Abstract
L83V–related variants of human papillomavirus (HPV) 16 E6, exemplified by the Asian-American variant Q14H/H78Y/L83V, were shown to be more prevalent than E6 prototype in progressing lesions and cervical cancer. We evaluated functions relevant to carcinogenesis for the E6 variants L83V, R10/L83V and Q14H/H78Y/L83V as well as the prototype in a model of human normal immortalized keratinocytes (NIKS). All E6 expressing NIKS equally abrogated growth arrest and DNA damage responses. Organotypic cultures derived from these keratinocytes demonstrated hyperplasia and aberrantly expressed keratin 5 in the suprabasal compartment. In contrast, differentiation and induction of apoptosis varied. The E6 variant rafts expressed keratin 10 in nearly all suprabasal cells while the prototype raft showed keratin 10 staining in a subset of suprabasal cells only. In addition, E6 variant NIKS expressing R10G/L83V and Q14H/H78Y/L83V were more prone to undergo cell-detachment-induced apoptosis (anoikis) than NIKS expressing E6 prototype. The combined differentiation and apoptosis pattern of high-risk E6 variants, especially of Q14H/H78Y/L83V, may reflect a phenotype beneficial to carcinogenesis and viral life cycle.
Keywords: Human papillomavirus 16, E6 prototype, E6 variant, keratinocyte, proliferation, differentiation, apoptosis
Background
Persistent infection with oncogenic human papillomavirus (HPV) types is a major risk factor in the development of a high-grade cervical lesion and progression to cancer. Among the HPV types associated with cervical cancer, HPV16 is the most prevalent type (zur Hausen, 1996, 2002). The properties governing viral persistence are largely unknown (Ferenzy & Franco, 2002). Accumulating epidemiological data suggest that viral genome variants, which diverge by about 2% within a given type and which differ geographically (Yamada et al., 1997), could contribute to viral pathogenicity presumably by altering its carcinogenic potential and immunogenicity (Bernard, 2005, 2006). The association between HPV16 variants with viral persistence and progression of cervical neoplasia was proven in numerous epidemiological studies (Berumen et al., 2001, del Refugio Gonzales-Losa et al., 2004; Grodzki et al., 2006; Hildesheim et al., 2001; Lee et al., 2008; Londesborough et al., 1996; Picconi et al., 2003; Sathish et al., 2005; Villa et al., 2000; Xi et al., 1995, 1997, 2006). In contrast, the European prototype HPV16, based on the Seedorf sequence (Seedorf et al., 1985), was linked to viral clearance and lesion regression (Grodzki et al., 2006). Another type of evidence indicated that the E6 open reading frame (ORF), preferentially over that of other HPV ORFs, is evolving under Darwinian selection (Chen et al., 2005). These findings were corroborated by our extensive surveys of E6, E7, L1, L2, E2 and the LCR of HPV16, where we showed that merely E6 variants are associated with disease development (Grodzki et al., 2006; Kämmer et al., 2002; Lee et al., 2008; Zehbe et al., 1998a, 1998b, 2001a, 2001b). Collectively, these findings underline the biological relevance of E6 variants and provide a strong justification for defining the underlying differences between E6 variants that contribute to their differential role in human disease.
To date, a limited number of investigations by us and others indicate that E6 variants confer differences in biochemical activities of E6 (Asadurian et al., 2007; Conrad-Stoppler et al.; 1996; Lichtig et al., 2006) as well as in differentiation (Conrad-Stoppler et al., 1996; Lichtig et al., 2006) and transformation properties (Chakrabarti et al., 2004) of human epithelial cells in tissue culture compared to the prototype HPV16 E6. Cellular homeostasis is finely tuned by controlling proliferation, differentiation and apoptosis. An abrogation or imbalance of these entities is implicated in carcinogenesis. To date, no study has addressed together all of these aspects of tissue homeostasis in the context of HPV16 E6 variants and cervical cancer. Herein, we carried out such a study using a human keratinocyte cell line that can undergo a normal cellular differentiation program when placed in organotypic culture. Noted differences in the current study pertaining to perturbation of differentiation and modulation of apoptosis may explain data obtained in previous epidemiological studies and provide insights for novel treatment strategies.
Results
Gene transfer of HPV16 E6 prototype and E6 variants into normal immortalized keratinocytes (NIKS)
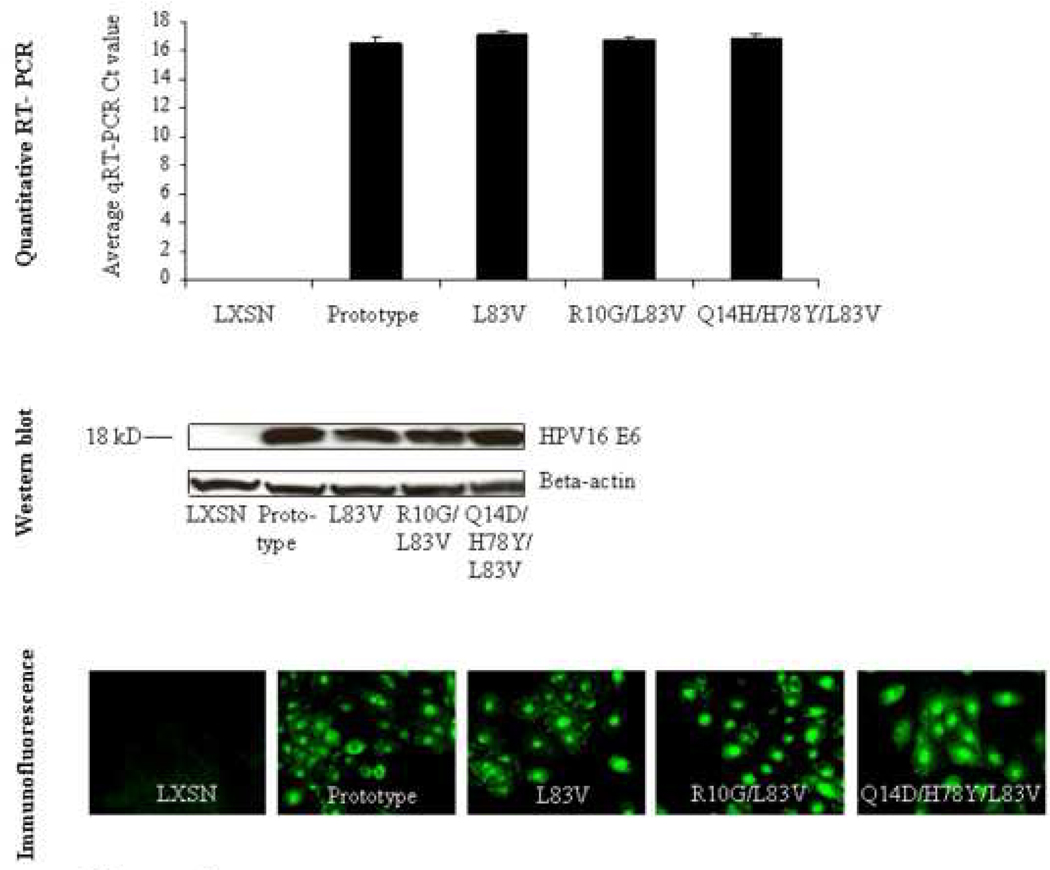
Individual E6 genes (prototype or variants) were cloned into the retroviral vector LXSN and transduced into normal immortalized keratinocytes (NIKS) (Allen-Hoffmann et al., 2000). After selection with G418, each E6 expressing culture was subjected to PCR followed by sequencing (Zehbe et al., 1998a) to confirm that only the expected variant was expressed by the recipient NIKS cell line. All E6 variants and the prototype were similarly expressed at the mRNA and protein level as determined by quantitative real time polymerase chain reaction (qRT-PCR) as well as by Western blotting using a monoclonal mouse-anti E6 antibody (clone 6F4; Lagrange et al., 2005) (Figure 1). To determine the sub-cellular localization of the E6 proteins by immunofluorescence the 6F4 clone was not applicable. Instead, an anti-HA tag antibody yielded specific, mostly nuclear and some cytoplasmic staining. Taken into consideration that E6 interacts with a wide variety of host cellular proteins both cellular locations would be expected.
Figure 1. Gene expression of HPV16 E6 variants and prototype in NIKS as defined by quantitative real time polymerase chain reaction (qRT-PCR) and Western blot.
For qRT-PCR, values represent raw qRT-PCR delta Ct values and standard deviations. cDNA input concentrations were determined using the Experion™ Automated Electrophoresis System (Bio-Rad, Mississauga, ON, Canada) as previously described (DeCarlo et al., 2008). For Western blot, input of 120 µg of total protein showed a band of the expected size for each E6 protein. The micrograph shows immunofluorescence against the HA-tag epitope. E6-specific staining appears in green.
HPV16 E6 variants and prototype similarly abrogate growth arrest and inhibit elevation of p53 induced by actinomycin D
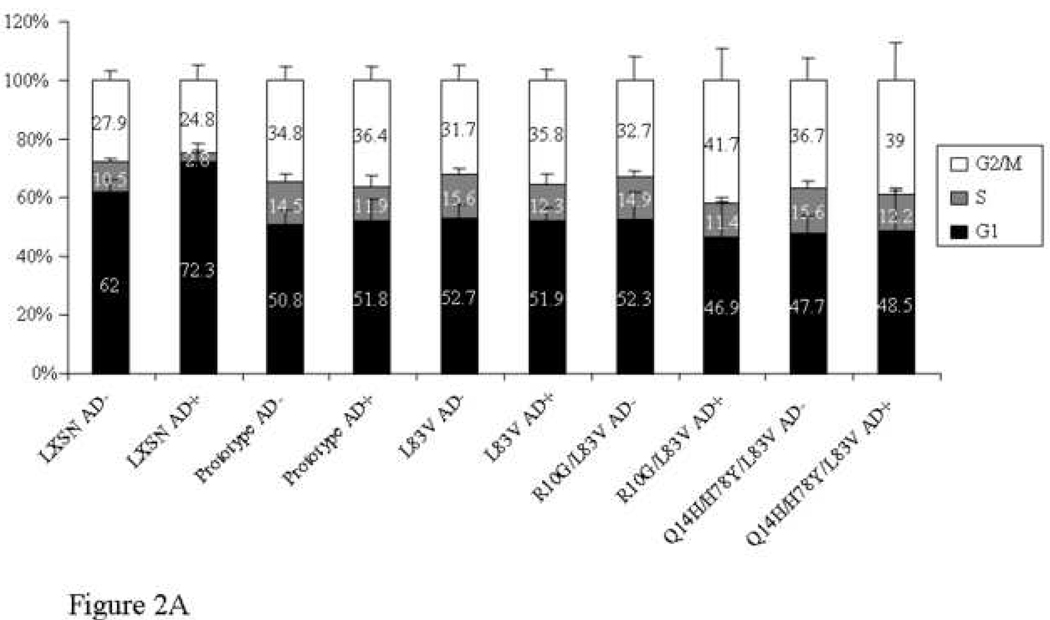
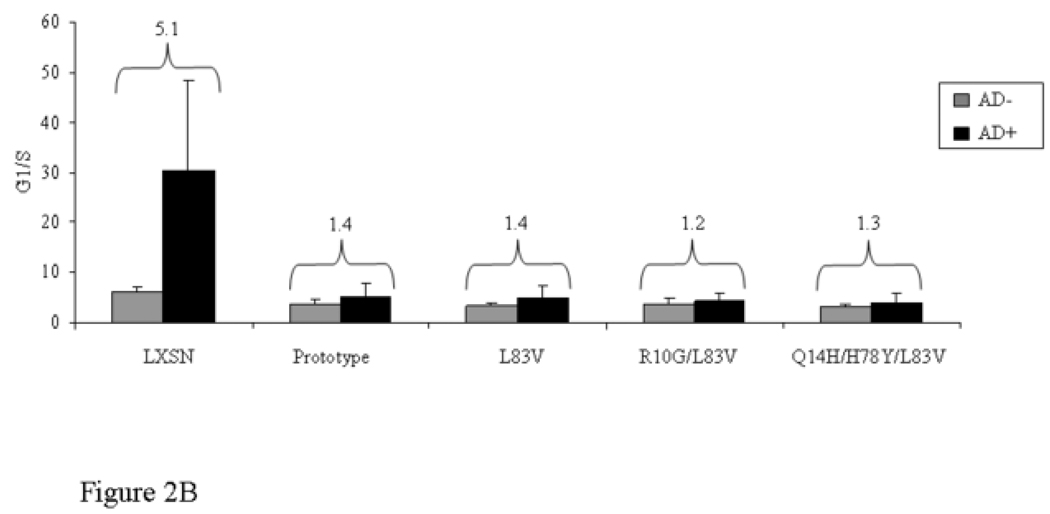
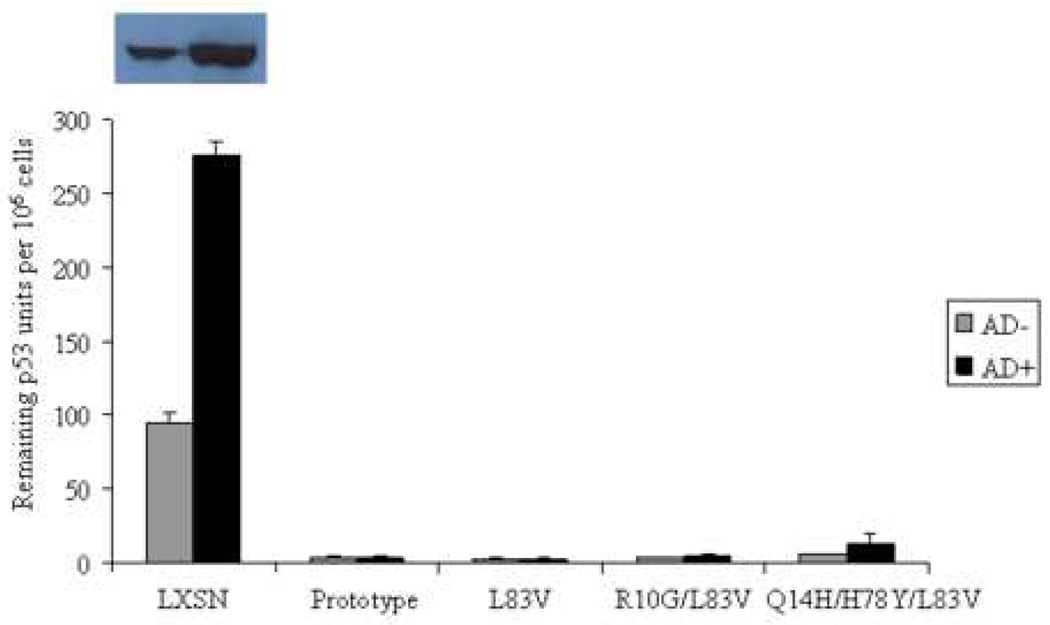
The ability to inhibit growth arrest and to abrogate DNA damage responses induced by p53 is crucial during HPV-associated carcinogenesis and thought to be largely mediated by high-risk HPV E6 proteins (Kessis et al., 1993). The G1 checkpoint requires functional p53. Low levels of actinomycin D (AD) cause strand breaks by interfering with topoisomerases resulting in growth arrest predominantly in G1 in normal cells (Trask et al., 1988). The capacity of the E6 protein to target p53 for degradation in vivo was shown to correlate with their ability to abrogate AD-induced growth arrest (Foster et al., 1994). We examined the ability of E6 prototype and E6 variants to override growth arrest and to reduce p53 levels induced by AD. The distribution of cells in the G1, S and G2/M phases and the G1/S ratios were determined with and without treatment of 0.5 nM AD for 24 h (Figure 2A–B). Treatment of control vector NIKS resulted in growth arrest as evidenced by an increase of cells in G1 phase and a reduction of cells in S phase (Figure 2A). The G1/S ratio differed significantly between treated and untreated cells (Figure 2B). In contrast, no significant change in G1/S ratio was observed in E6-transduced NIKS indicating that all E6 variant and the prototype cell lines could overcome the growth arrest induced by AD (Figure 2B). In the empty vector cells, the ratio increased five-fold, while it stayed close to 1 in the E6-transduced NIKS in the presence of AD (Figure 2B). In addition, all E6 cultures demonstrated an increased proportion of cells in G2/M after AD treatment (not significant; Figure 2A). Consistent with the above results, E6 variant and prototype but not empty vector NIKS significantly lowered the steady-state levels of p53 and were able to prevent the elevation of p53 after AD treatment as defined by ELISA and Western blot (Figure 3). These results suggest that the E6 protein can accommodate amino acid changes without significantly perturbing the activity of this protein in degrading p53 and overriding the growth arrest.
Figure 2. Cell cycle profile of HPV16 E6 prototype and variants.
Data with and without actinomycin D (AD) treatment (0.5 nM for 24 h) are shown. The distribution of cells in G1, S and G2/M phases obtained by flow cytometry is demonstrated in (A). Differences in G1/S ratio are depicted in (B). Mean values of at least three independent experiments are presented as average (mean) values ± standard deviation (SD). Significance was tested by a one-way ANOVA test. A P value of <0.05 was considered significant.
Figure 3. p53 levels of HPV16 E6 prototype and variants.
ELISA showing the remaining units of p53 quantified according to the number of cells after AD treatment (0.5 nM for 24 h). Mean values of at least three independent experiments are presented as average (mean) values ± standard deviation (SD). Significance was tested by a one-way ANOVA test. A P value of <0.05 was considered significant. The corresponding Western blot showed only p53-specific bands for empty vector NIKS. P53-negative results are not shown. After AD treatment the amount of p53 was increased approximately threefold as seen with ELISA.
HPV16 E6 variants and prototype selectively alter keratinocyte differentiation induced in organotypic keratinocyte raft cultures
Stratified squamous epithelium consists both of a poorly differentiated, proliferating basal compartment and a quiescent terminally differentiating suprabasal compartment. Expression of certain genes in these compartments are tightly regulated by several transcription factor families such as C/EBP, AP-2 and p63 (Koster et al., 2007) and are characterized by intermediate filament structures denoted keratins. While keratin pairs K5 and K14 are exclusively transcribed in the basal compartment, K1 and K10 are selectively expressed in the suprabasal compartment. To evaluate the HPV16 E6 variants in altering differentiation, we monitored E6-transduced NIKS when grown in organotypic raft cultures, which permits the normal differentiation/stratification program of human keratinocytes.
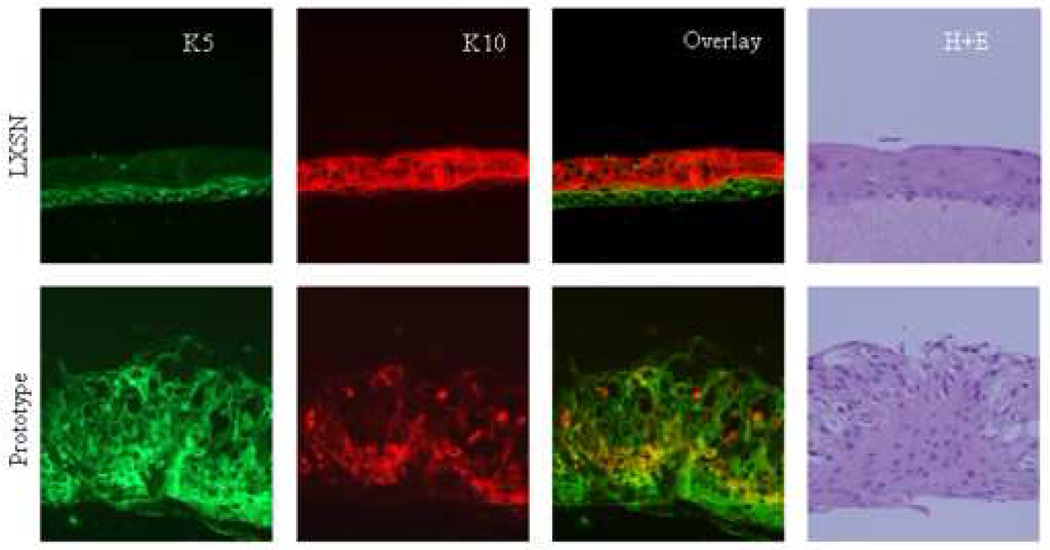
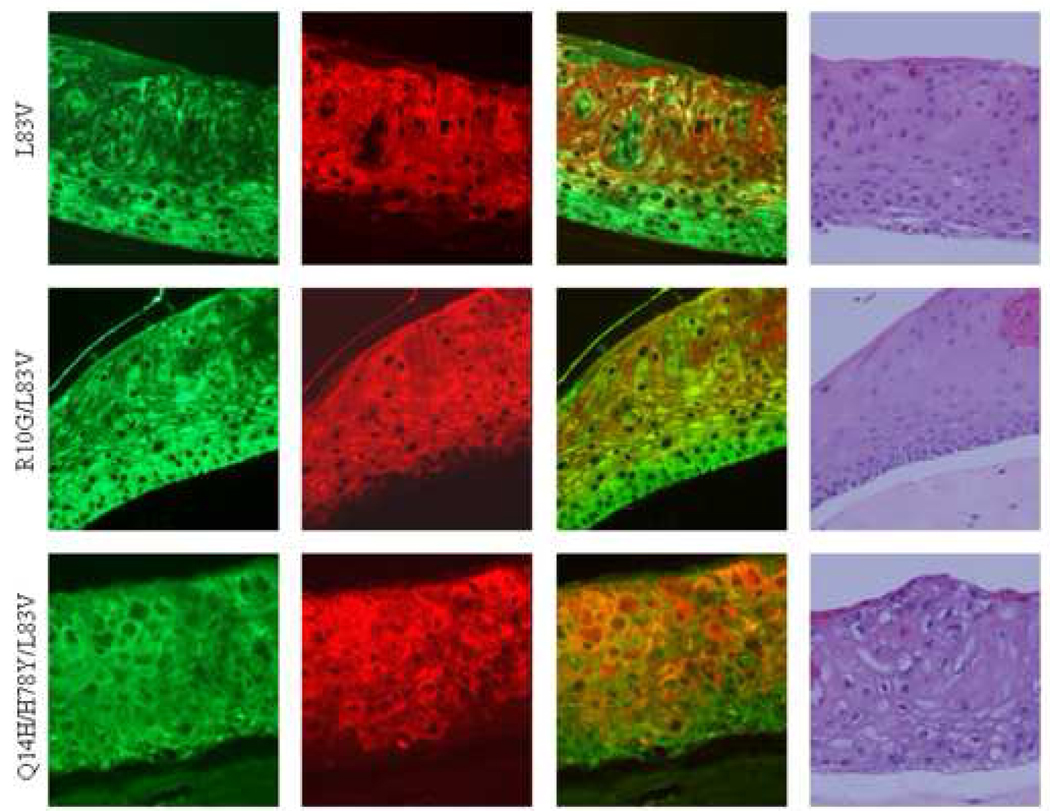
Haematoxylin and eosin staining of raft cultures revealed differences in histological architecture. The raft culture obtained from NIKS transduced with the empty vector showed ordered stratification and epithelial thickness comparable to normal skin as previously described (Allen-Hoffmann et al., 2000). A layer of undifferentiated basal cells and several layers of suprabasal cells could be distinguished (Figure 4A). NIKS transduced with the E6 variants strongly deviated from the normal histology. All cultures were hyperplastic with abnormal stratification, showed nuclear atypia and lacked polarity. The basal layer did not substantially differ from suprabasal layers in cell size or nuclear appearance. Nuclei were present in all layers but mitotic figures, usually observed in epithelial dysplasia, were absent (Figure 4A).
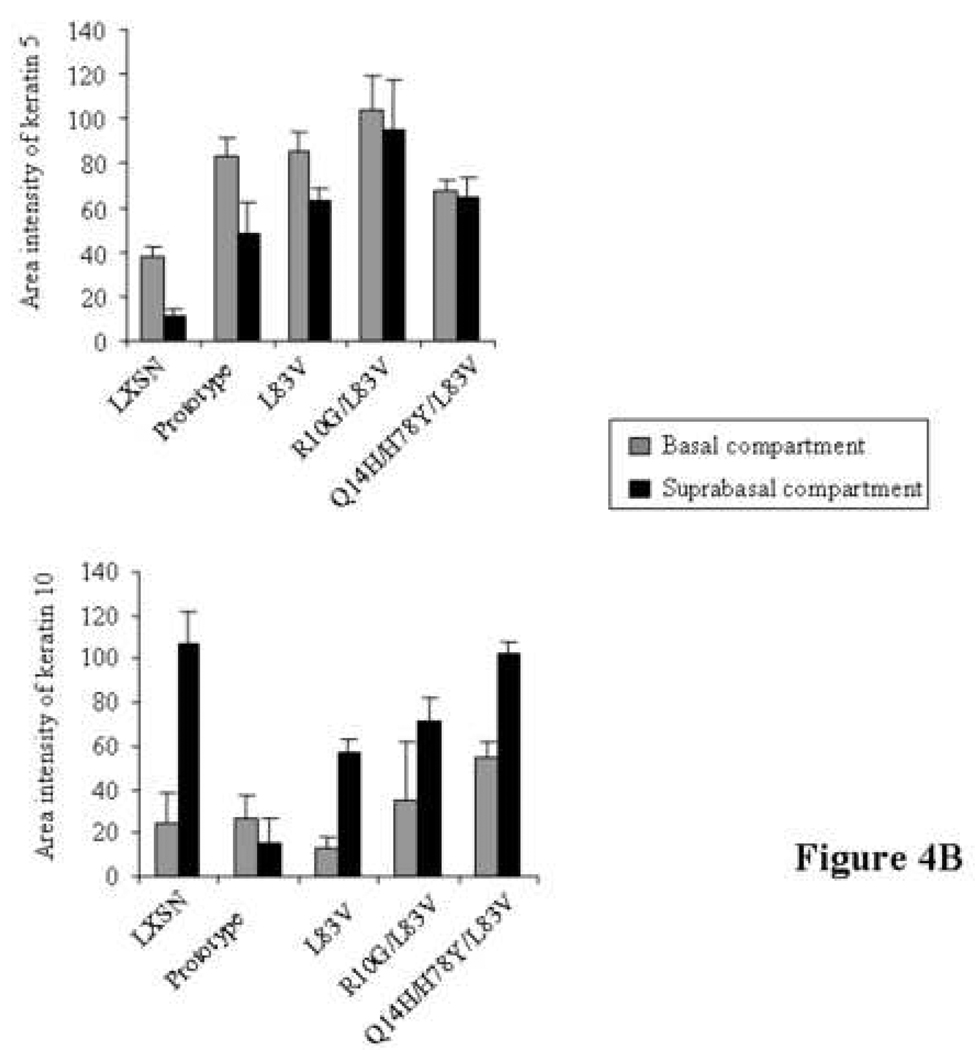
Figure 4. Keratinocyte differentiation induced in 3D raft cultures.
Micrographs including immunofluorescence for basal cell keratin marker K5 (green), suprabasal cell marker K10 (green), the overlay of both and H+E stainings are depicted in (A). Of note, rafts had been grown three times and sections were cut twice from each set of paraffin blocks to ensure reproducibility. Quantification diagrams of basal and suprabasal keratins of the above rafts are shown in (B). Calculations of K5 and K10 levels are shown for each HPV16 E6 genotype and the empty vector control raft as described in Material and methods. Values of area intensities for K5 and K10 in the basal and suprabasal regions from 4 sections are presented as average (mean) values ± standard deviation (SD). Significance was tested by a one-way ANOVA test. A P value of <0.05 was considered significant.
Next, we performed immunofluorescence using antibodies against basal cell keratins K5 and against the suprabasal cell differentiation marker K10 to examine whether K5 and K10 expression varied amongst E6 variants and prototype raft cultures (Figure 4A). In the vector control NIKS, expression of the basal cell type markers K5 was confined to the basal cell compartment showing staining in basal and parabasal cells. K10 was uniformly expressed in suprabasal cells of the NIKS control raft culture. Thus, control rafts demonstrated a complementary staining pattern of differentiation markers mostly reminiscent of normal skin. In contrast, the rafts expressing E6 variant or prototype proteins exhibited perturbed epithelial differentiation (Figure 4A). K5 was uniformly expressed in all layers within the raft epithelium, basal and suprabasal, independent of the E6 subtype. The average intensity of K5 staining in all E6 rafts was significantly higher than that of the vector (Figure 4B). The R10G/L83V culture exhibited the highest staining and compared to vector, prototype and Q14H/H78Y/L83V rafts the difference was statistically significant. Two differentiation patterns based on suprabasal cell type marker K10 were identified: one with uniform staining and one with sporadic staining within the suprabasal compartment (Figure 4A). The first category included the three E6 variants with K10 expressed in almost all cells within the suprabasal compartment. The second category contained the prototype with K10 staining only seen in a subset of cells within the suprabasal compartment. Overall area intensity of K10 staining in the suprabasal layer was lower in the prototype, L83V and R10G/L83V rafts as compared to the empty vector raft, with the difference being statistically significant (Figure 4B). The Q14H/H78Y/L83V raft showed a staining intensity comparable to that of the vector. The lowest K10 expression was seen in the prototype raft, which, compared to the expression intensity of all other rafts, is statistically significant. Collectively, these data suggest that E6 has the ability to abrogate the differentiation program in organotypic keratinocyte cultures and that E6 variants and the prototype selectively influence differentiation functions.
The ability of HPV16 E6 variants and prototype to modulate apoptosis induced by cell suspension in semisolid medium differs
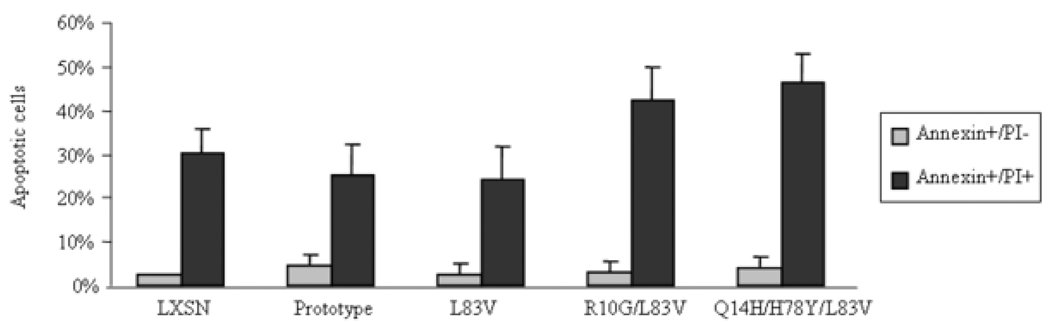
When detached from the basal membrane, keratinocytes are committed to terminal differentiation and eventually to programmed cell death in vivo. NIKS are permissive for cell-detachment-based apoptosis (anoikis), which is triggered by the extrinsic death receptor pathway (Marconi et al., 2004). This cascade can be mimicked in the laboratory by treating cells with semisolid medium. NIKS were exposed to serum-free medium made semisolid with methylcellulose. Incubation in suspension was carried out for 16 h. This time was chosen because prior experiments revealed that caspase-3 expression is highest at this time point (data not shown). Cell density had to be tightly controlled as it affected the results. NIKS kept in suspension underwent the signs of apoptosis as determined by annexin V-FITC labeling and flow cytometry analyses, based on the appearance of phosphatidylserine in the outer leaflet of the cell membrane. Apoptosis was induced in vector control as well as in E6 transduced NIKS (Figure 5). A small fraction, 2.4–5.6%, consisted of early apoptotic cells (annexinV+/PI-) with no significant difference in their proportion among the tested cultures. A larger fraction of cells, 24.3–54.5%, also stained with propidium iodide (annexinV+/PI+) indicating late apoptotic cells. With regard to late apoptotic cells, only Q14D/H78Y/L83V showed statistically significant results compared to vector control cells. Unexpectedly, levels of late apoptotic cells were higher in NIKS expressing this variant than in control cells. Similar but not significant results were obtained with R10G/L83V. The prototype and L83V NIKS on the other hand showed lower levels of late apoptotic cells than control NIKS but this finding was not statistically significant. Most importantly, significant differences in late apoptotic cell numbers were seen between E6 prototype and L83V versus R10G/L83V and Q14D/H78Y/L83V. These findings indicate that specific HPV16 E6 variants sensitize NIKS to suspension-induced extrinsic apoptosis.
Figure 5. Modulation of apoptosis by HPV16 E6 prototype and variants.
Using flow cytometry early (Annexin V-FITC+/propidium iodide−) and late apoptotic (Annexin V-FITC+/propidium iodide+), cells were calculated. Mean values of four independent experiments are presented as average (mean) values ± standard deviation (SD). Significance was tested by a one-way ANOVA test. A P value of <0.05 was considered significant.
Discussion
In the present study we have dissected the functional significance of three common variants isolated form Scandinavian women and the E6 prototype in a model of human keratinocytes. While all E6 proteins tested demonstrated similar abilities to abrogate cell cycle arrest and DNA damage responses, deregulation of differentiation and apoptosis were more variable. Note that this is the first investigation to compare the differentiation properties of naturally occurring HPV16 E6 variants in raft cultures providing a basis for future mechanistic studies. Here, we chose the model of NIKS because NIKS have the advantage over primary keratinocytes in that they do not rely upon HPV oncogenes for their long-term growth and therefore provide advantages in terms of establishing cell populations that can reproducibly be analyzed in organotypic culture. HA-tagged E6 proteins used in this investigation most likely did not impact on the biological activities of E6. For instance, resistance to apoptosis, altered expression of K10 and degradation of p53 that had been established for the E6 prototype protein earlier by other investigators (Filippova et al., 2007; Flores et al., 1999; Garnett et al., 2006; Scheffner et al., 1990) largely reflected our results obtained for the E6 prototype.
Activation of p53 and induction of p53-mediated growth-arrest are important cellular responses to DNA damage and disruption of these responses by E6 is believed to contribute to subsequent accumulation of genetic changes associated with cervical carcinogenesis (DiMaio & Liao, 2006; Mantovani & Banks, 2001). Consistent with our previous studies (Asadurian et al., 2007; Lichtig et al., 2006), we show that all tested variants were capable of strongly reducing steady-state levels of p53 in NIKS. The present study also shows that all variants were able to both suppress the elevation in levels of the p53 protein and override p53-mediated growth-arrest that normally is induced by actinomycin D (Kessis et al., 1993). The similarities in the modulation of DNA damage responses among the E6 variants, both in terms of suppression of p53 accumulation and overcoming growth arrest, suggest that these functions of E6 cannot be compromised to initiate the carcinogenic process.
Expression of high risk E6 and E7 oncoproteins was previously shown to induce alteration of keratinocyte differentiation in organotypic cultures (Flores et al., 2000; Halbert et al., 1992; Hudson et al., 1990; McCance, et al., 1988; Woodworth et al., 1992). The contribution of the individual proteins to this altered phenotype has not been completely defined. In several studies where E6 and E7 were expressed in the context of the whole genome or individually, E7 was found to be the major oncoprotein contributing to disorganized layer formation, and hyperplasia in organotypic cultures (Flores et al., 1999, 2000; Halbert et al., 1992; Ueno et al., 2006), whereas in other studies, a more significant contribution of E6 was identified (Hudson et al., 1990; Lee & Laimins, 2004). Moreover, HPV16 E6 was shown to inhibit terminal differentiation and stratification of primary human keratinocytes induced by calcium and serum in monolayer cultures (Alfandari et al., 1999; Sherman & Schlegel, 1996) and to induce epidermal hyperplasia in transgenic mice (Nguyen et al., 2006; Song et al., 1999). In the present study, the hyperplastic changes induced by HPV16 E6 variants and prototype in NIKS organotypic cultures recapitulated the effect of E6 observed in transgenic mice. In addition, we demonstrate deregulated expression of squamous cell differentiation proteins of all E6-expressing rafts. All E6 variant proteins and the prototype led to aberrant K5 expression in suprabasal cells. However, the variants but not the prototype simultaneously expressed K5 and K10 in all keratinocytes within the suprabasal compartment (Figure 4A). Replication of suprabasal keratinocytes aberrantly co-expressing K5 and K10 due to a premature migration of proliferating keratinocytes into the suprabasal compartment is a hallmark of early squamous cell carcinogenesis (Hansen et al., 2000, Huitfeldt et al., 1991; Ruiz et al., 2004). Interestingly, this phenotype is linked to the E6 variants rather than the prototype (Flores et al., 1999 and this study) and may reflect an advantage for the variants in promoting carcinogenesis. In addition, the observed phenotype may impact on the viral life cycle, which is tightly linked to the differentiation program of the cell and typically occurs during early carcinogenesis.
Suspension of normal keratinocytes into semisolid medium induces a rapid and substantial increase in the percentage of cells expressing suprabasal keratin proteins (Drozdoff & Pledger, 1993), e.g. of keratin 10 (Zehbe, unpublished data). This trigger also induces nucleosomal fragmentation, characteristic of apoptosis, in NIKS (Allen-Hoffmann et al. 2000). Using low calcium and serum-free semisolid medium, E6 prototype and L83V resisted apoptosis more than did the vector in the context of the NIKS cell line. Though this is in concordance with the notion that high-risk E6 proteins restrain keratinocytes from undergoing apoptosis, the difference was marginal and not statistically significant. Nevertheless both Q14H/H78Y/L83V and R10G/L83V demonstrated significantly higher levels of late apoptosis than E6 prototype and L83V, which seems unusual but is not unprecedented. Indeed, CaSki cells (R10G/L83V) more than SiHa cells (L83V) have been reported to undergo apoptosis when deprived of cell anchorage (Kikuchi et al., 1999). These data indicate that, under certain circumstances, HPV16 induces rather than inhibits apoptosis. The differential modulation of apoptosis observed in this investigation could form a basis for novel and individual treatment strategies. The biological significance of this finding could be a means for the virus to release viral particles thereby facilitating persistence via re-infection of the same host and transmission to new hosts. In line with this notion is the fact that caspase-3 has been found elevated during the productive stage of the viral life cycle (Moody et al., 2008). Additional studies are needed to characterize the process of apoptosis in terminally differentiating keratinocytes and its role in the HPV life cycle.
What could the underlying mechanism be for the differential phenotypes triggered by the various E6 proteins with respect to differentiation and apoptosis? The E6 oncoprotein targets cellular proteins through protein-protein interactions, which can result in loss or gain of function. Differential binding to and altered expression of E6 targets could be the reason for our observations. Possible candidates are previously reported cellular partners known to interact with E6 prototype such as tumor necrosis factor receptor 1 (Filippova et al., 2007), Fas-associated death domain protein (Garnett et al., 2006), caspase-8 (Filippova et al., 2002) and p73 (Park et al., 2001) from the death receptor pathway as well as histone acetyltransferase p300 (Patel et al., 1999), CREB-binding protein (Huang et al. 2002) and c-Myc (Veldman et al., 2003) involved in transcription during keratinocyte differentiation. Work exploring this possibility is in progress.
We postulate that the combined differentiation and apoptosis characteristics of high-risk E6 variants observed in this study confer a selective advantage for the virus to persist in the host impacting on its oncogenic potential. Our data may explain the high prevalence of certain HPV16 E6 variants in populations and in high-grade cervical disease worldwide, especially with respect to the Asian-American variant Q14H/H78Y/L83V (Berumen et al., 2001). Indeed, the risk of this variant to persist and cause lesion progression was reported to be 20-fold higher than that of prototype E6 (Berumen et al., 2001). While the current study has addressed certain aspects of deregulating cellular homeostasis in the presence of different HPV16 E6 proteins, the underlying mechanisms and pathways will have to be elucidated in a relevant infectious virus model using full-length HPV16 DNA.
Materials and methods
Human papillomavirus 16 E6 genotypes
We have chosen four HPV16 E6 species, which had previously been detected in clinical samples from Scandinavian women: the European species E6 prototype, L83V and R10G/L83V as well as the Asian-American variant Q14H/H78Y/L83V (Zehbe et al., 1998a, 2001a, 2001b).
Cloning of the E6 genotypes
Patient samples with the respective E6 variant were PCR-amplified with primers including the restriction site for EcoRI or BglII (underlined): 5’-GTTCATGTTTCAGGACCCACAG-3’ and 5’-AGATCTCAGCTGGGTTTCTCTACGTGT-3’ as described earlier (Lichtig et al. 2006). The E6 genes were cloned into the LXSN vector with the hemagglutinin (HA) tag sequence at the C-terminus followed by a stop codon to facilitate the delivery into the amphotropic packaging cell line Phoenix (Morgenstern & Land, 1990). The HA-tag approach was chosen for the current study because it was the only means to detect variant E6 proteins by immunofluorescence.
Retroviral gene transfer
High titer retroviral supernatants were generated by transient transfection of the Phoenix cell line (amphotropic virus). One day before transfection, 5 × 105 Phoenix cells were plated onto 10 cm Petri dishes. They were cultured in 10 ml DMEM supplemented with 10% FCS, 1% penicillin/streptomycin and 1% L-glutamine. Sub-confluent cells were transfected with 10 µg of the empty or with the recombinant retroviral vectors by calcium phosphate precipitation according to the manufacturer’s instructions (Clonetech, Heidelberg, Germany). Before transient transfection, 2.5 mM cloroquine in fresh medium was added to the cells for 15 min followed by the transfection cocktail. After 8 to 10 h the monolayer was washed with PBS and the medium was changed to DMEM without chloroquine. At 24 h post transfection, the medium was changed again and the viral supernatant was collected 24 h later. The virus-containing supernatant with the addition of 4 µM polybrene was immediately used to infect NIKS that were grown to 50% confluency in the presence of mitomycin-treated Swiss mouse 3T3 fibroblasts (CCL-92 from ATCC, Manassas, VA, USA) as previously described (Morgenstern & Land, 1990). After 3 h of incubation with the viral stock, fresh medium was added at the same volume as the viral supernatant. NIKS were selected in medium containing 150 µg G418/ml 48 h post infection.
Quantification of E6 expression
To ensure that equal amounts of E6 are expressed in each E6 culture E6 mRNA was quantified by qRT-PCR using TaqMan® chemistry and the ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA) as previously described (DeCarlo et al., 2008). HPV16 E6 specific primers were obtained as Custom TaqMan® Gene Expression Assays from Applied Biosystems.
Western blot analysis
To determine the levels of HPV16 E6 in each of the E6 variant cell lines, cells were trypsinized and subsequently lysed in ice-cold HNTG lysis buffer (50mM HEPES pH 7.5, 150mM NaCl, 1.1% Triton X-100, 1mM EGTA, 10% glycerol, 1mM PMSF, 1X PIC) for 20 min and centrifuged at 14,000 rpm for 20 min at 4°C in a micro centrifuge. One hundred and twenty µg of protein lysate were loaded onto a 15% SDS-PAGE mini-gel, transferred to a PVDF membrane and immunoblotted with monoclonal HPV16 E6 antibody (clone 6F4; Lagrange et al., 2005) at 5µg/mL. A goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Jackson Immunoresearch #115-036-071) was used either at 1:10.000 for E6 or at 1:50.000 for β-actin. Detection of E6 or β-actin was achieved by using the ECL Plus Western Detection Kit for horseradish peroxidase (Amersham, Piscataway, NJ, USA).
Immunofluorescence detection of HA-tagged E6 variants
NIKS (2 × 104/mL) were seeded on 8-well Lab-Tek chamber slides (LAB-TEK® Nalge Nunc Int., Naperville, IL, USA), for immunofluorescence. After 48 h NIKS were rinsed with PBS, air-dried, fixed with methanol at −20°C for 10 min, air-dried and processed immediately. Incubation with polyclonal rabbit anti-HA-tag (Y-11, Santa Cruz, Heidelberg, Germany) was performed at 4°C overnight followed by Alexa Fluor 488-donkey anti-rabbit (Molecular Probes, Mississauga, ON, Canada) (1:400 dilution) for 60 min at RT. Each step was followed by three washing steps in cold PBS for 5 min. Finally sections were rinsed with distilled water, air-dried and mounted in Vectashield mounting medium (Molecular Probes). For negative control staining, the primary antibody was omitted. Objects were photographed with a Nikon eclipse 80i microscope configured with a Nikon digital camera Dxm 1200c to image the NIKScells with 350, 488 and 594 lines for blue, green and red channel excitation, respectively. Exposure times were adjusted to each color channel and identical for each respective image.
Determination of cell cycle
For each E6 genotype and vector alone, 5 × 105 stably transduced NIKS were plated onto 10 cm Petri dishes and after 24 h were incubated with 0.5 nM actinomycin D (AD) for 24 h. After treatment, NIKS were harvested by trypsinization, washed in PBS and used for cell cycle or Western blot analysis. For cell cycle analysis, flow cytometry based on the method of Ormerod (Ormerod et al., 1987) was used. The cellular pellet was re-suspended in 70% ethanol while vortexing to prevent cell clumps. After ethanol fixation (30 min at 4°C), NIKS were rewashed in PBS and finally re-suspended in 0.2% Triton X-100/PBS and 250 µg propidium iodide (PI)/ml PBS. The suspension was then analyzed by flow cytometry on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA).
Determination of p53 protein levels by ELISA
After treatment with 0.5 nM AD NIKS were harvested by trypsinization, counted and 1 × 105cells/sample used per read-out. The cells were lysed and remaining levels of p53 were measured by Diaclone’s ready-to-use fully formatted ELISA kit (Diaclone, Besancon, France). The mean value/STD of 3 independent experiments using duplicates for each cell line were calculated with Excel.
Organotypic raft cultures and immunofluorescence
Normal human keratinocytes form well differentiated stratified squamous epithelia when placed in organotypic culture on collagen rafts raised to the air-liquid interface (Poumay & Coquette, 2007). NIKS, like normal keratinocytes, have retained a full differentiation program. They undergo squamous differentiation when cultivated in organotypic raft culture as defined by histology (Allen-Hoffmann et al., 2000). NIKS cells transduced with individual E6 variants or the prototype as well as NIKS with the empty vector as control were grown to 70% confluency and 5 × 106 cells were pipetted on collagen type I (Curacyte Discovery, Leipzig, Germany) containing primary fibroblasts. From day 2 on, EGF in the medium was exchanged for 1.88 mM Ca2+. Cultures were harvested after 14 days, fixed for 24 h in 4% PBS-buffered formaldehyde solution, processed through different grades of ethanol and embedded in paraffin. Five µ sections on positively charged slides were subjected to antigen retrieval in 6 mM citrate buffer using a pressure cooker (DAKO, Mississauga, ON, Canada) according to the manufacturer’s recommendations. Antigen double detection was performed with primary polyclonal rabbit anti cytokeratin 5 (Abcam, Cambridge, MA, USA) and primary monoclonal mouse anti keratin 10 (DAKO, Mississauga, ON, Canada) antibodies diluted 1:100 in Chemate antibody diluent (DAKO, Hamburg, Germany) and incubated overnight at 4°C. Alexa Fluor 488 and 594 donkey anti rabbit and mouse antibodies (Molecular Probes) and diluted 1:800 were used as secondary reagents. Incubation was stopped after 30 min at room temperature and slides were mounted in VectaShield mounting medium with DAPI (Molecular Probes) after 3 washing steps of PBS and 2 final washing steps of distilled water. For negative control staining, the primary antibodies were omitted. A Nikon eclipse 80i microscope configured with a Nikon digital camera Dxm 1200c was used to image the fluorescent-stained sections with 350, 488 and 594 lines for blue, green and red channel excitation, respectively. Exposure times were adjusted to each color channel and identical for each respective image. Overlays were obtained by using Photoshop CS3 extended.
Quantitative fluorescent analyses
The digital microscopy images were quantified and the average fluorescence intensity of each region of interest was calculated using the Multi Image Quantification Analysis System; Cytoview, Petach Tikva, Israel in the laboratory of Dr. Ilan Tsarfaty, Department of Human Microbiology at the Sackler School of Medicine (Golan et al., 2008). All color images from above had been saved in a Tag Image File Format (TIFF). Four images each of K5 and K10 staining were captured from different sections and used for quatitative analyses. The regions of interest, basal and suprabasal, in each image were defined manually according to the tissue morphology of the DAPI stain. These areas were defined for each section. To finally compare the relative levels of protein expression in each region, the average area intensity value was used.
Induction of apoptosis in semisolid medium
To prepare semisolid medium 3.37 g of methylcellulose was autoclaved with a magnetic stir bar in a 250 ml bottle, 100 ml of serum-free medium (Ham’s F12 and DMEM at a ratio of 3:1 as well as Ca2+ at a final concentration of 0.66 mM) was heated to 60°C, added to methylcellulose, stirred for 20 min at RT followed by adding an additional 100 ml medium and stirred at 4°C for 1 h. The semisolid medium was then centrifuged for 90 min at 10,000 rpm in 50 ml Falcon tubes to remove non-dissolved methylcellulose fibers. Approximately 2 × 106 NIKS were plated on 75 cm2 flasks and incubated for 72 h. Medium was changed every 24 h. NIKS were suspended in semisolid medium at a density of 1 × 106 cells/ml at 37°C and 5% C02 atmosphere for 16 h. Negative controls were adherent cells overlaid with semisolid medium. After incubation in semisolid medium, cells were washed three times with PBS, washed once in serum-containing medium and twice in PBS. Pellets were used for the Annexin V-FITC assay (Sigma) measured by flow cytometry.
Statistical analyses
Data of activity in the various functional assays are presented in the figures as average (mean) values + standard deviation (SD). Data were subjected to one-way ANOVA. Significance was accepted at P<0.05.
Acknowledgements
We are grateful to Dr. Ilan Tsarfaty from the Department of Human Microbiology at the Sackler School of Medicine for his assistance in tissue staining image analysis and to the staff from the Central Laboratory at the Thunder Bay Regional Health Sciences Centre for cutting and staining sections. This work was mainly funded by the Deutsche Forschungsgemeinschaft SFB 432-A9 and the Northern Cancer Research Foundation (both awarded to I Zehbe). Additional funding is acknowledged from the Israel’s Ministry of Science, Culture and Sport (MOST), Project No.1841 (awarded to L Sherman) and NIH grants CA098428 and CA022443 (both awarded to PF Lambert).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfandari J, Shnitman Magal S, Jackman A, Schlegel R, Gonen P, Sherman L. HPV16 E6 oncoprotein inhibits apoptosis induced during serum-calcium differentiation of foreskin human keratinocytes. Virology. 1999;257:383–396. doi: 10.1006/viro.1999.9675. [DOI] [PubMed] [Google Scholar]
- Asadurian Y, Kurilin H, Lichtig H, Jackman A, Gonen P, Tommasino M, Zehbe I, Sherman L. Activities of human papillomavirus 16 E6 natural variants in human keratinocytes. J. Med. Virol. 2007;79:1751–1760. doi: 10.1002/jmv.20978. [DOI] [PubMed] [Google Scholar]
- Allen-Hoffmann BL, Schlosser SJ, Ivarie CA, Sattler CA, Meisner LF, O’Connor SL. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Invest. Dermatol. 2000;114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- Bernard HU. The clinical importance of the nomenclature, evolution and taxonomy of human papillomaviruses. J. Clin. Virol. 2005;32 Suppl. 1:S1–S6. doi: 10.1016/j.jcv.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Bernard HU, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int. J. Cancer. 2006;118:1071–1076. doi: 10.1002/ijc.21655. [DOI] [PubMed] [Google Scholar]
- Berumen J, Ordonez RM, Lazcanao E, Salmeron E, Galvan SC, Estrada RA, Yunes E, Garcia-Carranca A, Gonzalez-Lira G, Madrigal-de-la-Campa A. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 2001;93:1325–1330. doi: 10.1093/jnci/93.17.1325. [DOI] [PubMed] [Google Scholar]
- Chakrabarti O, Veeraraghavalu K, Tergaonkar V, Liu Y, Androphy EJ, Stanley MA, Krishna S. Human papillomavirus type 16 E6 amino acid 83 variants enhance E6-mediated MAPK signaling and differentially regulate tumorigenesis by notch signaling and oncogenic Ras. J. Virol. 2004;78:5934–5945. doi: 10.1128/JVI.78.11.5934-5945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Terai M, Fu L, Herrero R, DeSalle R, Burk RD. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. J. Virol. 2005;79:7014–7023. doi: 10.1128/JVI.79.11.7014-7023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad-Stoppler M, Ching K, Stoppler H, Clancy K, Schlegel R, Icenogle J. Natural variants of the human papillomavirus type 16 E6 protein differ in their abilities to alter keratinocyte differentiation and to induce p53 degradation. J. Virol. 1996;70:6987–6993. doi: 10.1128/jvi.70.10.6987-6993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Refugio Gonzales-Losa M, Laviada Mier y, Teran MA, Puerto-Solis M, Garcia-Carranca A. Molecular variants of HPV16 type 16 among Mexican women with LSIL and invasive cancer. J. Clin. Virol. 2004;29:95–98. doi: 10.1016/s1386-6532(03)00094-5. [DOI] [PubMed] [Google Scholar]
- DeCarlo CA, Escott NG, Werner J, Robinson K, Lambert PF, Law RD, Zehbe I. Gene expression analysis of interferon kappa in laser capture microdissected cervical epithelium. Anal. Biochem. 2008;381:59–66. doi: 10.1016/j.ab.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Liao JB. Human papillomaviruses and cervical cancer. Adv. Virus Res. 2006;66:125–140. doi: 10.1016/S0065-3527(06)66003-X. [DOI] [PubMed] [Google Scholar]
- Drozdoff V, Pledger WJ. Commitment to differentiation and expression of early differentiation markers in murine keratinocytes in vitro are regulated independently of extracellular calcium concentrations. J. Cell Biol. 1993;123:909–919. doi: 10.1083/jcb.123.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczy A, Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11–16. doi: 10.1016/s1470-2045(01)00617-9. [DOI] [PubMed] [Google Scholar]
- Filippova M, Song H, Connolly JL, Dermody TS, Duerksen Hughes PJ. The human papillomavirus 16 E6 binds to tumor necrosis factor R1 and protects cells from TNF-induced apoptosis. J. Bio. Chem. 2002;277:21730–21739. doi: 10.1074/jbc.M200113200. [DOI] [PubMed] [Google Scholar]
- Filippova M, Johnson MM, Bautista M, Filipov V, Fodor N, Tungteakkhun SS, Williams K, Duerksen-Hughes J. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J. Virol. 2007;81:4116–4129. doi: 10.1128/JVI.01924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffman BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type16 (HPV-16) life cycle in an immortalized and human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. The human papillomavirus Type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 2000;74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Demers GW, Etscheid BG, Galloway DA. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J. Virology. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett TO, Filippova M, Duerksen Hughes PJ. Accelerated degradation of FADD and procaspase-8 in cells expressing human papillomavirus 16 E6 impairs TRAIL-mediated apoptosis. Cell Death Diff. 2006;13:1915–1926. doi: 10.1038/sj.cdd.4401886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan M, Hizi A, Resau JH, Yaal-Hohoshen N, Reichman H, Keydar I, Tsarfaty I. Human Endogenous Retrovirus (HERV-K) Reverse Transcriptase as a Breast Cancer Prognostic Marker Neoplasia. 2008;10:521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzki M, Besson G, Clavel C, Franceschi S, Arslan A, Birembaut P, Tommasino M, Zehbe I. Increased risk for cervical disease progression of French women infected with the human papillomavirus 16 E6-350G variant. Cancer Epidemiol. Biomarker Prev. 2006;15:820–822. doi: 10.1158/1055-9965.EPI-05-0864. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E6 and E7 genes of human papillomavirus Type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LA, Woodson RL, Holbus S, Strain K, Lo YC, Yuspa SH. The epidermal growth factor receptor is required to maintain the proliferative population in the basal compartment of epidermal tumors. Cancer Res. 2000;60:3328–3332. [PubMed] [Google Scholar]
- Hildesheim A, Schiffman M, Bromley C, Wacholder S, Herrero R, Rodriguez A, Bratti MC, Sherman ME, Scarpidis U, Lin QQ, Terai M, Bromley RL, Buetow K, Apple RJ, Burk RD. Human papillomavirus type 16 variants and risk of cervical cancer. J. Natl. Cancer Inst. 2001;93:315–318. doi: 10.1093/jnci/93.4.315. [DOI] [PubMed] [Google Scholar]
- Huang SM, McCance DJ. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol. 2002;76:8710–8721. doi: 10.1128/JVI.76.17.8710-8721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JB, Bedell MA, McCance DJ, Laimins LA. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus Type 18. J. Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitfeldt HS, Heyden A, Clausen OP, Thrane EV, Roop D, Yuspa SH. Altered regulation of growth and expression of differentiation-associated keratins in benign mouse skin tumors. Carcinogenesis. 1991;12:2063–2067. doi: 10.1093/carcin/12.11.2063. [DOI] [PubMed] [Google Scholar]
- Kämmer C, Tommasino M, Syrjanen S, Delius H, Hebling U, Warthorst U, Pfister H, Zehbe I. Variants of the long control region and the E6 oncogene in European human papillomavirus type 16 isolates: implications for cervical disease. Br. J. Cancer. 2002;86:269–273. doi: 10.1038/sj.bjc.6600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessis TD, Slebos RJ, Nelson WG, Kastan MB, Plunkett BS, Han SM, Lorincz AT, Hedrick L, Cho KR. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl.Acad.Sci. USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Yasumoto S. Retention of cell adhesion and growth capability in human cervical cancer cells deprived of cell anchorage. Jpn. J. Cancer Res. 1999;90:867–873. doi: 10.1111/j.1349-7006.1999.tb00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu. Rev. Cell Dev. Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Lagrange M, Charbonnier S, Orfanoudakis G, Robinson P, Zanier K, Masson M, Lutz Y, Trave G, Weiss E, Deryckere F. Binding of human papillomavirus 16 E6 to p53 and E6AP is impaired by monoclonal antibodies directed against the second zinc-binding domain of E6. J. Gen. Virol. 2005;86:1001–1007. doi: 10.1099/vir.0.80607-0. [DOI] [PubMed] [Google Scholar]
- Lee K, Magalhaes I, Clavel C, Birembaut P, Tommasino M, Zehbe I. Distribution of human papillomavirus 16 E6, L1, L2 and E2 gene variants in pre-malignant cervical disease. Virus Res. 2008;131:106–110. doi: 10.1016/j.virusres.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee C, Laimins LA. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus Type 31. J. Virol. 2004;78:12366–12377. doi: 10.1128/JVI.78.22.12366-12377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtig H, Algrisi M, Botzer LE, Abadi T, Verbitzkiy Y, Jackman A, Tommasino M, Zehbe I, Sherman L. HPV16 E6 natural variants exhibit different activities in functional assays relevant to the carcinogenic potential of E6. Virology. 2006;350:216–227. doi: 10.1016/j.virol.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Londesborough P, Ho L, Terry G, Cuzick J, Wheeler C, Singer A. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int. J. Cancer. 1996;69:364–368. doi: 10.1002/(SICI)1097-0215(19961021)69:5<364::AID-IJC2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Banks L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene. 2001;20:7874–7887. doi: 10.1038/sj.onc.1204869. [DOI] [PubMed] [Google Scholar]
- Marconi A, Atzei P, Panza C, Fila C, Tiberio R, Truzzi F, Wachter T, Leverkus M, Pincelli C. FLICE/caspase-8 activation triggers anoikis induced by beta1-integrin blockade in human keratinocytes. J Cell Sci. 2004;117:815–823. doi: 10.1242/jcs.01490. [DOI] [PubMed] [Google Scholar]
- McCance DJ, Kopan R, Fuchs E, Laimins LA. Human papillomavirus type16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. U S A. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. U S A. 2007;104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zang Z, Wang J, Tommasi di Vignano A, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod MG, Payne AW, Watson JV. Improved program for the analysis of DNA histograms. Cytometry. 1987;8:637–641. doi: 10.1002/cyto.990080617. [DOI] [PubMed] [Google Scholar]
- Park JS, Kim EJ, Lee JY, Sin HS, Namkoong SE, Um SJ. Functional inactivation of p73, a homologue of p53 tumour suppressor protein, by human papillomavirus E6 proteins. Int. J. Cancer. 2001;91:822–827. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1130>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picconi MA, Alonio LV, Sichero L, Mbayed V, Villa LL, Gronda J, Campos R, Teyssie A. Human papillomavirus type 16 variants in Quechua aboriginals from Argentina. J. Med. Virol. 2003;69:546–552. doi: 10.1002/jmv.10343. [DOI] [PubMed] [Google Scholar]
- Poumay Y, Coquette A. Modelling the human epidermis in vitro: tool for basic and applied research. Arch. Dermatol. Res. 2007;298:361–369. doi: 10.1007/s00403-006-0709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, Paramio JM, Vooijs M. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004;131:2737–2748. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- Sathish N, Abraham P, Peedicayil A, Sridharan G, Chandi G. HPV16 E6 sequence variations in Indian patients with cervical neoplasia. Cancer Lett. 2005;229:93–99. doi: 10.1016/j.canlet.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Wernes BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Seedorf K, Krammer G, Dürst M, Suhai S, Röwekamp WG. Human papillomavirus 16 DNA sequence. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Sherman L, Schlegel R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J. Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichero L, Ferreira S, Trottier H, Duarte-Franco E, Ferenczy A, Franco EL, Villa LL. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer. 2007;120:1763–1768. doi: 10.1002/ijc.22481. [DOI] [PubMed] [Google Scholar]
- Song S, Pitot HC, Lambert PF. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J. Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask DK, Muller MT. Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc. Natl. Acad. Sci. U S A. 1988;85:1417–1421. doi: 10.1073/pnas.85.5.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Sasaki K, Yoshida S, Kajitani N, Satsuka A, Nakamura H, Sakai H. Molecular mechanism of hyperplasia induction by human papillomavirus E7. Oncogene. 2006;25:4155–4164. doi: 10.1038/sj.onc.1209445. [DOI] [PubMed] [Google Scholar]
- Veldman T, Liu X, Yuan H, Schlegel R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci U S A. 2003;100:8211–8216. doi: 10.1073/pnas.1435900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa LL, Sichero L, Rahal P, Caballero O, Ferenczy A, Rohan T, Franco EL. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J. Gen. Virol. 2000;81:2959–2968. doi: 10.1099/0022-1317-81-12-2959. [DOI] [PubMed] [Google Scholar]
- Woodworth CD, Cheng S, Simpson S, Hamacher L, Chow LT, Broker TR, DiPaolo JA. Recombinant retroviruses encoding human papillomavirus type 18 E6 and E7 genes stimulate proliferation and delay differentiation of human keratinocytes early after infection. Oncogene. 1992;7:619–626. [PubMed] [Google Scholar]
- Xi LF, Demers GW, Koutsky LA, Kiviat NB, Kuypers J, Watts DH, Holmes KK, Galloway DA. Analysis of human papillomavirus type 16 variants indicates establishment of persistent infection. J. Infect. Dis. 1995;172:747–755. doi: 10.1093/infdis/172.3.747. [DOI] [PubMed] [Google Scholar]
- Xi LF, Koutski LA, Galloway DA, Kuypers J, Hughes JP, Wheeler CM, Holmes KK, Kiviat NB. Genomic variation of human papillomavirus type 16 and risk for high-grade cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 1997;89:796–802. doi: 10.1093/jnci/89.11.796. [DOI] [PubMed] [Google Scholar]
- Xi LF, Kiviat NB, Hildesheim A, Galloway DA, Wheeler CM, Ho J, Koutski LA. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J. Natl. Cancer Inst. 2006;98:1045–1052. doi: 10.1093/jnci/djj297. [DOI] [PubMed] [Google Scholar]
- Yamada T, Manos MM, Peto J, Greer CE, Munoz N, Bosch FX, Wheeler CM. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J. Virol. 1997;71:2463–2472. doi: 10.1128/jvi.71.3.2463-2472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehbe I, Wilander E, Delius H, Tommasino M. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 1998a;58:829–833. [PubMed] [Google Scholar]
- Zehbe I, Voglino G, Delius H, Wilander E, Tommasino M. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet. 1998b;352:1441–1442. doi: 10.1016/S0140-6736(05)61263-9. [DOI] [PubMed] [Google Scholar]
- Zehbe I, Voglino G, Wilander E, Delius H, Edler L, Klimek F, Andersson S, Tommasino M. p53 codon 72 polymorphism and various human papillomavirus 16 E6 genotypes are risk factors for cervical cancer development. Cancer Res. 2001a;61:608–611. [PubMed] [Google Scholar]
- Zehbe I, Tachezy R, Mytilineos J, Voglino G, Mikyskova I, Delius H, Gissmann L, Wilander E, Tommasino M. Human papillomavirus 16 E6 polymorphisms in cervical lesions from different European populations and their correlation with human leukocyte antigen class II haplotypes. Int. J. Cancer. 2001b;94:711–716. doi: 10.1002/ijc.1520. [DOI] [PubMed] [Google Scholar]
- Zur Hausen H. Papillomavirus infections - a major cause of human cancers. Biochim. Biophys. Acta. 1996;1288:55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]