Abstract
The proteomes that make up the collection of proteins in contemporary organisms evolved through recombination and duplication of a limited set of domains. These protein domains are essentially the main components of globular proteins and are the most principal level at which protein function and protein interactions can be understood. An important aspect of domain evolution is their atomic structure and biochemical function, which are both specified by the information in the amino acid sequence. Changes in this information may bring about new folds, functions and protein architectures. With the present and still increasing wealth of sequences and annotation data brought about by genomics, new evolutionary relationships are constantly being revealed, unknown structures modeled and phylogenies inferred. Such investigations not only help predict the function of newly discovered proteins, but also assist in mapping unforeseen pathways of evolution and reveal crucial, co-evolving inter- and intra-molecular interactions. In turn this will help us describe how protein domains shaped cellular interaction networks and the dynamics with which they are regulated in the cell. Additionally, these studies can be used for the design of new and optimized protein domains for therapy. In this review, we aim to describe the basic concepts of protein domain evolution and illustrate recent developments in molecular evolution that have provided valuable new insights in the field of comparative genomics and protein interaction networks.
Keywords: Protein domain, PDZ domain, systems biology, superfamily, molecular evolution, interactome.
INTRODUCTION
The protein universe is the collection of proteins of all biological species that exist or have once existed on Earth [1]. Our sampling and understanding of it began over half a century ago, when the first peptide and protein sequences were determined by Sanger [2, 3] and, subsequently, the sequencing of RNA and DNA [4-6]. In the meantime, the genome projects of the last decade have uncovered an overwhelming amount of sequence data and researchers are now starting to address a series of fundamental questions that should shed light onto protein evolution processes [7-10]. For instance, how many gene encoding sequences are present in one genome? How many sequences are repetitive and are these sequences similar in the various organisms on Earth? Which genes were involved in the large scale genome duplications that we see in animals?
A comparison of sequences for evolutionary insight is best achieved by looking at the structural and functional (sub)units of proteins, the protein domains. By convention, domains are defined as conserved, functionally independent protein sequences, which bind or process ligands using a core structural motif [11-13]. Examples of domain modes of actions in signaling cascades for instance, are to connect different components into a larger complex or to bind signaling-molecules [14, 15]. Protein domains can usually fold independently, likely due to their relatively limited size, and are well known to behave as independent genetic elements within genomes [16, 17]. The sum of these features makes protein domains readily identifiable from raw nucleotide and amino acid sequences and many protein family resources (e.g., Superfamily and SMART [see Table 1]) indeed fully rely on such sequence similarity and motif identifications [18, 19].
Table 1.
List of Public Resources and Databases Relevant to Domain Analysis
DOMAIN IDENTIFICATION, SEQUENCE ALIGNMENT AND PHYLOGENY
The algorithms that are used for domain identification are built around a set of simple assumptions that describe the process of evolution. In general, evolution is believed to form and mold genomes largely via three mechanisms, namely i) chemical changes through the incorporation of base analogs, the effects of radiation or random enzymatic errors by polymerases, ii) cellular repair processes that counter mutations, and iii) selection pressures that manifest themselves as the positive or negative influence that determines whether the mutation will be present in subsequent generations [20, 21]. By definition, each of these phenomena has its own rate, while their combined effect gives a certain probability for the change of one defined amino acid (or nucleotide) to another within a specific time interval.
Although already informative in its own right, mutation data can be significantly different among species due to dissimilar metabolisms, generation times, population sizes, lifestyles, reproductive strategies, or the lack of apparent polymerase-dependent proofreading such as in positive-stranded RNA viruses [22-25]. Consequently, substitution rates need therefore be calculated to correctly compare two or more sequences and hunt uncharted genomes for comparable domains. Particularly this last strategy, using general rate matrices like BLOSOM and PAM, is an elegant example of how new protein functions can be discovered [26-30]. Fast algorithms for pair-wise alignments can be found in the Basic Local Alignment Search Tool (BLAST), whereas multiple sequence alignments (MSAs, Fig. 1A) in which multiple sequences are compared simultaneously are commonly created with for example ClustalX and MUSCLE (see Table 1) [31-34].
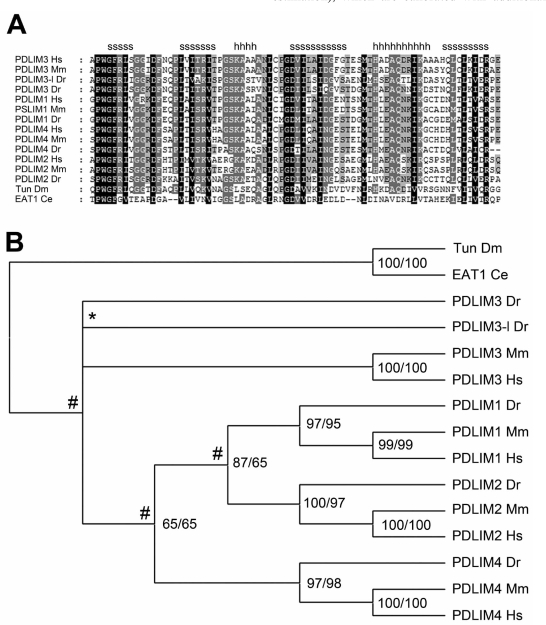
Fig. (1).
Example of sequence alignment and phylogeny. (A) This figure shows an example alignment of the PDZ domain with different shadings representing the amount of conservation (100, 75 or 50%) at a particular position in the sequence. (B) This tree is the phylogenetic presentation of the alignment in Fig. (1A). It was computed using Bayesian estimation and presents the best-supported topology for the alignment. Numbers indicate % support by the two methods used, while # indicates gene duplication events in the common ancestor and * marks a species-specific duplication event. For computational details, please see [42].
Close relatives, sharing an overall sequence identity above for example 50% and a set of functional properties, can also be grouped into families and subfamilies. In turn, these families share also evolutionary relationships with other domains and form together so-called domain superfamilies [18, 35]. Evolutionary distances between related domain sequences can easily be estimated from sequence alignments, provided that the correct rate assumptions are made. Subsequently, these can be used to compute the phylogenies of the domain that share an evolutionary history. These, often tree-like graphs (Fig. 1B), depend heavily on rate variation models, such as molecular clocks or relaxed molecular clocks (e.g., Maximum Likelyhood and Bayesian estimation), which are calibrated with additional evidence such as fossils and may therefore also provide valuable information on aspects like divergence times and ancestral sequences [36-38]. Commonly used phylogenetic analysis strategies are listed in Table 1.
A limitation of all inferred phylogenetic data is that it is directly dependent on the alignment and less so on the programs used to build the phylogenetic tree [39]. One of the shortcomings of automated alignments may thus derive from the fact that they commonly employ a scoring and penalty procedure to find the best possible alignment, since these parameters vary from species to species [22, 23], as mentioned above. Careful inspection of alignments is therefore advisable, even though software has been developed that combines the alignment procedure and phylogenetic analysis iteratively in one single program [40].
DOMAIN DIVERSIFICATION
Although sequence and phylogenetic analysis provide a relatively straightforward way for looking at domain divergence, comparison of solved protein structures has shown that protein tertiary organizations are much more conserved (>50%) than their primary sequence (>5%) [41]. For this reason, protein structures and their models provide significantly more insight into the relations of protein domains and how domain families diverged [16]. For example, the inactive guanylate kinase (GK) domain present in the MAGUK family was shown to originate from an active form of the GK domain residing in Ca2+ channel beta-subunits (CACNBs) through both sequence and structural comparison [42]. Furthermore, identification of functionally or structurally related amino acid sites in a fold sheds light on the complex, co-evolutionary dynamics that took place during selection [43].
As described above, the evolution of a protein domain is generally the result of a combination of a series of random mutations and a selection constraint imposed on function, i.e., the interaction with a ligand. The interaction between protein and ligand can be imagined as disturbances of the protein’s energy landscape, which in turn bring about specific, three-dimensional changes in the protein structure [44, 45]. Binding energies however, need not be smoothly distributed over the protein’s binding pocket as a limited number of amino acids may account for most of the free-energy change that occurs upon binding [45-47]. In these cases, new binding specificities (including loss of binding) may therefore arise through mutations at these hot spots. An example is a recent study of the PDZ domain in which it was shown that only a selected set of residues, and in particular the first residue of α-helix 2 (αB1), directly confers binding to a set of C-terminal peptides [48].
The folding of a domain is essentially based on a complex network of sequential inter-molecular interactions in time [49]. This has of course significant implications for domain integrity, particularly if one assumes that the core of a protein domain is and has to be largely structurally conserved. Indeed, even single mutations that arise in this area may easily derail the folding process, either because their free energy contribution influences residues in the direct vicinity or disturbs connections higher up in the intermolecular network [49]. It is therefore hypothesized that protein evolution took place at the periphery of the protein domain core, and that gradual changes via point mutations, insertions and deletions in surface loops brought about the evolutionary distance we see among proteins to date [21, 50-52].
However, distant sites also contribute to the thermodynamics of catalytic residues. This is achieved through a mechanism called energetic coupling, which is shaped by a continuous pathway of van der Waals interactions that ultimately influences residues at the binding site with similar efficiency as the thermodynamic hotspots [53, 54]. Indeed in such cases, evolutionary constraints are not placed on merely one amino acid in the binding pocket, but on two or more residues that can be shown to be statistically coupled in MSAs [54, 55]. In addition to contributions to binding, these principles also explain why the core of a domain structure will remain largely conserved, while at functionally related places residues can (rapidly) co-evolve with an overall neutral effect [56]. Of course, these aspects of co-evolution are also of practical consequence for structure prediction and rational drug design [43].
DOMAIN DUPLICATION
Through selective mutation, protein domains have been the tools of evolution to create an enormous and diverse assembly of proteins from likely an initially relatively limited set of domains. The combined data in GenBank and other databases now covers over 200.000 species with at least 50 complete genomes and this greatly facilitates genome comparisons [57-59]. Following such extensive comparisons, currently > 1700 domain superfamilies are recognized in the recent release of the Structural Classification of Proteins (SCOP) [60] and it has become clear that many proteins consist of more than one domain [17, 61, 62]. Indeed, it has been estimated that at least 70% of the domains is duplicated in prokaryotes, whereas this number may even be higher in eukaryotes, likely reaching up to 90% [35].
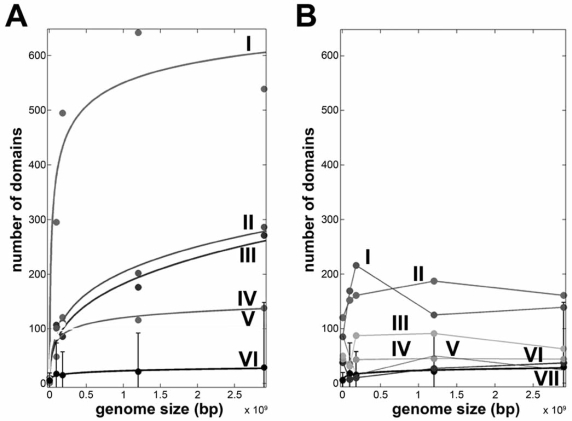
There are various mechanisms through which protein domain or whole proteins may have been duplicated. On the largest scale, whole genome duplication such as those seen in the vertebrate genomes duplicated whole gene families, including postsynaptic proteins, hormone receptors and muscle proteins, and thereby dramatically increased the domain content and expanded networks [42, 63, 64]. On the other end of the scale, domains and proteins have been duplicated through genetic mechanisms like exon-shuffling, retrotranspositions, recombination and horizontal gene transfer [65-67]. Since the genetic forces, like exon-shuffling and genome duplication vary among species, the total number of domains and the types of domains present fluctuate per genome. Interestingly, comparative analyses of genomes have shown that the number of unique domains encoded in organisms is generally proportional to its genome size [60, 68]. Within genomes, the number of domains per gene, the so-called modularity, is related to genome size via a power-law, which is essentially the relation between the frequency f and an occurrence x raised by a scaling constant k (i.e., f (x) ~ xk ) [69, 70]. A similar correlation is found when the multi-domain architecture is compared to the number of cell types that is present in an organism, i.e., the organism complexity or when the number of domains in a abundant superfamily is plotted against genome size (Fig. 2) [71, 72].
Fig. (2).
Selection on superfamily domain size. (A) Increase in superfamily domain size fitted to a power-law for kinase-like domains (I), Ankyrin-repeats (II), PDZ-like (III), voltage-gated potassium channels (IV), the catalytic domain of metalloproteases (V) and the average increase in superfamily size (VI). R2 value for each fit was at least 0.9. (B) Neutral or decreasing family sizes can be found for the MFS general substrate transporters (I), NAD(P)-binding Rossmann folds (II), Ribonucleases H (III), PLP-dependent transferases (IV), periplasmic binding proteins type II (V), ATPase domains of HSP90/topoisomerase II/histidine kinase-like folds (VI) and the average increase in superfamily size (VII) as in 2A.
DOMAIN SELECTION
Given the amount of domain duplication and apparent selection for specific multi-domain encoding genes in, for example, vertebrates, it may come as little surprise that not all domains have had the same tendency to recombine and distribute themselves over the genomes [68, 73]. In fact, some are highly abundant and can be found in many different multi-domain architectures, whereas others are abundant yet confined to a small sample of architectures or not abundant at all [68, 70]. Is there any significant correlation between the propensity to distribute and the functional roles domains have in cellular pathways?
Some of the most abundant domains can be found in association with cellular signaling cascades and have been shown to accumulate non-linearly in relation to the overall number of domains encoded or the genome size [70]. Additionally, the on-set of the exponential expansion of the number of abundant and highly recombining domains has been linked to the appearance of multicellularity [70]. A reoccurring theme among these abundant domains is the function of protein-protein interaction and it appears that particularly these, usually globular domains, have been particularly selected for in more complex organisms [70]. This positive relation is underlined by the association of these abundant domains with disease such as cancer and gene essentiality as the highly interacting proteins that they are part of have central places in cascades and need to orchestrate a high number of molecular connections [74, 75]. Their shape and coding regions, which usually lie within the boundaries of one or two exons, make them ideally suited for such a selection, since domains are most frequently gained through insertions at the N- or C-terminus and through exon shuffling [76-78].
From a mutational point of view, protein-protein interaction domains are different from other domains as well and this appears to be particularly true for the group of small, relatively promiscuous domains like SH3 and PDZ. These domains are promiscuous in the sense that they both tend to physically interact with a large number of ligands [79, 80] and are prone to move through the genome to recombine with many other domains. It has been found that particularly these domains evolve more slowly than non-promiscuous domains [70]. This likely stems from the fact that they are required to participate in many different interactions, which makes selection pressures more stringent and the appearance of the branches on phylogenetic trees relatively short and more difficult to assess when co-evolutionary data in terms of other domains in the same gene family or expression patterns is limited [42, 63]. Non-promiscuous domains on the other hand can quite easily evade the selection pressure by obtaining compensatory mutations either within themselves or their specific binding partner [70].
The overall phenomenon that the number of protein domains and their modularity increases as the genome expands has not been linked to a conclusive biological explanation yet. A rationale for the increase in interactions and functional subunits, however, may derive from the paradoxical absence of correlation between the number of genes encoded and organism complexity, the so-called G-value paradox [81]. There is indeed evidence that domains involved in the same functional pathway tend to converge in a single protein sequence, which would make pathways more controllable and reliable without the need for supplementary genes [73]. Additionally, the number of different arrangements found in higher eukaryotes is, given the vast scale of unique domains present, relatively limited. This in turn implies that evolutionary constraints have played an important role in selecting the right domain combinations and the right order from N- to C-terminus in multi-domain proteins [13, 82]. In fact, the ordering and co-occurrence of domains was demonstrated to hold enough evolutionary information to construct a tree of life similar to those based on canonical sequence data [70]. Furthermore, the increased use of alternative splicing and exon skipping in higher eukaryotes likely supplied a novel way of proteome diversification by restricting gene duplication and stimulating the formation of multi-domain proteins [83, 84]. In plants, however, the latter notion is not supported since both mono- and dicots show limited alternative splicing and a more extensive polyploidy [85-87].
THE EVOLUTION OF DOMAIN INTERACTION NETWORKS
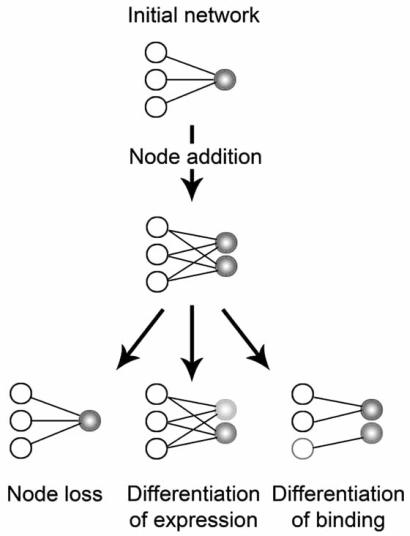
It is clear that some of the above characteristics are underappreciated in the phylogenetic analysis of linear amino acid sequences. Moreover, the effects of evolution extend even further than these aspects and entail transcriptional and translational regulation, intramolecular domain-domain interactions, gene modifications and post-translational protein modifications [88-96]. New methods are thus being developed to take into account that when sequences evolve, their close and distant functional relationships evolve in parallel. Correlations of mutations have already been found between residues of different proteins [97, 98] and compensating mutational changes at an interaction interface were shown to recover the instability of a complex [99]. These observations are evidence for the current evolutionary models for the protein-protein interaction (PPIs) networks that are being constructed through large-scale screens [100-102]. In these, a gene duplication or domain duplication (depending on the resolution of the network) implies the addition of a node, while the deletion of a gene or domain reduces the amount of links in the network (Fig. 3). In the next step, extensive network rewiring may take place, driven by the effect of node addition or node loss in the network (i.e., the duplicability or essentiality of a domain/protein) and mutations in the domain-interaction interface [67, 74, 103-105].
Fig. (3).
Evolutionary models for protein-protein interactions. The evolution of protein networks is tightly coupled to the addition or deletion of nodes. Additionally, events that introduce mutations in binding interfaces of proteins may result in the addition or loss of links in the network. Node addition may take place through e.g., domain duplication or horizontal gene transfer, while rewiring of the network is mediated by point mutations, alternative splice variants and changes in gene expression patterns.
Beyond mutations at the domain and protein level, regulation of protein expression provides another vital mechanism through which protein networks can evolve. Microarray studies are now well under way to map genome-wide expression levels of related and non-related genes under a variety of conditions [91, 94-96]. For example, transcriptional comparisons have investigated aging [106] and pathogenicity [107]. Unfortunately, given the highly variable nature of gene expression and the fact that different species may respond different to external stimuli, such comparisons can only be performed under strictly controlled research conditions. To date most studies have therefore focused on the embryogenesis, metamorphosis, sex-dependency and mutation rates of subspecies [94, 108-111]. Other studies have revealed valuable information on promoter types and duplication events [91-94].
To overcome the limitations mentioned in the previous paragraph, the analysis of co-expression data has been developed to supplement the direct comparison of individual gene expression changes [95]. In this procedure, a co-expression analysis of gene pairs within each species precedes the cross-comparison of the different organisms in the study. This approach thus primarily focuses on the similarity and differences of the orthologous genes within network, and is therefore ideally suited for the study of protein domain evolution and has already revealed that species-specific parts of an expression network resulted via a merge of conserved and newly evolved modules [95, 112, 113].
CONCLUDING REMARKS
Finding evolutionary relationships protein domains is mostly based on orthology and thus commonly performed on best sequence matches. Identifying these and categorizing them depends largely on multiple sequence alignments and this will in most cases give good indications for function, fold and ultimately evolution. However, this approach usually discards apparent ambiguities that arise from species-specific variations (e.g., due to population size, metabolism or species-specific domain duplications or losses) and may therefore introduce significant biases [114]. Biases may also derive from the method of alignment, the rate variation model used to infer the phylogeny, and the sample size used to build the alignment [39, 40, 115]. Care should therefore be taken to not regard orthology as a one-to-one relationship, but as a family of homologous relations [91], to select for appropriate analysis methods [39, 115] and extend comparative data to protein interactions and expression profiles [91]. Indeed, as our wealth of biological information expands, our systems perspective will improve and provide us with an opportunity to reveal protein domain evolution at the level network organization and dynamics. Large-scale expression studies are beginning to show us evolutionary correlations between gene expression levels and timings [94, 106, 107, 112, 116], while others demonstrate spatial differences between paralogs or (partial) overlap between interaction partners [117-120]. Indeed, when we are able to map the spatiotemporal aspects of inter- and intra-molecular interactions we will begin to fully understand the versatile power of evolution that shaped the protein universe and life on Earth [118].
FUNDING
AV is supported by The Netherlands Organization for Scientific Research (NWO) through Toptalent grant 021.001.037.
REFERENCES
- 1.Ladunga I. Phylogenetic continuum indicates "galaxies" in the protein universe: perliminary results on the natural group structures of proteins. J. Mol. Evol. 1992;34:358–375. doi: 10.1007/BF00160244. [DOI] [PubMed] [Google Scholar]
- 2.Bailey K, Sanger F. The chemistry of amino acids and proteins. Annu. Rev. Biochem. 1951;20:103–130. doi: 10.1146/annurev.bi.20.070151.000535. [DOI] [PubMed] [Google Scholar]
- 3.Sanger F. Some peptides from insulin. Nature. 1948;162:491. doi: 10.1038/162491a0. [DOI] [PubMed] [Google Scholar]
- 4.Adams JM, Jeppesen PG, Sanger F, Barrell BG. Nucleotide sequence from the coat protein cistron of R17 bacteriophage RNA. Nature. 1969;223:1009–1014. doi: 10.1038/2231009a0. [DOI] [PubMed] [Google Scholar]
- 5.Sanger F, Donelson JE, Coulson AR, Kössel H, Fisher D. Use of DNA polymerase I primed by a synthetic oligonucleotide to determine a nucleotide sequenc of phage fl DNA. Proc. Natl. Acad. Sci. USA. 1973;70:1209–1213. doi: 10.1073/pnas.70.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF. The Genome Sequence of Drosophila melanogaster. Science. 2000;287:2185. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 8.Crosby MA, Goodman JL, Strelets VB, Zhang PL, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.Weinstock GM, Robinson GE, Gibbs RA, Worley KC, Evans JD, Maleszka R, Robertson HM, Weaver DB, Beye M, Bork P. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castagnoli L, Costantini A, Dall'Armi C, Gonfloni S, Montecchi-Palazzi L, Panni S, Paoluzi S, Santonico E, Cesareni G. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567:74–79. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- 12.Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle WF. The multiplicity of domains in proteins. Annu. Rev. Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann K. The modular nature of apoptotic signaling proteins. Cell. Mol. Life Sci. 1999;55:1113–1128. doi: 10.1007/s000180050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 16.Orengo CA, Thornton JM. Protein families and their evolution: a structural perspective. Annu. Rev. Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 18.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponting CP, Schultz J, Milpetz. F, Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nuceic Acids Res. 1999;27:229–32. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat. Rev. Gent. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- 21.Bin Qian RAG. Distribution of indel lengths. Proteins Struct. Funct. Genet. 2001;45:102–104. doi: 10.1002/prot.1129. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg MS, Kumar S. Heterogeneity of nucleotide frequencies among evolutionary lineages and phylogenetic inference. Mol. Biol. Evol. 2003;20:610–621. doi: 10.1093/molbev/msg067. [DOI] [PubMed] [Google Scholar]
- 23.Vingron M, Waterman MS. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J. Mol. Biol. 1994;235:1–12. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
- 24.Bromham L. Who do species vary in their rate of molecular evolution. Biol. Lett. 2009;5:401–404. doi: 10.1098/rsbl.2009.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckerle LC, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Path. 2010;6:e1000896–0. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotech. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 28.Attwood TK. The role of pattern databases in sequence analysis. Briefings in Bioinformatics. 2000;1:45–49. doi: 10.1093/bib/1.1.45. [DOI] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. PHYLIP version 3.63. Deptartment of Genetics, University of Washington, Seattle. 2004 [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson WR. Comparison of methods for searching protein sequence databases. Protein Sci. 1995;4:1145–1160. doi: 10.1002/pro.5560040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apic G, Gough J, Teichmann SA. An insight into domain combinations. Bioinformatics. 2001;17:S83–89. doi: 10.1093/bioinformatics/17.suppl_1.s83. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 37.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 38.Springer MS. Mammalian evolution and biomedicine: new views from phylogeny. Biol. Rev. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Filipski A. Multiple sequence alignment: In pursuit of homologous DNA positions. Genome Res. 2007;17:127–135. doi: 10.1101/gr.5232407. [DOI] [PubMed] [Google Scholar]
- 40.Lunter G, Miklos I, Drummond A, Jensen J, Hein J. Bayesian coestimation of phylogeny and sequence alignment. BMC Bioinformatics. 2005;6:83. doi: 10.1186/1471-2105-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.te Velthuis A, Admiraal J, Bagowski C. Molecular evolution of the MAGUK family in metazoan genomes. BMC Evol. Biol. 2007;7:129. doi: 10.1186/1471-2148-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Codoner FM, Fares MA. Why should we care about molecular coevolution. Evol. Bioinform. Online. 2008;4:29–38. [PMC free article] [PubMed] [Google Scholar]
- 44.Freire E. The propagation of binding interactions to remote sites in proteins: analysis of the binding of the monoclonal antibody D1.3 to lysozyme. Proc. Natl. Acad. Sci. USA. 1999;96:10118–10122. doi: 10.1073/pnas.96.18.10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luque I, Freire E. Structural stability of binding sites: consequences for binding affinity and allosteric effects. Proteins. 2000;(Suppl 4):63–71. doi: 10.1002/1097-0134(2000)41:4+<63::aid-prot60>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 46.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 47.Atwell S, Ultsch M, De Vos AM, Wells JA. Structural plasticity in a remodeled protein-protein interface. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 48.Tonikian R, Sazinsky S, Currell B, Yeh J, Reva B, Held H, Appleton B, Evangelista M, Wu Y, Xin X, Chan A, Seshagiri S, Lasky L, Sander C, Boone C, Bader G, Sidhu S. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239–0. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luque I, Leavitt S, Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? . Annu. Rev. Biophys. Biomol. Struct. 2002;31:235–256. doi: 10.1146/annurev.biophys.31.082901.134215. [DOI] [PubMed] [Google Scholar]
- 50.Benner SA, Cohen MA, Gonnet GH. Empirical and structural models for insertions and deletions in the divergent evolution of proteins. J. Mol. Biol. 1993;20:1065–1082. doi: 10.1006/jmbi.1993.1105. [DOI] [PubMed] [Google Scholar]
- 51.Pascarella S, Argos P. Analysis of insertions/deletions in protein structures. J. Mol. Biol. 1992;224:461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- 52.Panchenko A, Madej T. Structural similarity of loops in protein families: toward the understanding of protein evolution. BMC Evol. Biol. 2005;5:10. doi: 10.1186/1471-2148-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Todd MJ, Freire E. The effect of inhibitor binding on the structural stability and cooperativity of the HIV-1 protease. Proteins. 1999;36:147–156. doi: 10.1002/(sici)1097-0134(19990801)36:2<147::aid-prot2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 54.Lockless SW, Ranganathan R. Evolutionary conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 55.Neher E. How frequent are correlated changes in families of protein sequences? . Proc. Natl. Acad. Sci. USA. 1994;91:98–102. doi: 10.1073/pnas.91.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 1970;4:579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- 57.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, Maskeri B, Hansen NF, Schwartz MS, Weber RJ, Kent WJ, Karolchik D, Bruen TC, Bevan R, Cutler DJ, Schwartz S, Elnitski L, Idol JR, Prasad AB, Lee-Lin SQ, Maduro VVB, Summers TJ, Portnoy ME, Dietrich NL, Akhter N, Ayele K, Benjamin B, Cariaga K, Brinkley CP, Brooks SY, Granite S, Guan X, Gupta J, Haghighi P, Ho SL, Huang MC, Karlins E, Laric PL, Legaspi R, Lim MJ, Maduro QL, Masiello CA, Mastrian SD, McCloskey JC, Pearson R, Stantripop S, Tiongson EE, Tran JT, Tsurgeon C, Vogt JL, Walker MA, Wetherby KD, Wiggins LS, Young AC, Zhang LH, Osoegawa K, Zhu B, Zhao B, Shu CL, De Jong PJ, Lawrence CE, Smit AF, Chakravarti A, Haussler D, Green P, Miller W, Green ED. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 58.Premzl M, Gready JE, Jermiin LS, Simonic T, Marshall Graves JA. Evolution of vertebrate genes related to prion and shadoo proteins--clues from comparative genomic analysis. Mol. Biol. Evol. 2004;21:2210–2231. doi: 10.1093/molbev/msh245. [DOI] [PubMed] [Google Scholar]
- 59.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreeva A, Howorth D, Chandonia J-M, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf YI, Grishin NV, Koonin EV. Estimating the number of protein folds and families from complete genome data. J. Mol. Biol. 2000;299:897–904. doi: 10.1006/jmbi.2000.3786. [DOI] [PubMed] [Google Scholar]
- 62.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.te Velthuis AJW, Isogai T, Gerrits L, Bagowski CP. Insights into the molecular evolution of the PDZ-LIM family and indentification of a novel conserved protein motif. PLoS ONE. 2007;2:e189. doi: 10.1371/journal.pone.0000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc. Natl. Acad. Sci. USA. 2009;106:11913–11918. doi: 10.1073/pnas.0812138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol. Biol. Evol. 2008;25:559–67. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- 66.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 67.Wagner A. How the global structure of protein interaction networks evolves. Proc. Biol. Sci. 2003;270:280–284. doi: 10.1098/rspb.2002.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 69.Cohen-Gihon I, Lancet D, Yanai I. Modular genes with metazoan-specific domains have increased tissue specificity. Trends Genet. 2005;21:210–213. doi: 10.1016/j.tig.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 70.Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 72.Tordai H, Nagy A, Farkas K, Bányai L, Patthy L. Modules, multidomain proteins and organismic complexity. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 73.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interaction from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 74.Jeong H, Mason SP, Barabasi AL, Oitvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 75.Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- 76.Wiener J, Beaussart F, Bornberg-Bauer E. Domain deletions and substitutions in the modular protein evolution. FEBS J. 2006;273:2037–2047. doi: 10.1111/j.1742-4658.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 77.Patthy L. Genome evolution and the evolution of exon-shuffling-a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 78.Liu M, Walch H, Wu S, Grigoriev A. Significant expansion of exon-bordering protein domains during animal proteome evolution. Nucleic Acids Res. 2005;33:95–105. doi: 10.1093/nar/gki152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basdevant N, Weinstein H, Ceruso M. Thermodynamic basis for promiscuity and selectivity in protein-protein interactions: PDZ domains, a case study. J. Am. Chem. Soc. 2006;128:12766–12777. doi: 10.1021/ja060830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agrawal V, Kishan KV. Promiscuous binding nature of SH3 domains to their target proteins. Protein Pept. Lett. 2002;9:185–193. doi: 10.2174/0929866023408760. [DOI] [PubMed] [Google Scholar]
- 81.Betran E, Long M. Expansion of genome coding regions by acquisition of new genes. Genetica. 2002;115:65–80. doi: 10.1023/a:1016024131097. [DOI] [PubMed] [Google Scholar]
- 82.Bashton M, Chothia C. The geometry of domain combination in proteins. J. Mol. Biol. 2002;315:927–939. doi: 10.1006/jmbi.2001.5288. [DOI] [PubMed] [Google Scholar]
- 83.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;7:773–781. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 85.Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat. Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- 86.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr. Opin. Plant. Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 87.Severing EI, van Dijk AD, Stiekema WJ, van Ham RC. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics. 2009;10:154. doi: 10.1186/1471-2164-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tavares GA, Panepucci EH, Brunger AT. Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. Mol. Cell. 2001;8:1313–1325. doi: 10.1016/s1097-2765(01)00416-6. [DOI] [PubMed] [Google Scholar]
- 89.McGee AW, Bredt DS. Identification of an Intramolecular Interaction between the SH3 and Guanylate Kinase Domains of PSD-95. J. Biol. Chem. 1999;274:17431–17436. doi: 10.1074/jbc.274.25.17431. [DOI] [PubMed] [Google Scholar]
- 90.Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrman M, Clausen T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. USA. 2009;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tirosh I, Bilu Y, Barkai N. Comparative biology: beyond sequence analysis. Curr. Opin. Biotechnol. 2007;18:371–377. doi: 10.1016/j.copbio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 92.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 93.Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 94.Hooper SD, Boue S, Krause R, Jensen LJ, Mason CE, Ghanim M, White KP, Furlong EEM, Bork P. Identification of tightly regulated groups of genes during Drosophila melanogaster embryogenesis. Mol. Syst. Biol. 2007;3:72. doi: 10.1038/msb4100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:294–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 96.Bergmann S, Ihmels J, Barkai N. Similarities and Differences in Genome-Wide Expression Data of Six Organisms. PLoS Biol. 2004;2:e9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burger L, van Nimwegen E. Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method. Mol. Syst. Biol. 2008;4:165. doi: 10.1038/msb4100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein-protein interaction. J. Mol. Biol. 1997;271:511–535. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 99.Mateu MG, Fersht AR. Mutually compensatory mutations during evolution of the tetramerization domain of tumor suppressor p53 lead to impaired hetero-oligomerization. Proc. Natl. Acad. Sci. USA. 1999;96:3595–3599. doi: 10.1073/pnas.96.7.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gavin A, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A, Cruciat C, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier M, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 101.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sørensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 102.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 103.Prachumwat A, Li WH. Protein function, connectivity, and duplicability in yeast. Mol. Biol. Evol. 2006;23:30–39. doi: 10.1093/molbev/msi249. [DOI] [PubMed] [Google Scholar]
- 104.Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Res. 2004;14:1310–1314. doi: 10.1101/gr.2300204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fraser HB. Modularity and evolutionary constraint on proteins. Nat. Genet. 2005;37:351–352. doi: 10.1038/ng1530. [DOI] [PubMed] [Google Scholar]
- 106.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin C-S, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 107.Jeon J, Park S, Chi M, Choi J, Park J, Rho H, Kim S, Goh J, Yoo S, Choi J, Park J, Yi M, Yang S, Kwon M, Han S, Kim BR, Khang CH, Park B, Lim S, Jung K, Kong S, Karunakaran M, Oh H, Kim H, Kim S, Park J, Kang S, Choi W, Kang S, Lee Y. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 108.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 109.Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 110.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 111.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 112.Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Conservation and coevolution in the scale-free human gene coexpression network. Mol. Biol. Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- 113.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blouin C, Butt D, Roger AJ. Impact of taxon sampling on the estimation of rates of evolution at sites. Mol. Biol. Evol. 2005;22:784–791. doi: 10.1093/molbev/msi065. [DOI] [PubMed] [Google Scholar]
- 116.Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome. Res. 2008;18:242–251. doi: 10.1101/gr.6887408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ott EB, Te Velthuis AJW, Bagowski CP. Comparative analysis of splice form-specific expression of LIM Kinases during zebrafish development. Gene Expr. Patterns. 2007;7:620–629. doi: 10.1016/j.modgep.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 118.Bork P, Serrano L. Towards cellular systems in 4D. Cell. 2005;121:507–509. doi: 10.1016/j.cell.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 119.Yadav RK, Girke T, Pasala S, Xie M, Reddy G. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]