Abstract
Despite the availability of dozens of animal genome sequences, two key questions remain unanswered: First, what fraction of any species' genome confers biological function, and second, are apparent differences in organismal complexity reflected in an objective measure of genomic complexity? Here, we address both questions by applying, across the mammalian phylogeny, an evolutionary model that estimates the amount of functional DNA that is shared between two species' genomes. Our main findings are, first, that as the divergence between mammalian species increases, the predicted amount of pairwise shared functional sequence drops off dramatically. We show by simulations that this is not an artifact of the method, but rather indicates that functional (and mostly noncoding) sequence is turning over at a very high rate. We estimate that between 200 and 300 Mb (∼6.5%–10%) of the human genome is under functional constraint, which includes five to eight times as many constrained noncoding bases than bases that code for protein. In contrast, in D. melanogaster we estimate only 56–66 Mb to be constrained, implying a ratio of noncoding to coding constrained bases of about 2. This suggests that, rather than genome size or protein-coding gene complement, it is the number of functional bases that might best mirror our naïve preconceptions of organismal complexity.
What fraction of a genome confers biological function, as opposed to the remaining proportion that has had no biological effect and thus has not been subject to selection? While the complement of (functional) protein-coding sequence has been estimated in many organisms (e.g., 1.06% of the human genome; Church et al. 2009), it has been more challenging to identify functional sequence that fails to encode protein (Mouse Genome Sequencing Consortium 2002). Even the more simple task of estimating the size of this fraction, or more precisely, the genomic fraction that is under evolutionary constraint and is thereby inferred to confer function to the organism, has proven particularly contentious (Chiaromonte et al. 2003; Pheasant and Mattick 2007).
Methods to detect constraint do so by comparing genomic sequence and therefore show greatest power to identify “shared” constrained sequence, and lower power to reveal sequence whose function is “lineage-specific.” Analyzing species at various divergences thus offers an opportunity to investigate the dynamics of genome evolution: Is the functional fraction largely shared and evolving slowly by accumulating a low rate of point mutations, or does, instead, rapid sequence turnover of lineage-specific functional sequence play an important role? While protein-coding genes appear to evolve predominantly in the first mode, it is readily apparent that lineage-specific sequence occurs abundantly in most genomes. Instances where functional sequence has been gained, and erstwhile functional sequence has been lost, have been identified in mammals (Dermitzakis and Clark 2002; Smith et al. 2004; Odom et al. 2007; Kunarso et al. 2010), flies (Ludwig et al. 2000; Bergman and Kreitman 2001; Moses et al. 2006), and yeast (Borneman et al. 2007). Although convincing, these examples represent a very small fraction of the functional complement of each genome, and argue neither for nor against the ubiquity of functional sequence turnover.
A second key question is whether the genomes of different species contain different amounts of functional sequence, and whether this measure is related to organismal complexity. For example, it is clear that both the genome size and the number of genes present in a genome fail to reflect at least naïve preconceptions of organismal complexity (Gregory 2005; Ponting 2008). While varying proportions of nonfunctional (“junk”) DNA, often in the form of transposed repetitive elements (TEs), may explain the large variation in genome size across species, the relatively stable number of protein-coding genes suggests the possibility that our naïve notion of complexity is fundamentally incorrect, and that many species are in fact of comparable complexity, in a sense yet to be defined. Alternatively, it may be that much of the apparent differences in complexity between species are encoded by a varying amount of noncoding regulatory sequence, regulating a fairly stable core of protein-coding genes.
Addressing these two questions requires accurate estimates of the amount of functional, yet noncoding, sequence in genomes from across the metazoan subkingdom. Several groups have developed comparative genomic methods to estimate this quantity. For example, an early estimate of the genomic fraction of human constrained sequence was obtained from alignments of human and mouse genome assemblies, and suggested that approximately αsel = 5% of the human genome has been subject to selective constraint (Chiaromonte et al. 2003). (Here, we adopt from Chiaromonte et al. the symbol αsel as the estimated fraction of a genome that has been subject to selective constraint and thus may be considered functional. In addition, we define g as the full extent of the euchromatic sequence of a genome, and gsel = g × αsel as the amount of sequence that has been subject to purifying selection.) This estimate of αsel was obtained by contrasting nucleotide conservation inside and outside of ancestral repeats (ARs, TEs whose insertion predates the species' last common ancestor) while taking account of the known regional variation in nucleotide substitution rates. Subsequently, other substitution-based approaches, taking advantage of multiple genome sequence alignments, yielded similar results (Margulies et al. 2003; Cooper et al. 2005; Siepel et al. 2005).
All such estimates of αsel have shown a strong dependence on the parameterization of the underlying neutral substitution model, and as neutral substitutions are difficult to model (Clark 2006), the resulting estimates have wide confidence intervals. For example, the initial approach by Chiaromonte et al. (2003) indicated αsel as being between 2.3% and 7.9% of the human genome, depending on which values of model parameters were chosen. The attendant uncertainty in the final estimates makes it difficult to use this or similar methods to quantify lineage-specific constrained sequence.
More recently, three analyses have estimated αsel by taking advantage of the 1% of the human genome that has been scrutinized within the pilot phase of the ENCODE project (The ENCODE Project Consortium 2007). These yielded higher αsel estimates of between 5% and 12% (Asthana et al. 2007; Garber et al. 2009; Parker et al. 2009) with the spread of αsel values being again dependent upon the values of model parameters that were chosen. With one algorithm constraint was identified within 45% of ARs (Parker et al. 2009). Estimates of αsel in ENCODE regions may also be upwardly biased, since only some of ENCODE's regions were randomly selected, while others were chosen because of their functional content.
For invertebrates estimates of αsel have also been imprecise, in the main because their small genomes often contain only a meager amount of neutrally evolving sequence on which to tune a neutral model (Peterson et al. 2009). Estimates of αsel for Drosophila range between ∼40% and 70% (Andolfatto 2005; Siepel et al. 2005; Halligan and Keightley 2006; Keith et al. 2008), while one study indicated that 18%–37% of the Caenorhabditis elegans genome is under selective constraint (Siepel et al. 2005).
As alluded to above, methods for inferring quantities of functional DNA rest upon the hypothesis that in functional sequence most nucleotide changes are detrimental, causing such changes to be purged from the species' populations, which results in evolutionarily conserved sequence. Methods for quantifying constrained sequence typically contrast interspecies levels of sequence conservation within a sequence of interest and within matched putatively neutrally evolved sequence, typically ARs. While the deletion of conserved sequence identified in this manner does not always result in an overt phenotype (Ahituv et al. 2007; Visel et al. 2009), it has been shown that selection rather than mutational cold-spots are responsible for the low rate of mutation accumulation (Drake et al. 2006). The outlined approach has been further criticized for overlooking sequence that is lineage-specific or that exhibits only weak conservation (Dermitzakis and Clark 2002), for tacitly assuming, rather than demonstrating, the neutrality of ARs, and for overlooking sequence that has evolved by positive, rather than negative, selection (Pheasant and Mattick 2007).
Here, we estimate the quantities of functional DNA that are shared between species pairs at various divergences. This allows us to investigate the dependence of this quantity on species divergence, thus partially addressing lineage specificity. An earlier study using the same method demonstrated that ARs are predominantly neutrally evolving (Lunter et al. 2006), thereby addressing the second concern, and the present study confirms these findings. By continuing to overlook potentially positively selected sequence our estimates of the amount of functional sequence are expected to remain slightly conservative.
The approach presented here (based on the neutral indel model; Lunter et al. 2006) uses indel mutations, rather than single-nucleotide substitutions, to estimate αsel. Although indel events occur approximately eightfold less often than substitution mutations (Lunter 2007; Cartwright 2009), their impact upon functional sequence may well be more profound than that exerted by single-nucleotide substitutions. Indels may induce, for example, frame shifts in coding regions and secondary structure changes in RNAs, suggesting that stronger purifying selection may often act upon them. This will compensate for their lower mutation rate when indels are exploited in approaches to detecting evolutionary constraint. In contrast to many substitution-based methods that require fitting an explicit background model to neutrally evolving sequence, the present method has a single free parameter (the indel rate) which can be trained from the full data, without the requirement of first identifying the neutral fraction.
Here, we estimate αsel values for diverse mammalian species and for birds, teleost fish, and fruit flies. We show that the neutral indel model estimates gsel for closely related pairs as being up to threefold higher than for more distantly related species, a result that is a feature of the data rather than being an inherent bias of the method. This suggests a substantial rate of “turnover” of otherwise constrained sequence. Finally, we show that, despite their comparable protein-coding gene complement, vertebrate (mammalian or avian) genomes harbor substantially more functional sequence than invertebrate (Drosophila and C. elegans) genomes, as a result of a larger complement of functional noncoding sequence.
Results
Comparison of mouse and rat
The neutral indel model predicts that for neutrally evolving genomic sequence the lengths of “inter-gap segments” (IGS) between adjacent indel events follow a geometric distribution (Lunter et al. 2006). This prediction holds regardless of the size of the indels, or whether they are insertions or deletions, but requires indel rates to be uniform across the genome. Purifying selection purging indels from the genome will cause a fraction of IGS to become longer than expected under the neutral model, and the excess of these long IGS provides an estimate of the total length of sequence from which indels have been purged (see Supplemental Text 1 and Lunter et al. 2006 for further details).
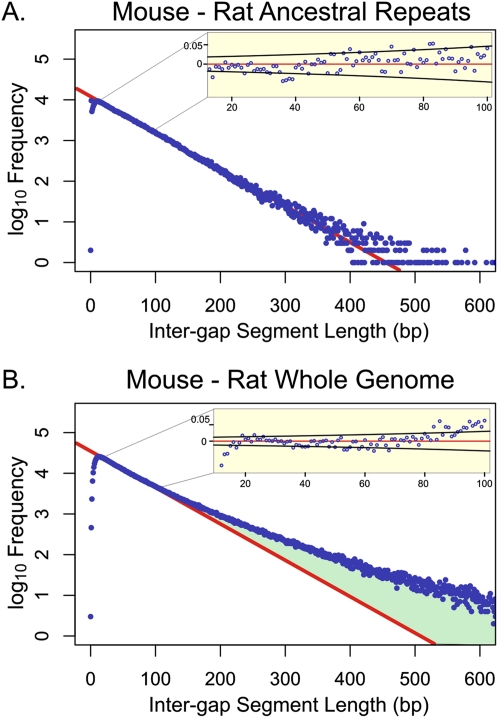
We started by considering genome-wide alignments between mouse and rat, species that diverged ∼13–19 million years ago (Mya) (Douzery et al. 2003). Our previous application of the model was limited to a three-way comparison of mouse, human, and dog (Lunter et al. 2006), which share a more ancient last common ancestor ∼97 Mya (Murphy et al. 2007). When limiting our comparisons to mouse–rat ARs, we observed, as seen previously for mouse–human ARs, the IGS frequency histogram to be very well approximated by a geometric distribution, as predicted by the neutral indel model (Fig. 1A). This provides support for the hypothesis that the vast majority of mouse or rat TEs have evolved neutrally since their insertion at least 13–19 Mya. The neutral indel model predicted a negligible proportion of the 413-Mb mouse–rat ancestral ARs to be subject to purifying selection on indels (Table 1). Similarly, minimal amounts of conserved sequence (0.74–0.85 Mb; 0.4%–0.5%) were observed in the 173 Mb of human–mouse AR sequence, commensurate with an earlier estimate (Lowe et al. 2007).
Figure 1.
Representative genomic distributions of IGS lengths in mouse–rat alignments. Frequencies of IGS (blue) are shown on a log10 scale for AR regions (A) and whole-genome sequences (B) with G+C contents of 0.415–0.425. The red line represents the prediction of the neutral indel model, a geometric distribution of IGS lengths calibrated over IGS ∼15–80 bp in length. For mouse–rat AR sequence, the observed data accurately fit the predictions of the neutral indel model, with no deviation from the model apparent within this interval (inset shows residuals, and 95% confidence bounds in black based on a Bernoulli model). For whole-genome alignments, the data fit accurately for IGS 10–100 bp in length. Beyond 100 bp, there is an excess of longer IGS (green), representing sequence which contains fewer indels than would be predicted under the neutral indel model. The underrepresentation of short IGS (<10) is due to “gap attraction,” an artifact of the alignment process (Lunter et al. 2008). Histograms for the 19 remaining G+C bands are provided as Supplemental Figure 1.
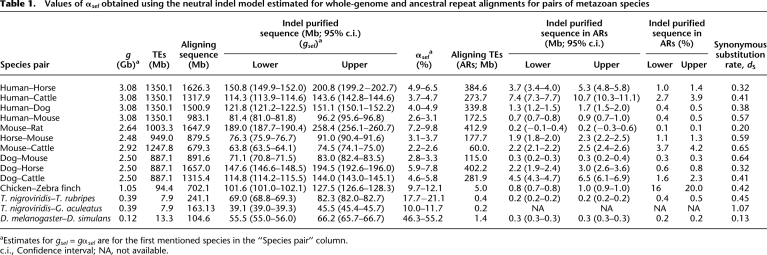
Table 1.
Values of αsel obtained using the neutral indel model estimated for whole-genome and ancestral repeat alignments for pairs of metazoan species
aEstimates for gsel = gαsel are for the first mentioned species in the “Species pair” column.
c.i., Confidence interval; NA, not available.
Turning next to whole-genome alignments of mouse and rat (Fig. 1B), we found that the number of IGS over 70 bp in length greatly exceeds the prediction under neutrality. In all, the IGS that are unaccounted for by the neutral model cover gsel + Δ = 328 Mb, where Δ represents the expected amount of “neutral overhang” that forms part of most IGS spanning a conserved element (Lunter et al. 2006). By estimating upper and lower bounds for Δ (Lunter et al. 2006) we obtain estimates for gsel of between 189 and 258 Mb (αsel = 7.2%–9.8%; Fig. 1B). This estimate is over twofold higher than our previous estimate of gsel, obtained using the identical approach, for functional sequence present in alignments of human, mouse, and dog sequence (78.8–100.0 Mb; Fig. 2), consistent with the notion that a much smaller amount of functional sequence is shared between all three species than is shared between rat and mouse.
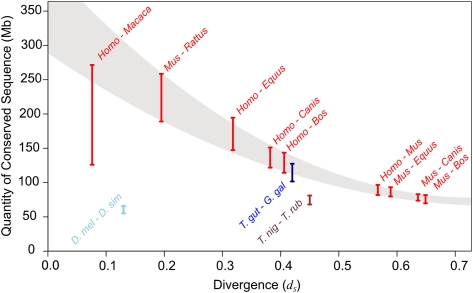
Figure 2.
Quantities of constrained sequence (gsel) estimated across a range of diverse metazoan species' pairs. Estimates of constrained sequence in eutherian mammalian (red), avian (dark blue), teleost fish (brown), and fruit fly (light blue) species' pairs. For mammalian estimates, a dramatic drop-off in estimates of conservation is associated with increasing divergence between species' pairs, which is not seen in simulations (Fig. 3). The indicative sweep (shaded) suggests that the true quantity of functional material in mammalian genomes may be around 300 Mb (10% of the human genome). The range for human and macaque represents several estimates with varying parameters for the calibration of the neutral model. Consequently, these values may underestimate the true level of constraint. Our highest estimate of conserved sequence in mammals is between mouse and rat, for which we estimate 189.0–258.4 Mb of functional sequence.
Neutral indel model comparisons across eutherian mammals
We next considered alignments of genomic sequence from further pairs of mammals for seven eutherian mammals, namely human, rhesus macaque, mouse, rat, cattle, horse, and dog. Divergences between species pairs are quantified in terms of both their median rate of synonymous substitutions per synonymous site dS and their lineages' estimated date of divergence. For example, human and macaque (median dS = 0.075) diverged 25 Mya (Gibbs et al. 2007), whereas laurasiatherians (for example, cattle, horse, and dog) and euarchontoglires (mouse, rat, macaque, and human), which last shared a common ancestor ∼90 Mya (Murphy et al. 2007), are related by median dS values between 0.32 and 0.65.
Estimated amounts of indel-purified (and by implication functional) sequence present within ARs were low for most species pairs, spanning between 0.2 Mb and 5.3 Mb (0.1%–1.4% of AR sequence; Table 1). The notable exceptions to this were seen for alignments involving the cattle genome, which were associated with elevated estimates of indel rates specifically within TEs; resolution of whether these elevated estimates reflect assembly errors or else unusual biology that is specific to the bovid lineage will require additional sequence data (see Supplemental Text 2). For most sets of AR alignments IGS frequency distributions were, once again, well approximated by the geometric distribution expected from the model. For alignments that paired a rodent genome sequence with a non-rodent genome sequence, we used the TE annotations for the non-rodent species, because TEs are less well annotated within the rodent genome sequences owing to their rapid evolution. Regardless of which TE annotations were used, estimates of constrained sequence remained essentially constant. The IGS distribution for human and macaque AR alignments was unexpectedly found to contain peaks, but these reflect an artifact arising from sequence and assembly error, as we demonstrate elsewhere (Meader et al. 2010), as well as being a consequence of Alu TEs containing a pair of relatively hypermutable poly-A tracts physically separated by ∼150 bp (Batzer and Deininger 2002). For this reason we were unable to estimate the amount of constrained sequence in primate ARs, and for the primate–primate comparisons we only considered aligned non-TE sequence.
Genome-wide comparisons for these eutherian mammals resulted in estimated values of gsel from 63.8 to 74.5 Mb for the most distantly related species pair (cattle–mouse) to 189–258 Mb for the least diverged pairs (mouse–rat; Fig. 2; Table 1). The neutral indel model thus consistently predicts gsel as being threefold higher in closely related eutherian species than those that are more distantly related.
Analyses of simulated genome sequence alignments
Next, we considered whether this unexpected variation in gsel might reflect an artifact of the neutral indel model. To this end we evolved simulated genomes from initially identical pairs of 200 Mb in size, each for the same amount of time, with constant rates of substitution and of insertion/deletion events, and subsequently aligned them (see Methods). In each simulated genome, 50% of sequence was annotated as “TE” sequence, to serve as known neutrally evolving control sequence. Five percent (10 Mb) of each genome was annotated as constrained sequence, which in the simulations was refractory to indel mutations to various degrees. We were mostly concerned with any dependence of the estimated fraction of conserved sequence with evolutionary distance. Nevertheless, to assess robustness of the various assumptions, we additionally investigated a range of other parameters, including (1) “cryptic” indel rate variation in neutral sequence (i.e., rate variation that is not accounted for by G+C content), (2) the length distribution and clustering characteristic of conserved sequence, and (3) the probability of indel fixation within them. For each parameter we chose initial values based on our knowledge of (known) functional elements; for instance, the rate of indel fixation within exons is ∼10% of the rate observed in neutrally evolving sequence (Brandstrom and Ellegren 2007).
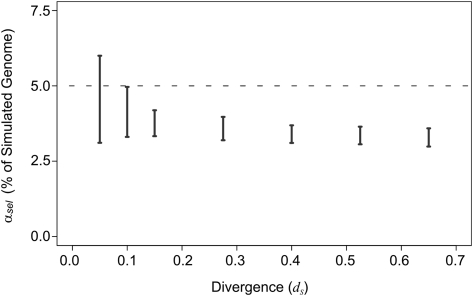
By varying each model parameter across a wide range, while keeping others fixed, we assessed its influence on αsel when analyzing these simulated genome pairs using the neutral indel model (see Supplemental Text 3). Of all combinations of parameters, only two caused an overestimation of the true amount of conserved sequence within the simulated genome. First, only when the simulated divergence drops below dS = 0.1 does the upper-bound αsel estimate (but not the lower-bound estimate) exceed the true value (Fig. 3). Second, only when we include in our simulations an exceptionally high level of “cryptic” indel rate variation do both upper- and lower-bound estimates of αsel exceed the true value. However, in this case the same simulations show that we would also see high levels of predicted constrained sequence in ARs, which we fail to see in real data (see Supplemental Text 3). Consequently, our simulations indicate that both the upper- and lower-bound estimates of αsel are expected to be conservative estimates of the true proportion of sequence under purifying selection.
Figure 3.
Estimates of constrained sequence (black bars) in simulated genomes. Simulated genomes contained 5% of constrained sequence (broken line). Constrained sequence rejects 90% of indel events. The neutral indel model consistently underestimates the true quantity of conserved sequence for genome pairs with more than one substitution per neutral base. Only at a divergence of 0.1 does the upper-bound estimate approach the true quantity of constrained sequence. Over divergences of 0.15 to 0.65, the reduction in estimates of constraint is minimal. This is in contrast to observations in alignments of real mammalian genome assemblies, for which there is a 2.2-fold difference over the same evolutionary range (Fig. 2).
Analysis of ENCODE pilot regions
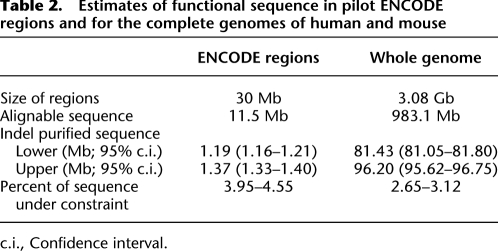
Recent studies have estimated αsel values for the phylogenetically deep multiple alignments of ENCODE pilot regions which cover ∼1% of the human genome (The ENCODE Project Consortium 2007). Such values may serve as genome-wide estimates only if the ENCODE regions are representative of the genome as a whole. However, half of ENCODE pilot regions were chosen at random, while the other half were targeted because they encompass genes of particular interest. It is thus possible that ENCODE regions possess unusually high fractions of constrained sequence. When we applied the neutral indel model to the 11.5 Mb of human ENCODE pilot sequence that aligns to the mouse genome assembly, αsel was estimated to be 3.95%–4.55%, which is ∼50% higher than the human–mouse genome-wide prediction (αsel = 2.64%–3.13%; Table 2). We thus conclude that ENCODE regions are a biased sample of the entire genome sequence, and that estimates of αsel derived from them will tend to overestimate the true genome-wide αsel value.
Table 2.
Estimates of functional sequence in pilot ENCODE regions and for the complete genomes of human and mouse
c.i., Confidence interval.
Comparisons between non-eutherian vertebrates
We next turned to the second question of this study, namely whether genomes from diverse metazoan phyla harbor similar amounts of functional sequence. To address this, we considered the aligned genomes of two avian species (the zebra finch, Taeniopygia guttata, and chicken, Gallus gallus) and two pufferfish species (Takifugu rubripes and Tetraodon nigroviridis). The known genomes of other non-eutherian species are too divergent for accurate and extensive alignment of their neutrally evolved regions to allow application of the neutral indel model. Each pair of these birds or fish is, by contrast, closely related (median dS values of 0.42 and 0.45, respectively).
The neutral indel model estimates gsel to be between 101.6 and 127.5 Mb for the two avian species. This range of gsel falls just within the range observed between human and dog (gsel = 121.8–151.1 Mb), whose divergence (median dS value of 0.38) is similar to that for these two birds. In a close parallel to our observations in eutherian mammalian genomes, we find that shared TEs show an exceedingly good fit to the neutral indel model, and we estimate that only 0.78–0.95 Mb of constrained sequence is present within ARs, ∼1% of all TE sequence in chicken (94.4 Mb). We conclude that, as for eutherian mammals, avian TEs evolve predominantly neutrally.
For the two pufferfish species (median dS value of 0.45), we estimate gsel to be between 69.0 and 82.3 Mb. Thus, despite a comparable divergence, the pufferfish share much less functional sequence than is shared between zebra finch and chicken. The data again show a remarkably good fit to the model, similar to the cases of mammalian and avian genomes; for example, only 0.16–0.18 Mb of ARs (44.3%–50.2%) exhibit evidence of constraint between the pufferfish species. For stickleback (Gasterosteus aculeatus) and tetraodon, a more divergent pair of teleost fish (median dS = 1.07), slightly lower estimates of gsel of 41.1 to 45.5 Mb were obtained. Thus, less constrained sequence is observed for more distantly related fish, just as we found for more diverged mammals.
Comparison between Drosophila fruit flies
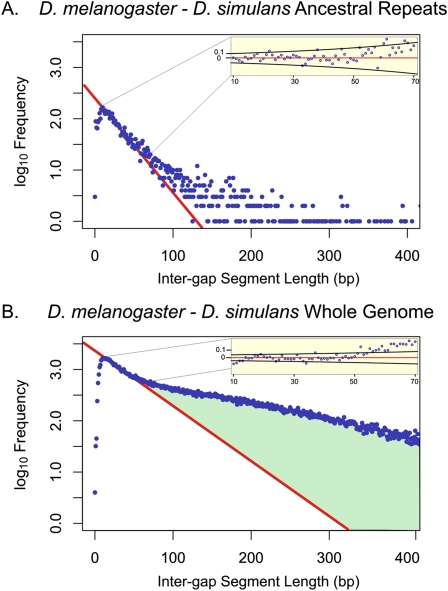
Finally, to assess quantities of constrained material in non-vertebrate metazoan species, we applied the neutral indel model to whole-genome alignments of the fruit fly species D. melanogaster and D. simulans. Of the ∼140-Mb D. melanogaster genome sequence assembly (including both euchromatic and heterochromatic sequence), 104.6 Mb is alignable with that of the D. simulans genome. In contrast to the vertebrate sequences we considered, only a small amount (13.3 Mb) of the D. melanogaster genome consists of TEs, of which 1.42 Mb are aligned between assemblies.
In contrast to all other species pairs we considered, the IGS histogram for flies does not contain a well-defined neutral regime. Presumably this reflects the compactness of the fruit fly genome from which, apparently, much neutrally evolving sequence has been purged. This presents us with the difficulty of calibrating the neutral expectation of the model from data that are likely to be composed, in part, of functional sequence. For the whole-genome analysis, the neutral regime was estimated to be short IGS of 15–55 bp in length, and we calibrated the neutral indel model using this interval (Fig. 4B). With this calibration, the resulting estimates of gsel lay between 55.5 Mb and 66.2 Mb (αsel = 47.1%–55.2%), similar to a previous estimate (Andolfatto 2005). Drosophila genome sequence that is predicted by the model to be functional was found to be evolving approximately three times more slowly than putative neutral sequence (see Supplemental Text 4), supporting the notion that this sequence indeed largely consists of functional sequence. Within the small fraction of fruit flies' ARs, 0.29–0.32 Mb of sequence was predicted, from a deficit of indels, to be functional (Fig. 4A). Compared to equivalent estimates for vertebrates, this represents a small amount, yet a large proportion (29.6%–34.5%) of ARs.
Figure 4.
Representative genomic distribution of IGS lengths in D. melanogaster and D. simulans alignments Frequencies of IGS (blue) lengths shown on a log10 scale for AR regions (A) and whole-genome sequences (B) with a G+C content of 0.495–0.445. The predictions of the neutral indel model are shown in red. In contrast to AR sequence for mouse and rat (Fig. 1), a relatively large proportion (20%–23%) of the small number of ancient fruit fly transposons appear to be under constraint, although the absolute quantity of sequence remains low (0.29–0.32 Mb). Similarly, for whole-genome sequence, we estimate that 55.5–66.2 Mb (46%–55%) of the genome is subject to constraint regarding indels. The difference in the predictions of the neutral indel model for whole-genome and AR sequence indicates that functional sequence may contribute to Drosophila short IGS.
Estimates of αsel were also obtained from alignments of D. melanogaster and D. sechellia, a sibling species of D. simulans, resulting in similar figures (αsel = 48.7%–58.7%). As noted, functional Drosophila sequence is likely to contribute to the short IGS portion of the frequency distribution (Fig. 4B) over which the model is calibrated. Our gsel estimates thus should be regarded as lower-bound estimates. Nevertheless, we note that even in the extreme case of αsel = 100%, our gsel estimates for eutherians would be 2.2-fold greater than for fruit flies.
Discussion
gsel values for diverse animals
We applied an evolutionary method across the metazoan phylogeny that estimates the amount of constrained DNA that is shared between pairs of species. Our main findings are, first, that mammalian genomes contain greater amounts of putative functional bases than genomes of fish and fruit flies, and second, that as the divergence between mammalian species increases, the predicted amount of pairwise shared functional sequence drops off dramatically, approximately halving in 90 million yr since the last common ancestor of laurasiatherians and euarchontoglires (Fig. 2; Table 1).
Our findings now indicate 260 Mb (the amount of constrained sequence shared between mouse and rat) as our best estimate of the total amount of constrained sequence in rodents and, by extrapolation, in other eutherian mammals. This is in contrast to previous much lower estimates of the amount of constrained sequence for mammalian genomes (Chiaromonte et al. 2003; Lunter et al. 2006). For sequence pairs that include human, the highest estimate we obtain is 200 Mb (that between human and horse). Estimates from human and rhesus macaque alignments were hindered by the relatively high proportion of indels between assemblies that represent errors of sequence or assembly (Meader et al. 2010). Nevertheless, we obtained upper-bound estimates of gsel for these primates in the range 197–271 Mb (see Supplemental Text 5). Based on these results, and extrapolating the apparent dependence of pairwise constrained sequence with divergence, our results suggest that between 200 and 300 Mb (6.7%–10.0%) of the human genome is under functional constraint. This estimate was arrived at as follows. First, the amount of human genome under functional constraint is at least 200 Mb, the upper-bound estimate for human and horse made in a divergence regime associated with conservative estimations, according to our simulations. Second, the indicative higher estimate of 300 Mb was obtained by extrapolating the trend for lower-bound estimates involving human (see Fig. 2).
Our findings indicate that the total amounts of constrained sequence in mammalian genomes substantially exceed those of the pufferfish, when considering species pairs whose divergences are similar (human–dog, chicken–zebra finch and Tetraodon–fugu; Fig. 2). These conclusions remain even when it is considered that the human and mouse euchromatic sequences are more complete than those for the pufferfish. Our estimates of gsel for pairs of Drosophila fruit flies will be less accurate than those for vertebrates because of the lower fraction of neutrally evolving sequence in these genomes (Ometto et al. 2005). Nevertheless, these estimates and, indeed, the full extent of their genomes (118 Mb and 100 Mb for the D. melanogaster and C. elegans genomes, respectively) imply that these invertebrate genomes harbor considerably less constrained sequence than genomes from mammals and other vertebrates.
A marked contrast between mammals on the one hand, and nematodes and fruit flies on the other, is the amount of noncoding constrained sequence that appears to be present in their genomes, both in absolute terms and as a proportion of protein-coding genes. For instance, we estimate the human genome to harbor 170–270 Mb of noncoding constrained sequence, or five to eight times the amount of protein-coding DNA (32.6 Mb; Church et al. 2009). In contrast, the D. melanogaster genome contains 21.8 Mb protein-coding sequence (Taft et al. 2007), and we estimate that it contains an additional 35–45 Mb of constrained noncoding sequence, ∼1.5–2 times its complement of protein-coding DNA. It is suggestive that the complement of protein-coding genes between these two species, of apparently very different organismal “complexity,” is fairly similar, while the amount of noncoding constrained sequence differs by at least twofold, and possibly over fourfold, between these species. This is compatible with the notion that much of the organismal complexity of mammals, and by implication much of the interspecific differences, are encoded in the non-protein-coding functional complement rather than in protein-coding sequence (King and Wilson 1975).
Turnover of functional sequence
Our second key finding is that, as the divergence between mammalian or fish species increases, the predicted amount of pairwise shared and putatively functional sequence drops off dramatically (Fig. 2; Table 1). It is clear that most constrained sequence is not perfectly conserved, and an increased divergence implies a larger number of fixed indels within conserved sequence, which might possibly reduce the estimate. Nevertheless, we have performed extensive simulations of constrained sequence that is partly refractory to indels, and these show no evidence for a significant drop-off in αsel with increasing divergence. Rather, our estimates of the amount of indel-refractory sequence, particularly the lower-bound estimate, consistently appear to be conservative, and nearly independent of the divergence between the species, across a wide range of divergences.
Without evidence to the contrary, we must assume that all mammals contain within their genomes similar amounts of functional sequence. How can this null model be reconciled with our observation of a decreased amount of shared conserved sequence between more divergent species? One possibility is to propose a dynamic equilibrium involving a spectrum of conservation, from a core of highly conserved DNA (including most of the protein-coding genes and some ancient regulatory sequence; Bejerano et al. 2004; Woolfe et al. 2005) that is shared across most of the mammals, to functional sequence that is being “turned over” at various rates. Here, turnover may refer to different processes. One possibility is the acquisition of sequence with novel function, either through random fortuitous change to previously nonfunctional sequence, or through duplication and mutation of previously functional sequence. These processes will by necessity be matched by roughly equal amounts of loss of such sequence by (slightly) deleterious changes, including deletions, as previously described from a study of mammalian regulatory sequence (Dermitzakis and Clark 2002). The changes required to instill function in such sequence need not be great, and a modest number of fixation events could easily bring a much larger region of functional sequence under purifying selection. A second possible process is the retention of equivalent functions of orthologous sequence, despite substantial DNA changes, as described in Drosophila (Ho et al. 2009). The existence of turnover of functional sequence is supported by several recent studies that indicate that a lack of sequence constraint does not necessarily imply a lack of function (Ludwig et al. 2000; Bergman and Kreitman 2001; Dermitzakis and Clark 2002; Moses et al. 2006; Borneman et al. 2007; Odom et al. 2007). An early study looking at substitution patterns for eight mammals within a single 1.8-Mb gene also found that the inferred proportion of constrained sequence increased with decreasing divergence, with the greatest contribution from noncoding sequence, and estimated the total fraction of constrained sequence at 10% (Smith et al. 2004). Although the authors stressed the large uncertainty in this estimate, the agreement of our present conclusions, obtained with an orthogonal approach and with whole-genome data, is striking.
In summary, we have presented evidence for the existence of substantial amounts of functional and mostly noncoding nucleotides that are specific to subclades of the mammalian phylogeny. Determining the biological function of primate-specific conserved elements will require extensive investigations of greater numbers of primate genomes but also, more importantly, the development of experimental tools that reveal the molecular basis of their function.
Methods
Sequences and annotation
Genome sequence data were obtained from UCSC Genome Informatics at http://genome.ucsc.edu (Santa Cruz). For mammalian genomes, these were for human (Homo sapiens, hg18), macaque (Macaca mulatta, rheMac2), mouse (Mus musculus, both mm8 and mm9), rat (Rattus norvegicus, rn4), dog (Canis familaris, canFam2), horse (Equus caballus, equCab1), and cattle (Bos taurus, bosTau4) genome assemblies. For non-mammalian species, assemblies used were for the zebra finch (Taeniopygia guttata, taeGut2), chicken (Gallus gallus, galGal3), pufferfish (Tetraodon nigroviridis, tetNig1, and Takifugu rubripes, fr2), stickleback (Gasterosteus aculeatus, gasAcu1), and three fruit flies: Drosophila melanogaster (dm2), Drosophila simulans (droSim1), and Drosophila sechellia (droSec1). Sets of BLASTZ whole-genome alignments were acquired from UCSC Genome Informatics for each of the species' pairs considered. For mouse, the mm8 genome assembly was used in all instances, with the exception of alignments with cattle, where the later mm9 genome assembly was used.
The repetitive portion of each genome was identified using annotations from RepeatMasker (http://www.repeatmasker.org). The locations of 30 Mb of pilot ENCODE regions in the human genome were also acquired from UCSC Genome Informatics.
IGS length histograms
Inter-gap segments (IGS) are defined as gap-delimited (ungapped) segments of aligned sequence from genome assemblies of two species. Segments that were excluded, for example in analyses considering ARs only, were excised from alignments and resultant flanking alignment blocks were artificially joined. Where assembly gaps (Ns) were present in either of the two genome sequences, the aligned regions were excised and the flanking sequences joined to form one contiguous alignment.
The neutral indel model provided a fit to the observed histogram of IGS counts against ungapped alignment block length by weighted linear regression on the log frequencies, with weights derived from the expected sampling error per length bin (modeled as a binomial distribution) in log-space. The length intervals over which this regression was performed were determined by maximizing the coefficient of determination over a range of IGS length intervals. This procedure was performed independently for each of 20 genomic subsets partitioning the genome into subsets of approximately equal G+C content, as measured on 250 bp windows. For fruit flies, pufferfish, and alignments specific to mammalian ENCODE regions, the number of G+C subsets was reduced to 5 to account for the reduced amount of aligned sequence available. Limits were placed on the length intervals we considered so that the regression would be over an interval beginning with IGS 10–25 bp in length, and ending with IGS 40–100 bp in length (with the exception of the human and macaque analysis, see below); within these constraints an interval was chosen to maximize the model's explained variance (R2). The interval limits prevented the regression from fitting to frequencies of shorter IGS where counts are reduced as a result of the alignment artifact “gap attraction” (Holmes and Durbin 1998; Lunter et al. 2008), and longer IGS, where counts are inflated by a contribution of longer IGS due to functional sequence; they also ensured that the regression interval chosen was never very small, in which case an artificially high R2 statistic would be expected. The resulting regression line represents the expected counts under the neutral indel model. To estimate αsel we accumulated the difference between the observed and expected IGS counts for longer IGS lengths, starting from the smallest IGS lengths that exceeded the predictions of the neutral indel model while accounting for “neutral overhang” sequence (Lunter et al. 2006; Supplemental Text 1).
Simulations of genome evolution
Two-hundred-mega-base genome sequences were simulated in 5-kb blocks with G+C content based upon 20 equally populated bins, reflecting the known G+C distribution of the human genome sequence. A total of 5% of each simulated genome was annotated as being functional with the lengths of functional elements drawn from a gamma distribution with default scale parameter θ = 60 and shape parameter k = 2. Clustering of functional sequence was simulated by adjusting the probability (0 to 0.95, default value 0.5) that functional elements were closely followed by a second functional segment. Where functional segments were clustered, these were separated by intervening neutral sequence whose length was drawn from a gamma distribution (default: θ = 15, k = 2). Use of alternative parameter values had only a limited effect on the neutral indel model to estimate functional sequence (data not shown). Half of the simulated genome was annotated as containing “TE” sequence, which differed in no way from the remaining nonconserved sequence, but was used to identify known neutrally evolving sequence.
Identical simulated genome sequence was then evolved twice each to half the evolutionary distance given by the neutral substitution rate (which is assumed to be well approximated by dS, the number of synonymous substitutions per synonymous site in coding sequence). Substitutions were modeled using the HKY85 model (transition/transversion ratio = 2.0). Functional regions were allowed to accept (“fix”) only 50% of substitutions. Indel mutation rates varied with G+C content, according to previous rate estimates from alignments of human, mouse, and dog (Lunter et al. 2006). These rates were scaled so that one indel mutation occurred for every eight substitutions in the median G+C category. Indel acceptance in constrained sequence varied from 0% to 20%; however, for most simulations an acceptance rate of 10% in functional sequence was employed, based upon observations from protein coding sequence (Brandstrom and Ellegren 2007). Indel lengths were drawn from a geometric distribution (Pr [length = n] = [1 − 0.7]0.7n). Indel probabilities were initially constant within each G+C bin. However, in order to model indel rate variation locally within G+C bins, indel rates were drawn uniformly from an interval taken symmetrically around the mean rate, plus or minus a set percentage (0%–50%), and the rate applied to the entire 5-kb block.
Estimation of neutral substitution rates
Estimates of dS for the divergence of a species pair were obtained by taking the median dS value for all one-to-one orthologous genes within the Ensembl Compara database with a dS value ≤ 1.0. These values were very similar to substitution rates estimated in other studies (Cannarozzi et al. 2007). The exceptions to this were with the synonymous substitution rate of D. melanogaster and D. simulans, for which a dS of 0.13 was used (Haddrill et al. 2005), and for the teleost fish T. nigroviridis and G. aculeatus for which no data were available from the Ensembl database. For this pair, we identified orthologous protein coding sequence using the PHYOP pipeline (Goodstadt and Ponting 2006) and determined synonymous substitution rates using PAML (Yang 2007), the median value of which was dS = 1.07.
Acknowledgments
We thank the UK Medical Research Council (C.P.P., G.L., S.M.), the Biotechnology and Biological Sciences Research Council (C.P.P., G.L.) (BB/F007590/1), and The Wellcome Trust (G.L.) (075491/Z/04) for funding.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108795.110.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM 2007. Deletion of ultraconserved elements yields viable mice. PLoS Biol 5: e234 doi: 10.1371/journal.pbio.0050234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Asthana S, Roytberg M, Stamatoyannopoulos J, Sunyaev S 2007. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol 3: e254 doi: 10.1371/journal.pcbi.0030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bergman CM, Kreitman M 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res 11: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M 2007. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Brandstrom M, Ellegren H 2007. The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) genome: A high frequency of deletions in tandem duplicates. Genetics 176: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schneider A, Gonnet G 2007. A phylogenomic study of human, dog, and mouse. PLoS Comput Biol 3: e2 doi: 10.1371/journal.pcbi.0030002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright RA 2009. Problems and solutions for estimating indel rates and length distributions. Mol Biol Evol 26: 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Weber RJ, Roskin KM, Diekhans M, Kent WJ, Haussler D 2003. The share of human genomic DNA under selection estimated from human-mouse genomic alignments. Cold Spring Harb Symp Quant Biol 68: 245–254 [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG 2006. Genomics of the evolutionary process. Trends Ecol Evol 21: 316–321 [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15: 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG 2002. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: Conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Douzery EJ, Delsuc F, Stanhope MJ, Huchon D 2003. Local molecular clocks in three nuclear genes: Divergence times for rodents and other mammals and incompatibility among fossil calibrations. J Mol Evol 57: S201–S213 [DOI] [PubMed] [Google Scholar]
- Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, et al. 2006. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38: 223–227 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X 2009. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25: i54–i62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Goodstadt L, Ponting CP 2006. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput Biol 2: e133 doi: 10.1371/journal.pcbi.0020133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR 2005. Animal Genome Size Database. http://www.genomesize.com
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P 2005. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol 6: R67 doi: 10.1186/gb-2005-6-8-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD 2006. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MC, Johnsen H, Goetz SE, Schiller BJ, Bae E, Tran DA, Shur AS, Allen JM, Rau C, Bender W, et al. 2009. Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet 5: e1000709 doi: 10.1371/journal.pgen.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes I, Durbin R 1998. Dynamic programming alignment accuracy. J Comput Biol 5: 493–504 [DOI] [PubMed] [Google Scholar]
- Keith JM, Adams P, Stephen S, Mattick JS 2008. Delineating slowly and rapidly evolving fractions of the Drosophila genome. J Comput Biol 15: 407–430 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D 2007. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci 104: 8005–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567 [DOI] [PubMed] [Google Scholar]
- Lunter G 2007. Probabilistic whole-genome alignments reveal high indel rates in the human and mouse genomes. Bioinformatics 23: i289–i296 [DOI] [PubMed] [Google Scholar]
- Lunter G, Ponting CP, Hein J 2006. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput Biol 2: e5 doi: 10.1371/journal.pcbi.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Rocco A, Mimouni N, Heger A, Caldeira A, Hein J 2008. Uncertainty in homology inferences: Assessing and improving genomic sequence alignment. Genome Res 18: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Blanchette M, Haussler D, Green ED 2003. Identification and characterization of multi-species conserved sequences. Genome Res 13: 2507–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meader S, Hillier LW, Locke D, Ponting CP, Lunter G 2010. Genome assembly quality: Assessment and improvement using the neutral indel model. Genome Res 20: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB 2006. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol 2: e130 doi: 10.1371/journal.pcbi.0020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res 17: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L, Stephan W, De Lorenzo D 2005. Insertion/deletion and nucleotide polymorphism data reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics 169: 1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH 2009. Local DNA topography correlates with functional noncoding regions of the human genome. Science 324: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Hare EE, Iyer VN, Storage S, Conner L, Papaj DR, Kurashima R, Jang E, Eisen MB 2009. Big genomes facilitate the comparative identification of regulatory elements. PLoS ONE 4: e4688 doi: 10.1371/journal.pone.0004688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS 2007. Raising the estimate of functional human sequences. Genome Res 17: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Ponting CP 2008. The functional repertoires of metazoan genomes. Nat Rev Genet 9: 689–698 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Brandstrom M, Ellegren H 2004. Evidence for turnover of functional noncoding DNA in mammalian genome evolution. Genomics 84: 806–813 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29: 288–299 [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA 2009. Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi: 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]