Abstract
The combining of genome-wide association (GWA) data across populations represents a major challenge for massive global meta-analyses. Genotype imputation using densely genotyped reference samples facilitates the combination of data across different genotyping platforms. HapMap data is typically used as a reference for single nucleotide polymorphism (SNP) imputation and tagging copy number polymorphisms (CNPs). However, the advantage of having population-specific reference panels for founder populations has not been evaluated. We looked at the properties and impact of adding 81 individuals from a founder population to HapMap3 reference data on imputation quality, CNP tagging, and power to detect association in simulations and in an independent cohort of 2138 individuals. The gain in SNP imputation accuracy was highest among low-frequency markers (minor allele frequency [MAF] < 5%), for which adding the population-specific samples to the reference set increased the median R2 between imputed and genotyped SNPs from 0.90 to 0.94. Accuracy also increased in regions with high recombination rates. Similarly, a reference set with population-specific extension facilitated the identification of better tag-SNPs for a subset of CNPs; for 4% of CNPs the R2 between SNP genotypes and CNP intensity in the independent population cohort was at least twice as high as without the extension. We conclude that even a relatively small population-specific reference set yields considerable benefits in SNP imputation, CNP tagging accuracy, and the power to detect associations in founder populations and population isolates in particular.
In the next generation of genome-wide association studies (GWAS), large consortia combine GWA results from platforms that have different single nucleotide polymorphism (SNP) marker resolutions and the capability for copy number polymorphism (CNP) discovery and genotyping (Cooper et al. 2008). The challenge of having data of different SNP resolutions across GWA studies can be overcome by genotype imputation: the inference of missing and unobserved data from the local linkage disequilibrium (LD) structure of a high-resolution reference sample (Li and Abecasis 2006; Marchini et al. 2007). Similarly, information on CNPs can be inferred from the variation of SNPs that correlate with CNP signal intensities in a reference set genotyped with a high-density array with good CNP coverage (Locke et al. 2006; McCarroll et al. 2006; Redon et al. 2006). The strategy of inferring CNPs from SNP data may assist future CNP studies by allowing for the determination of CNPs using a specific set of tagging SNPs instead of costly CNP detection and genotyping pipelines.
In studies using samples of European origin, the SNP imputation and CNP-tagging reference panel has typically been the 60 CEPH (Utah residents with ancestry from Northern and Western Europe, abbreviation: CEU) samples from HapMap Phase 2 (The International HapMap Consortium 2003, 2007). Recently, additional HapMap populations were genotyped with two commercially available chips, the Illumina Infinium Human1M-single and Affymetrix Genome-Wide Human SNP Array 6.0 (The International HapMap 3 Consortium 2010). This new HapMap3 data set of 1184 samples offers better genomic information due to its increased sample size and the SNP and CNP data from the high-density genotyping arrays. The design of HapMap3 allows for the extension of the reference set into additional populations with different LD structures. Some of the most unique LD patterns in the world arise in founder populations (Service et al. 2006), many of which also display unique health risks (Peltonen et al. 1999; Orton et al. 2008). Given these characteristics, does it make sense for a founder population to have its own reference set and, if so, how would this impact our ability to detect disease variants? Here, we use a local reference set from the genetically distinct Finnish founder population (Supplemental Fig. 1; Service et al. 2006; Jakkula et al. 2008) to evaluate the accuracy, coverage, and power of SNP imputation and CNP tagging relative to that of the less-specific HapMap3 European samples CEU and TSI (from Tuscans in Italy).
Results
Imputation quality
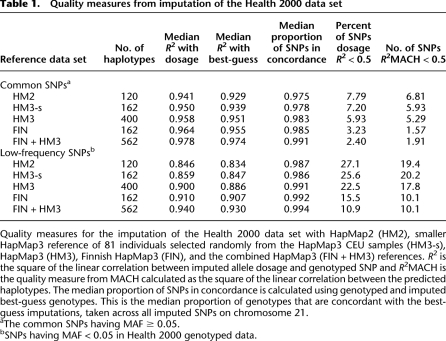
To study the SNP imputation quality, we used the Health 2000 study data (H2000; for details see Methods and http://www.nationalbiobanks.fi), which was genotyped with the Illumina 610-Quad BeadChip. We masked SNPs that were not included in the previous generation Illumina HumanCNV370 BeadChip (310,906 SNPs after quality control). The masked SNPs were imputed using five phased reference sets: HapMap2 CEU (HM2, N = 60), HapMap3 CEU + TSI founders (HM3, N = 200), Finnish HapMap3 (FIN, N = 81), the smaller HapMap3 reference set of 81 individuals randomly selected from HM3 CEU (HM3-s, N = 81), and combined HapMap3 (FIN + HM3, combining CEU + TSI + FIN, N = 281) (Fig. 1). We illustrate the results by imputing data for the complete chromosome 21. Figure 2 and Table 1 show the median proportion of concordant SNPs between the genotyped and imputed best-guess genotype and the medians of the squared linear correlations (R2) between the genotyped 610K SNPs with the imputed allele dosage genotypes or best-guess genotypes. Specifically, the R2 values presented measure how well-correlated the imputed genotypes are to the true genotypes and, therefore, they represent the factor by which linear regression statistics of association will be reduced, on average, when the imputed data is used in place of directly typed SNPs. Using HM2 reference, the median R2 between imputed allele dosages and true genotypes was 0.941 for common and 0.846 for low-frequency SNPs. HM3 showed slightly higher R2 values and the FIN reference set offered even better R2 despite having less haplotypes than HM3 (Table 1; Fig. 2). When comparing the imputation quality of FIN to HM3-s, both having the same number of individuals, the gain of quality when using FIN was even larger. The strength of the correlation was further increased when using the pooled haplotypes, FIN + HM3 (R2 = 0.978 for common and 0.940 for low-frequency SNPs). The pooled reference set performed particularly well for low-frequency alleles (MAF < 5%) (Supplemental Fig. 2). We were unable to compare the imputation of rare alleles (MAF < 0.5%) due to the low N of the FIN data set.
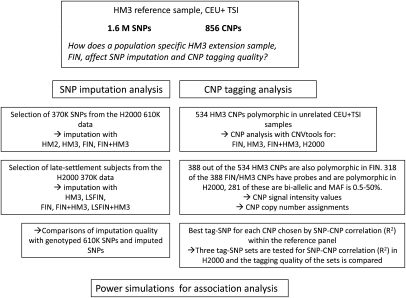
Figure 1.
Study flow. The setting and flow of the study for both imputation and CNV tagging. (CEU) CEPH (Utah residents with ancestry from Northern and Western Europe); (TSI) Tuscans in Italy; (CNP) copy number polymorphism; (SNP) single nucleotide polymorphism; (FIN) Finnish HapMap3; (LSFIN) late-settlement subset of the FIN data set; (H2000) Health 2000 data set.
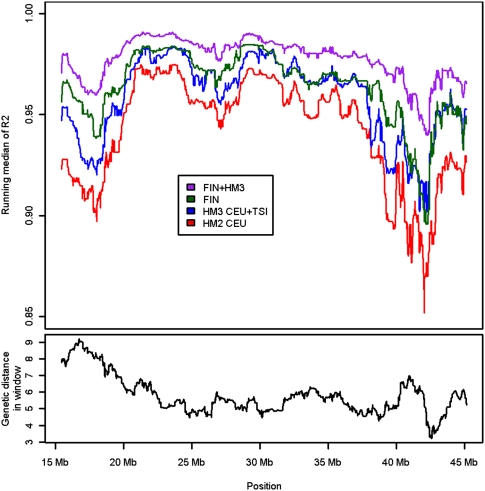
Figure 2.
Running median of squared linear correlations from the H2000 imputations. The top of the plot shows the running median of squared linear correlations between imputed allele dosages and the genotyped SNPs on chromosome 21 in the H2000 imputation with HapMap2 (HM2), HapMap3 (HM3), Finnish HapMap3 (FIN), and the combined reference, FIN + HM3. At bottom are the genetic distances, based on the recombination map of HapMap2 trios, within the windows in which the medians of R2s have been calculated.
Table 1.
Quality measures from imputation of the Health 2000 data set
Quality measures for the imputation of the Health 2000 data set with HapMap2 (HM2), smaller HapMap3 reference of 81 individuals selected randomly from the HapMap3 CEU samples (HM3-s), HapMap3 (HM3), Finnish HapMap3 (FIN), and the combined HapMap3 (FIN + HM3) references. R2 is the square of the linear correlation between imputed allele dosage and genotyped SNP and R2MACH is the quality measure from MACH calculated as the square of the linear correlation between the predicted haplotypes. The median proportion of SNPs in concordance is calculated using genotyped and imputed best-guess genotypes. This is the median proportion of genotypes that are concordant with the best-guess imputations, taken across all imputed SNPs on chromosome 21.
aThe common SNPs having MAF ≥ 0.05.
bSNPs having MAF < 0.05 in Health 2000 genotyped data.
To address the impact of population bottlenecks within the isolated Finnish population (Peltonen et al. 1999), we studied a subset of 429 individuals from the H2000 study originating from the “late-settlement” population isolate of Northeastern Finland (Nevanlinna 1972; Varilo et al. 2000). While our results suggest that the late-settlement combined HapMap3 (LSFIN + HM3) reference set provides better imputation quality than the original HM3 set, the quality is highest when using the FIN + HM3 set (Supplemental Table 1). Using a “general population” reference set seems to be sufficient for good imputation quality of subisolate populations, which have been separated by additional, more recent bottlenecks from the original founder population.
In order to investigate the source of the gain in the H2000 imputation quality when using the FIN and FIN + HM3 reference set instead of HM3, we fitted a linear model that explained the variability of the R2-increase, calculated as the difference between the R2s divided by the R2 of the HM3 imputed data, by the recombination rate and relative MAF difference between HM3 and FIN as a surrogate for genetic drift. The relative difference in MAFs was calculated as the absolute difference between the FIN and HM3 MAFs divided by the HM3 MAF. The analysis showed that the best imputation quality gain was achieved for variants in genomic regions where the recombination rate is high and for variants with high relative MAF difference (Supplemental Table 2; Supplemental Fig. 3A,B). The recombination rates were matched by the position from the HapMap combined recombination map (ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2008-03_rel22_B36/rates/). To further characterize this general observation, we selected nine SNPs having R2 increases higher than five and examined the LD structure around those SNPs (Supplemental Table 3). Of these nine SNPs, four were located directly within recombination hotspots, which were defined as areas having the top 5% of cM/Mb rates along chromosome 21 according to the HapMap combined recombination map. A further four were located in the immediate (<20 kb) vicinity of one or more hotspots. R2 increase of one SNP could not be explained by local recombination rates.
CNP tagging
Parallel to the SNP imputation analysis, we assessed the best combination of reference panels for choosing CNP tag-SNPs for H2000, our independent Finnish data set. We chose a tag-SNP for each CNP in the FIN, HM3, and FIN + HM3 reference panels, independently. We then measured the correlation coefficient R between the H2000 genotypes at each tag-SNP and the corresponding CNP. The median SNP-CNP R2 in the H2000 data was 0.412 (min. 0.000, max. 0.960) with the HM3 tag-SNPs, 0.424 (min. 0.000, max. 0.942) with the FIN tag-SNPs, and 0.448 (min. 0.000, max. 0.960) with the FIN + HM3 tag-SNPs. Although the overall R2 distribution was similar for all three tag-SNP sets, there was a subset of CNPs whose CNP-tagging accuracy in the H2000 data was largely impacted by the choice of the tag-SNP reference panel (Fig. 3).
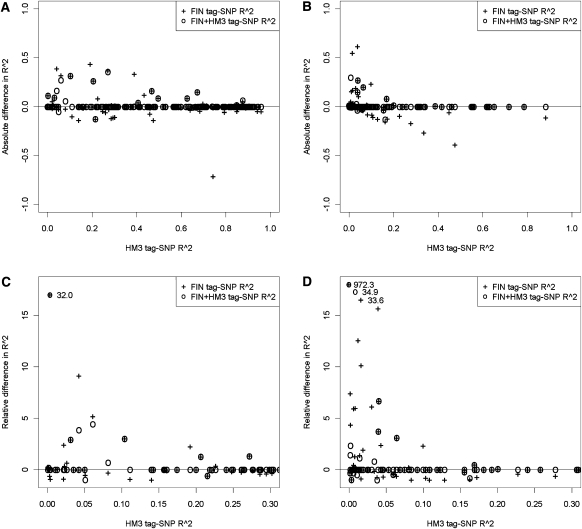
Figure 3.
Comparison of the correlations (R2) between CNP signal and tag-SNP genotype in the H2000 data. Three tag-SNPs were tested for each CNP; the tag-SNP with the highest R2 value between SNP genotypes and CNP signal in HM3 data, FIN data, and FIN + HM3 data. Data is plotted here separately for biallelic CNPs of common (MAF ≥ 5%, N = 192) (A,C) and low-frequency (MAF between 0.5% and 5%, N = 89) (B,D). In top plots, FIN + HM3 tag-SNP and FIN tag-SNP sets are compared with HM3 tag-SNPs by plotting the FIN + HM3 and FIN differences with the HM3 tag-SNP against the HM3 tag-SNP R2 value, all from the H2000 correlation analysis. A negative difference indicates that the HM3 tag-SNP has a stronger correlation with the CNP signal in the H2000 data. If the difference is positive, then the correlation is better when the CNP tag-SNP for H2000 is selected using FIN + HM3 or FIN data. At bottom we show the relative differences in the R2 values ([FIN or FIN + HM3 R2–HM3 R2]/HM3 R2), plotted against the HM3 tag-SNP R2 value. We focus on the CNPs where the HM3 tag-SNP R2 value is <0.3, since this is the area where the highest relative differences are possible. One observation in C and three in D are of significantly larger values than the rest of the data; these are shown in the top left region of the plot with their corresponding values next to them. (HM3) HapMap3 reference set; (FIN) Finnish HapMap3; (FIN + HM3) combined reference.
A total of 9.6% (N = 27) of the CNPs were tagged with at least moderately higher R2 values (0.05 cut-off for difference) using FIN tag-SNPs compared with the corresponding HM3 tag-SNP (Fig. 3A,B; Supplemental Table 4). Concurrently, however, for 8.5% (N = 24) of the CNPs, the tagging accuracy was reduced when FIN tag-SNPs were used instead of HM3 tag-SNPs (cut-off for difference 0.05). The tag-SNP set based on FIN + HM3 reference data was more robust in tagging H2000 CNPs; FIN + HM3 tag-SNPs with at least moderately higher R2 than HM3 tag-SNP were identified for 5.3% (N = 15) of CNPs (Fig. 3B), while only 1.8% (N = 5) of FIN + HM3 tag-SNPs performed worse than the HM3 tag-SNP. As expected, most of the large R2 differences between the FIN or FIN + HM3 and the HM3 tag-SNP set occurred for CNPs where the correlation between the HM3 tag-SNP and the corresponding CNP was low (Fig. 3). For low-frequency CNPs, the gain in tagging accuracy seemed mostly to come from CNPs where R2 for HM3 tag-SNP was <0.05. For common CNPs, the gain was more evenly distributed among the HM3 R2 values.
There were 12 CNPs in the H2000 data for which the FIN + HM3 R2 values were at least twice as high as the HM3 R2 values (Fig. 3C,D; Supplemental Table 4). There were no CNPs whose R2 worsened by the same degree when using FIN + HM3 tag-SNPs. We more closely examined these 12 events where the CNP tagging accuracy gain in the H2000 data was highest when using the FIN + HM3 reference, as opposed to that of HM3 alone. On 10 occasions the correlation between any of the three tag-SNPs and CNP signal was similar in the HM3 data, but within the FIN data there was a large difference in the correlation between SNP genotypes and CNP signals for the tag-SNP suggested by the HM3 data, and the best SNP based on the FIN data. This best FIN SNP was identified and chosen as the tag-SNP for the H2000 correlation test when the FIN + HM3 data was used for tag-SNP selection. In two events, the tagging accuracy was poor with all three tag-SNPs, and the R2 gain may be explained by chance.
Association power simulation
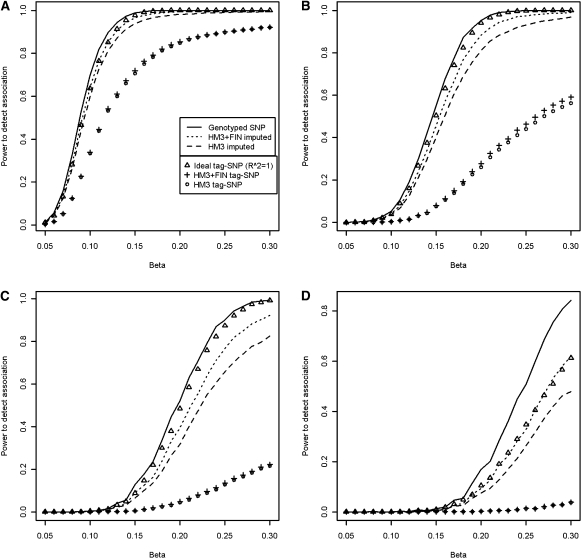
Improvements in SNP imputation quality and CNP-tagging accuracy can potentially increase the power to detect association between a SNP or CNP and a phenotype. We simulated the effect of the SNP imputation quality and CNP-tagging accuracy gain in association analysis (Fig. 4) and found that the change in power to detect association for SNPs was most evident among SNPs with allele frequencies ranging from 0% to 3% (Fig. 4D), and among CNPs with allele frequencies of 5%–10% (Fig. 4B). The gain in power to detect association for SNPs in the MAF range of from 0% to 3% was highest for the effect size, beta, of 0.3; the power increased from 0.48 to 0.62 when the Finnish HapMap3 sample set was added to the HM3 reference set. Similarly, in the MAF range of from 3% to 5%, beta of 0.26 showed the peak power increase of 0.70 to 0.82 when the FIN was added. The gain in imputation power was more modest for CNPs, for which the highest increase in power, from 0.56 to 0.59, was observed in the MAF range of from 5% to 10%, with a beta of 0.3. The percentage of variance explained by the simulated genotyped SNPs (R2 = 1) ranged in the lowest MAF range from 0.016 to 0.267 and in the highest MAF range from 0.102 to 3.657.
Figure 4.
Association power simulation results. Effect of the difference in R2 distributions in terms of power to detect an association is plotted separately for four different allele frequency ranges. SNPs and CNPs having a minor allele frequency between 10% and 50% (A), 5% and 10% (B), 3%–5% (C), and 0%–3% (D), including the upper limits. We have used an effective population size of 10,000 and ran 40,000 simulations for both SNPs and CNPs. The minor allele frequencies and squared linear correlations have been sampled jointly from the empirical distribution. (HM3) HapMap3 reference set; (FIN) Finnish HapMap3; (FIN + HM3) combined reference.
Discussion
In summary, we have used a reference set from one of the best-characterized founder populations, the Finns, to show that increases in imputation accuracy, CNP tagging quality, and even in the overall power to detect association can be achieved by densely genotyping a relatively small population-specific high-density reference panel. Importantly, these gains are more pronounced when imputing low-frequency single-nucleotide variants, which are of interest as the search for variants involved in complex diseases moves toward the less-frequent variants being identified by the 1000 Genomes Project (http://www.1000genomes.org) and other large scale sequencing efforts (McCarthy et al. 2008). Furthermore, our CNP tag-SNP analysis suggested that for a subset of CNPs, population-specific reference data can identify tag-SNPs with considerably higher tagging accuracy in the target population. We have demonstrated the value of a population-specific reference set in one founder population, but see no reason why this strategy would not be beneficial for studies in other special populations worldwide. The patterns of the Finnish population growth and genetic bottlenecks are well-characterized (Peltonen et al. 1999), and this information can be used to evaluate the advantages of population-specific reference panels in other special populations. If such reference sets are combined and made publicly accessible, they would also enhance our understanding of genomic variation and their relation to traits in various founder populations.
Methods
Finnish HapMap3 sample set
The population-specific reference data set, FIN, consists of 81 individuals genotyped with the Illumina Human 1M-Duo chip and Affymetrix Genome-Wide Human SNP Array 6.0 chip. Detailed quality control of this data set was similar to one used in HM3 (The International HapMap 3 Consortium 2010). In quality control 209,354 SNPs were removed for not meeting the following criteria: (1) was monomorphic, (2) had a calling rate < 95%, or (3) the P-value from Fisher's exact test for Hardy-Weinberg equilibrium was <1 × 10−6. As a result, 1,163,280 SNPs comprised the final FIN data set. Individuals in this data set were collected as two separate samples. Forty individuals were collected from the Finnish capital area, which is genetically representative of the general population, and 41 individuals from the late-settlement area (LSFIN), a Finnish subisolate. The haplotypes of the cleaned data set were phased following the same protocol as the HapMap3 samples of Utah residents with ancestry from Northern and Western Europe (abbreviation CEU) and the Tuscan samples from Italy (abbreviation TSI). The HapMap3 sample set (HM3) and FIN were combined to form a combined HapMap3 reference, FIN + HM3. Only SNPs that passed quality control in both HM3 and FIN were included in the combined set. HM3 CEU + TSI data included 200 unrelated founder individuals, which had passed genotyping quality control in HapMap3 release 2 (http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2009-02_phaseIII/HapMap3_r2/hapmap3_r2_b36_fwd.consensus.qc.poly.info).
Finnish test data
The H2000 data used in this study is a subset of 2212 individuals taken from the whole national Health2000 survey. These 2212 individuals are metabolic syndrome cases and their matched controls, of which 2173 have been genotyped with the Illumina 610K chip. The chip simultaneously genotypes 598,203 SNPs, of which 52,645 were excluded for having a calling rate lower than 95%, MAF < 2%, or HWE P-value < 1 × 10−4. All individuals had genotyping frequencies greater than 95%, and thus, no one needed to be removed on this basis. Thirty-five individuals were removed for having too high relatedness (π > 0.2), which is indicative of possible sample contamination, and for having non-European ancestry. After these quality-control procedures, 550,284 SNPs and 2138 individuals were available for study. From this data we took a subset of 5196 SNPs on chromosome 21, which were present on the Illumina 370K chip, to use in imputation. Using the H2000 data, it was also possible to study the imputation quality among individuals from the subisolate region. For this, we selected 429 individuals from the late settlement of Finland region (Nevanlinna 1972; Varilo et al. 2000) (Late-settlement data set) based on the multidimensional scaling of the genome (Purcell et al. 2007).
Phasing
The phasing of FIN samples was performed using the recently published algorithm IMPUTE v2 (Howie et al. 2009). The method, recently used for phasing HapMap3 samples (The International HapMap 3 Consortium 2010), uses a hidden Markov model (HMM) structure for the conditional distribution of haplotypes similar to that of IMPUTE (Marchini et al. 2007). In turn, each genotype is phased using haplotypic information from the reference panel as well as from the current haplotype estimates for the rest of the new data. CEU TRIOS from the HapMap3 data set were used as a reference panel for increased accuracy (44 CEU TRIOS, 176 phased haplotypes). The phasing was performed for the SNPs that overlap with FIN and HM3. The genetic maps used are available online (ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2008-03_rel22_B36/rates/). For both HM3 and FIN samples, each chromosome was phased in chunks of ∼8000 SNPs, with an overlap window of 200 SNPs on each side to ensure correct binding of the phased chunks into a single haplotype per chromosome. IMPUTE v2 itself accounts for edge effects by internally using 250-kb windows at each side of a chunk. The algorithm was run for 110 iterations, of which the first 10 burn-in phase iterations were discarded. Informed selection of 120 conditioning states per genotype per iteration was used. Details on this can be found in the main IMPUTE v2 paper (Howie et al. 2009).
Imputation
The imputation was performed with MACH 1.0.16 (Li and Abecasis 2006; Li et al. 2009), which is a HMM-based imputation program. The main idea behind this algorithm is to estimate the missing genotype by using the haplotype structure and frequency of the haplotypes and alleles in the reference data. Because of the large number of individuals in the H2000 data, the imputation was performed in two steps. In the first step we used 200 random individuals from the sample to predict the recombination and error maps. Using these maps, the data was then imputed in two batches of ∼1000 individuals. The numbers of metabolic syndrome cases and controls, as well as females and males in each batch, were checked to ensure that they were approximately even. For the imputation of the H2000 data, we used five different phased reference sets; HapMap2 Utah residents with ancestry from Northern and Western Europe (N = 60), HM3 (N = 200), a smaller HM3 sample of 81 individuals randomly selected from the HapMap3 CEU sample set (N = 81), FIN (N = 81), and FIN + HM3 (N = 281) (Fig. 1).
Because of the lower number of individuals in the Late-settlement sample set, the imputation was done in one step only, which estimates the maps and imputes the data simultaneously. The reference sets used for the imputation of Late-settlement sample sets were HM2, HM3, FIN, FIN + HM3, the subset of the FIN from the late-settlement area (LSFIN, N = 41), and the combined data set of HM3 samples and the late-settlement reference (N = 241).
CNP genotyping
The population-specific HM3 extension study sample, FIN, was genotyped on the same two platforms as the HM3 CEU + TSI individuals: Illumina Human 1M-Duo beadchip and Affymetrix Genome-Wide Human SNP Array 6.0. In addition to SNP probes, these contain 35,969 (Illumina beadchip) and 946,000 (Affymetrix SNP array) intensity-only probes targeting genomic copy number variation. The Illumina Human610-quad beadchip, which was the whole-genome genotyping method for the H2000 study sample, contains 620,901 probes, of which 21,890 are nonpolymorphic. Probe signal-intensity data of all SNP and CNP probes was exported for CNP analysis from raw data files using the BeadStudio software version 3.2 (http://solexa.co.uk/downloads/BEADSTUDIODataSheet.pdf) for the Illumina beadchips and the Affymetrix Power Tools (APT 1.10.2, http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx) Software Package for the Affymetrix SNP Arrays.
We attempted to genotype 856 CNP regions, which were recently identified using HapMap3 samples (The International HapMap 3 Consortium 2010), in four datasets: our Finnish HapMap3 sample, FIN; our Finnish population study sample, H2000; HapMap3 samples of European origin, CEU + TSI; and a combined reference sample of FIN and HM3 CEU + TSI. Probe signal intensity data were analyzed with CNVtools (Barnes et al. 2008). The probe signal intensities for each CNP region were summarized into a single measure for each sample. The summary method was either the first principal component analysis (PCA) method or the PCA method and linear discriminant function (LDF), depending on which one of the methods gave the best cluster separation (Q). Summarized CNP signals were used for testing correlation between SNP genotypes and CNPs in the downstream analysis. CNVtools also assigned a categorical CNP copy number estimate for each sample and CNP. We used these estimates to determine whether a CNP was polymorphic in the data set. We tested whether using the categorical CNP copy numbers yielded results in downstream analyses concordant to those of summarized signals. The two measurements yielded concordant results: Both identified a larger subset of CNPs for which tagging was improved compared with the subset where it worsened. In this study we have concentrated on the results from the copy number signal analysis, because uncertainties in copy number class assignments are avoided and an increased number of CNP loci are available for analysis, since CNPs that fail genotyping due to poor cluster separation can still be analyzed using signal intensities. However, we also used the categorical copy number cluster assignments to test the effect of applying stricter quality control criteria for the CNPs. These criteria required a Hardy-Weinberg equilibrium statistic (Pearson's χ2) < 15, <10% missing copy number assignment data, and a good quality copy number cluster separation (Q > 4). In our downstream analysis, the full CNP set and the QC+ showed similar results—both identified a larger subset of CNPs for which tagging was improved, compared with the subset where it worsened—and we have thus maximized the amount of data in our analyses by using all available CNP data.
Out of the original 856 HM3 CNPs, 322 were not polymorphic in the CEU + TSI samples and were thus excluded from our analyses. In the FIN analysis, 388 of the 534 CNP regions were estimated to be polymorphic. The Illumina 1M chip used in FIN genotyping is a more recent version than the one used in HM3 CEU + TSI genotyping, and some of the probes were not found on both chip versions. We excluded data for probes that were not common to both genotyping platforms before summarizing the probe signal intensity data in the CNP regions. In total, 862 probes out of 12,370 were excluded in FIN and 268 probes out of 11,776 were excluded in HM3, leaving 11,508 probes for CNP genotyping analysis in 388 CNP regions.
The Finnish H2000 sample set was also analyzed with CNVtools. The probe signal data for H2000 was, however, from the Illumina Human610-quad beadchip that only has probes in 745 of the 856 HM3 CNP regions. A total of 663 of these regions were found to be polymorphic in the CNVtools analysis. For CNP tag-SNP comparisons, the 663 H2000 CNPs was further reduced to 318 CNPs polymorphic in both FIN and HM3 analyses, and from that to 192 common (MAF ≥ 5%) and 89 low-frequency (MAF between 0.5% and 5%) biallelic CNPs (Fig. 1). CNPs' minor allele frequencies were calculated from the original HapMap3 data (The International HapMap 3 Consortium 2010).
Correlation tests between SNP genotypes and CNP intensity signals
We created three sets of CNP tag-SNPs for testing SNP genotype—CNP signal correlations in the H2000 data set. The first tag-SNP set was obtained by testing for the correlation between the summarized CNP signal intensities and SNP genotypes within the FIN reference sample using the cor()-function of R statistical software (R Development Core Team 2008; http://www.R-project.org), version 2.7.1. For each CNP, SNPs from the same recombination block were tested and the SNP with the highest correlation coefficient R was chosen as the tag-SNP. Recombination blocks were defined by recombination hot spots determined by data retrieved from ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2006-10_rel21_phaseI+II/hotspots/ that was converted to NCBI build 36 coordinates prior to analyses. The second tag-SNP set was created in the same manner using the HM3 reference sample. The third tag-SNP set was based on the FIN + HM3; CNP signals were first summarized separately in the two reference sample sets, then the distribution of the summarized signals was scaled with the scale()-function of R in both data sets, after which the HM3 and FIN SNP and CNP data were combined for testing for correlation between SNP genotype and CNP signals in order to identify the best tag-SNP for each CNP. Next, we analyzed the correlation between CNP signals and SNP genotypes in the imputed H2000 data; for each CNP, we calculated the correlation coefficient R and R2 between the CNP signal and SNP genotypes of the FIN tag-SNP, HM3 tag-SNP, and FIN + HM3 tag-SNP. We increased the number of SNPs in the analysis by including imputed SNP genotypes. The H2000 SNP data was imputed from the Illumina 610K resolution to HapMap3 SNP resolution with IMPUTE v2 (Howie et al. 2009) using the FIN + HM3 as the imputation reference panel. SNPs with an imputation R2 < 0.5 were excluded from all CNP tag-SNP analysis.
Association power simulation
We simulated the effect of the difference in R2 distributions on the power to detect association. The number of individuals in the simulations was 10,000 in both SNP and CNP simulations. For each linear-regression effect sizes, betas, from 0.05 to 0.30 we ran 40,000 iterations. In each of those iterations we (1) sampled a MAF–R2 pair jointly from the empirical distributions, (2) simulated the genotype/tagging SNP using the sampled MAF, (3) created a phenotype using the simulated genotype and beta, (4) changed the simulated genotype/tagging SNP so that the correlation between the simulated and the changed genotype/tagging SNP was smaller or equal to R2, and (4) fitted a new linear model with the changed genotype. In order to simulate SNP MAF distributions as appropriately as possible, MAFs were selected from the H2000 data set, which has the biggest number of observations available for SNP MAF calculations. For CNPs, a similar approach was used and the MAFs were obtained from the original HM3 data set, in which CNP genotyping included a high number of probes in the CNP regions, a larger number of samples in the CNP signal clustering compared with FIN, and CNP signals were successfully assigned to categorical copy number clusters.
Acknowledgments
This work was supported by the Wellcome Trust (WT089062/Z/09/Z, WT089061/Z/09/Z). L.P., S.R., and A.P. are supported by the Center of Excellence for Complex Disease Genetics of the Academy of Finland (grant nos. 213506 and 129680). L.P. is supported by the Biocentrum Helsinki Foundation and The Nordic Center of Excellence in Disease Genetics. V.S. is supported by the Academy of Finland (grant no. 129494), the Finnish Foundation for Cardiovascular Research, and the Sigrid Juselius Foundation. K.K. is supported by the Academy of Finland (grant no. 125973) and the Orion-Farmos Research Foundation. L.M. is supported by the EPSRC. V.A. is supported by the Finnish Cultural Foundation. We thank Peter Wagner for language revision of this manuscript.
Author contributions: Senior investigators S.R., L.P., A.P., M.D., and V.S. planned and managed the project. S.R. and K.K. led the data analysis. I.S., K.K., V.A., M.I., C.B., and L.M. prepared the data and performed the data analyses. I.S. and K.K. wrote the manuscript (with significant contributions from other authors).
Footnotes
[Supplemental material is available online at http://www.genome.org. The data are available at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega), under accession no. EGAS0000000030 and at ftp://ftp.fimm.fi/pub/FIN_HAPMAP3.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106534.110.
References
- Barnes C, Plagnol V, Fitzgerald T, Redon R, Marchini J, Clayton D, Hurles ME 2008. A robust statistical method for case-control association testing with copy number variation. Nat Genet 40: 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA 2008. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 40: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529 doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium 2003. The International HapMap Project. Nature 426: 789–796 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkula E, Rehnström K, Varilo T, Pietiläinen OP, Paunio T, Pedersen NL, deFaire U, Jarvelin MR, Saharinen J, Freimer N, et al. 2008. The genome-wide patterns of variation expose significant substructure in a founder population. Am J Hum Genet 83: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Abecasis GR 2006. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet S79: 2290 [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G 2009. Genotype imputation. Annu Rev Genomics Hum Genet 10: 387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCaroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, et al. 2006. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet 79: 275–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906–913 [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Pardis CS, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. 2006. Common deletion polymorphisms in the human genome. Nat Genet 38: 86–92 [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN 2008. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369 [DOI] [PubMed] [Google Scholar]
- Nevanlinna HR 1972. The Finnish population structure. A genetic and genealogical study. Hereditas 71: 195–236 [DOI] [PubMed] [Google Scholar]
- Orton NC, Innes AM, Chudley AE, Bech-Hansen NT 2008. Unique disease heritage of the Dutch-German Mennonite population. Am J Med Genet 146A: 1072–1087 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T 1999. Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8: 1913–1923 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: A toolset for whole genome association and population-based linkage analysis. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, et al. 2006. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 38: 556–560 [DOI] [PubMed] [Google Scholar]
- Varilo T, Laan M, Hovatta I, Wiebe V, Terwilliger JD, Peltonen L 2000. Linkage disequilibrium in isolated populations: Finland and a young sub-population of Kuusamo. Eur J Hum Genet 8: 604–612 [DOI] [PubMed] [Google Scholar]