906. doi: 10.1111/j.1365-313X.2006.02922.x. [DOI] [PubMed] [Google Scholar]
- 71.Apuya N R, Yadegari R, Fischer R L, Harada J J, Goldberg R B. RASPBERRY3 gene encodes a novel protein important for embryo development. Plant Physiol. 2002;129:691–705. doi: 10.1104/pp.004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albert S, Després B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M. The EMB506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J. 1999;17:169–179. doi: 10.1046/j.1365-313x.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- 73.Latvala-Kilby S, Kilby N. Uncovering the post-embryonic role of embryo essential genes in Arabidopsis using the controlled induction of visibly marked genetic mosaics: EMB506, an illustration. Plant Mol. Biol. 2006;61:179–194. doi: 10.1007/s11103-006-6268-6. [DOI] [PubMed] [Google Scholar]
- 74.Mooney B P, Miernyk J A, Randall D D. Cloning and characterization of the dihydrolipoamide S-acetyltransferase subunit of the plastid pyruvate dehydrogenase complex (E2) from Arabidopsis. Plant Physiol. 1999;120:443–452. doi: 10.1104/pp.120.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi J K, Yu F, Wurtele E S, Nikolau B J. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol. 1995;109:619–625. doi: 10.1104/pp.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thelen J J, Mekhedov S, Ohlrogge J B. Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 2001;125:2016–2028. doi: 10.1104/pp.125.4.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 2008;147:573–584. doi: 10.1104/pp.108.116194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao D, Froehlich J E, Zhang H, Cheng C-L. The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant J. 2003;33:107–118. doi: 10.1046/j.1365-313x.2003.016011.x. [DOI] [PubMed] [Google Scholar]
- 80.Song H, Zhao R, Fan P, Wang X, Chen X, Li Y. Overexpression of AtHsp90.2 , AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta. 2009;229:955–964. doi: 10.1007/s00425-008-0886-y. [DOI] [PubMed] [Google Scholar]
- 81.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hörmann F, Küchler M, Sveshnikov D, Oppermann U, Li Y, Soll J. Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 2004;279:34756–34762. doi: 10.1074/jbc.M402817200. [DOI] [PubMed] [Google Scholar]
- 83.Sancenón V, Puig S, Mira H, Thiele D J, Peñarrubia L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 2003;51:577–587. doi: 10.1023/a:1022345507112. [DOI] [PubMed] [Google Scholar]
- 84.Yu B, Wakao S, Fan J, Benning C. Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 2004;45:503–510. doi: 10.1093/pcp/pch064. [DOI] [PubMed] [Google Scholar]
- 85.Xu C, Yu B, Cornish A J, Froehlich J E, Benning C. Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3- phosphate acyltransferase. Plant J. 2006;47:296–309. doi: 10.1111/j.1365-313X.2006.02790.x. [DOI] [PubMed] [Google Scholar]
- 86.Székely G, Ábrahám E, Csépl0151 , Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, Koncz C, Szabados L. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- 87.Singh S A, Christendat D. Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway. Biochemistry. 2006;45:7787–7796. doi: 10.1021/bi060366+. [DOI] [PubMed] [Google Scholar]
- 88.Cairns N G, Pasternak M, Wachter A, Cobbett C S, Meyer A J. Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006;141:446–455. doi: 10.1104/pp.106.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jez J M, Cahoon R E, Chen S. Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. J. Biol. Chem. 2004;279:33463–33470. doi: 10.1074/jbc.M405127200. [DOI] [PubMed] [Google Scholar]
- 90.Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple tran-scription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 2005;41:15–30. doi: 10.1111/j.1365-313X.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- 91.Xiong Y, DeFraia C, Williams D, Zhang X, Mou Z. Characterization of Arabidopsis 6-Phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiol. 2009;50:1277–1291. doi: 10.1093/pcp/pcp070. [DOI] [PubMed] [Google Scholar]
- 92.Baud S, Wuillème S, Dubreucq B, Almeida A d, Vuagnat C, Lepiniec L, Miquel M, Rochat C. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 2007;52:405–419. doi: 10.1111/j.1365-313X.2007.03232.x. [DOI] [PubMed] [Google Scholar]
- 93.Routaboul J-M, Benning C, Bechtold N, Caboche M, Lepiniec L. The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol. Biochem. 1999;37:831–840. doi: 10.1016/s0981-9428(99)00115-1. [DOI] [PubMed] [Google Scholar]
- 94.Zhang M, Fan J, Taylor D C, Ohlrogge J B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009;21:3885–3901. doi: 10.1105/tpc.109.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaup M T, Froese C D, Thompson J E. A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol. 2002;129:1616–1626. doi: 10.1104/pp.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gómez L D, Baud S, Graham I A. The role of trehalose-6-phosphate synthase in Arabidopsis embryo development. Biochem. Soc. Trans. 2005;33:280–282. doi: 10.1042/BST0330280. [DOI] [PubMed] [Google Scholar]
- 97.Blázquez M A, Santos E, Flores C l, Martínez2010Zapater J M, Salinas J, Gancedo C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 1998;13:685–689. doi: 10.1046/j.1365-313x.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- 98.Wall M K, Mitchenall L A, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA. 2004;101:7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franzmann L, Patton D A, Meinke D W. In vitro morphogenesis of arrested embryos from lethal mutants of Arabidopsis thaliana. Theor. Appl. Genet. 1989;77:609–616. doi: 10.1007/BF00261231. [DOI] [PubMed] [Google Scholar]
- 100.Kobayashi K, Otegui M S, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell. 2007;19:1885–1897. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arimura S-i, Takusagawa S, Hatano S, Nakazono M, Hirai A, Tsutsumi N. A novel plant nuclear gene encoding chloroplast ribosomal protein S9 has a transit peptide related to that of rice chloroplast ribosomal protein L12. FEBS Lett. 1999;450:231–234. doi: 10.1016/s0014-5793(99)00491-3. [DOI] [PubMed] [Google Scholar]
- 102.Bollenbach T J, Lange H, Gutierrez R, Erhardt M, Stern D B, Gagliardi D. RNR1, a 3'-5' exoribonuclease belonging to the RNR superfamily, catalyzes 3' maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucl. Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kishine M, Takabayashi A, Munekage Y, Shikanai T, Endo T, Sato F. Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis. Plant Mol. Biol. 2004;55:595–606. doi: 10.1007/s11103-004-1507-1. [DOI] [PubMed] [Google Scholar]
- 104.Apuya N R, Yadegari R, Fischer R L, Harada J J, Zimmerman J L, Goldberg R B. The Arabidopsis embryo mutant schlepperless has a defect in the Chaperonin-60α gene. Plant Physiol. 2001;126:717–730. doi: 10.1104/pp.126.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Viitanen P V, Schmidt M, Buchner J, Suzuki T, Vierling E, Dickson R, Lorimer G H, Gatenby A, Soll J. Functional characterization of the higher plant chloroplast chaperonins. J. Biol. Chem. 1995;270:18158–18164. doi: 10.1074/jbc.270.30.18158. [DOI] [PubMed] [Google Scholar]
- 106.Xu C, Fan J, Froehlich J E, Awai K, Benning C. Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell. 2005;17:3094–3110. doi: 10.1105/tpc.105.035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu C, Fan J, Riekhof W, Froehlich J E, Benning C. A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 2003;22:2370–2379. doi: 10.1093/emboj/cdg234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang X, Zhang X, Yang S. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 2009;19:1205–1216. doi: 10.1038/cr.2009.84. [DOI] [PubMed] [Google Scholar]