Abstract
The human transcription factor TP53 is a pivotal roadblock against cancer. A key unresolved question is how the p53 protein selects its genomic binding sites in vivo out of a large pool of potential consensus sites. We hypothesized that chromatin may play a significant role in this site-selection process. To test this, we used a custom DNA microarray to measure p53 binding at approximately 2000 sites predicted to possess high-sequence specificity, and identified both strongly bound and weakly bound sites. When placed within a plasmid, weakly bound sites become p53 responsive and regain p53 binding when stably integrated into random genomic locations. Notably, strongly bound sites reside preferentially within genomic regions whose DNA sequence is predicted to encode relatively high intrinsic nucleosome occupancy. Using in vivo nucleosome occupancy measurements under conditions where p53 is inactive, we experimentally confirmed this prediction. Furthermore, upon p53 activation, nucleosomes are partially displaced from a relatively broad region surrounding the bound p53 sites, and this displacement is rapidly reversed upon inactivation of p53. Thus, in contrast to the general assumption that transcription-factor binding is preferred in sites that have low nucleosome occupancy prior to factor activation, we find that p53 binding occurs preferentially within a chromatin context of high intrinsic nucleosome occupancy.
The TP53 transcription factor and tumor-suppressor gene is one of the genes most frequently mutated in human cancer (Aylon and Oren 2007; Vousden and Prives 2009). Since TP53's tumor-suppressor function is largely based on its sequence-dependent transcription-factor activity, extensive efforts have been directed toward identifying the binding specificities of the p53 protein and the sites bound by it in vivo. Furthermore, much effort was dedicated to the development of tools that will enable prediction of DNA sites bound by p53 (Riley et al. 2008). Although binding to numerous sites has been experimentally validated (Cawley et al. 2004; Krieg et al. 2006; Wei et al. 2006; Kaneshiro et al. 2007; Smeenk et al. 2008), many sites with predicted high-sequence specificity are not bound by p53. The differential binding of these putative high-affinity sites is the subject of this study.
We set out to identify factors that serve as an additional layer of regulation on p53's DNA binding. More specifically, we hypothesized that the chromatin context of the site may be involved in p53 site selection, such that high-affinity sites may be accessible or nonaccessible to p53, depending on their chromatin organization. The fundamental subunit of chromatin is the nucleosome core particle, composed of 147 bp of DNA wrapped in nearly two superhelical turns around the histone octamer (Richmond and Davey 2003). Although it is able to wrap all genomic sequences, the histone octamer preferentially occupies specific DNA sequences in vivo, where the basis for this preferred occupancy depends in part on the DNA sequence (Segal and Widom 2009). Since the genomic positions of nucleosomes influence the ability of proteins to bind DNA, the intrinsic nucleosome DNA sequence preferences may thus be important for the binding of transcription factors (TFs) to their cognate sites. In yeast, functional binding sites for TFs exhibit low nucleosome occupancy in vivo (Lee et al. 2004; Sekinger et al. 2005; Yuan et al. 2005; Segal et al. 2006; Field et al. 2008), and sites for most TFs exhibit this low occupancy even when nucleosomes are reconstituted on purified genomic DNA in vitro in the absence of TFs (Kaplan et al. 2009). This encoding of low nucleosome occupancy has been suggested to facilitate the binding of TFs to their relevant cognate sites. We thus speculated that as in yeast, genomic regions surrounding p53-bound sites might possess low intrinsic (DNA-dependent) nucleosome occupancy. To test this idea, we measured p53 binding and, separately, nucleosome occupancy, at a large number of putative high-affinity sites using a custom-designed DNA array. Notably, we found that p53-bound sites reside preferentially within DNA regions that encode high nucleosome occupancy, and which are experimentally shown to be nucleosome-rich in vivo. Upon p53 activation, this high nucleosome occupancy is reduced in many of the bound sites, a reduction that is rapidly reversed when p53 activity subsides.
Results
Identification of strongly and weakly bound p53 sites
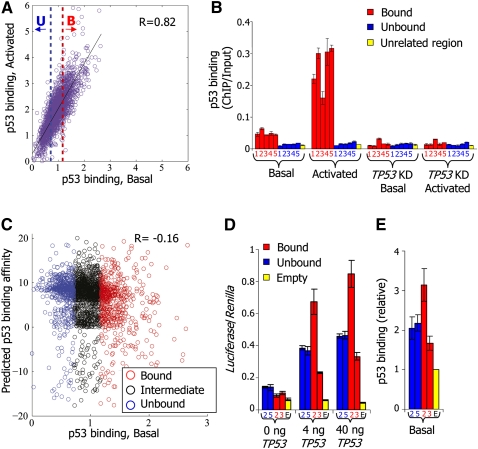
To study why p53 binds some high-affinity sites in vivo significantly more than others, we identified approximately 2000 putative high-affinity sites genome wide using previously reported p53-binding specificity predictions (Wei et al. 2006), and designed a custom tiling microarray at high resolution (10 bp) around these predicted sites and around another set of experimentally identified p53-bound regions (Wei et al. 2006; Kaneshiro et al. 2007; Riley et al. 2008). We then measured p53 binding at these sites by hybridizing the array with DNA obtained from p53 chromatin immunoprecipitation (ChIP) in nonstressed MCF7 cells expressing endogenous wild-type p53 (basal state), as well as in MCF7 cells in which p53 had been activated by exposure to the DNA damaging agent, Neocarzinostatin (NCS). In agreement with earlier studies (Espinosa and Emerson 2001; Kaeser and Iggo 2002; Shaked et al. 2008), we found a high correlation over sites between the binding strength of p53 under basal condition and upon activation, with higher levels of binding in the activated condition (Fig. 1A, R = 0.82). That p53-binding signals exhibit a continuous behavior and that they roughly linearly scale from the basal to the activated condition suggests that these sites are only partially bound, and that weaker binding reflects a higher proportion of unsaturated sites within a population of cells.
Figure 1.
p53 sequence specificity is not a sufficient predictor of p53 binding in vivo. (A) For each of the putative p53-binding sites around which we measured p53 binding with a high-resolution tiling array, shown is the log-ratio between p53 ChIP and input-sonicated genomic DNA, averaged across the probes that tile the 500-bp region centered around the site. For each site, binding is shown for nonstressed MCF7 cells expressing endogenous wild-type p53 (p53 binding, Basal, x-axis), as well as for MCF7 cells in which p53 had been activated by exposure to the DNA damaging agent Neocarzinostatin (p53 binding, Activated, y-axis). The blue-dotted line marks the arbitrary binding cutoff in the basal condition below which we define sites as unbound (U, lowest 25% of the sites). The red-dotted line marks the cutoff above which we define sites as bound (B, highest 25% of the sites). The correlation between binding in basal and activated conditions across all sites is indicated. (B) Small-scale validation of p53 binding. ChIP was performed with antibodies against p53, followed by quantification by qPCR of ChIP vs. input-sonicated genomic DNA for five bound and five unbound sites, among which are the well-described sites at the CDKN1A (bound2), GADD45A (bound4), and MDM2 (bound5) genes, as defined in A (identifier numbers and full details of the sites are provided in Supplemental Table 1), as well as for one unrelated control region. Shown are the average and standard deviation (calculated from duplicate qPCR reactions) of p53-binding values at each site, for nonstressed MCF7 cells (Basal), MCF7 cells in which p53 was activated by Neocarzinostatin (Activated), MCF7 cells with shRNA-mediated stable TP53 knockdown (TP53KD, Basal), and MCF7 cells with stable TP53 knockdown treated with Neocarzinostatin (TP53KD, Activated). (C) p53-binding affinity is a poor predictor of p53 binding. For each of the putative p53-binding sites at which we measured p53 binding, shown is its predicted affinity for p53 using a log-ratio of a model of p53-binding specificities (Wei et al. 2006) to genome background (y-axis) and its measured binding in nonstressed MCF7 cells (p53 binding, Basal, x-axis). The correlation between predicted binding affinity and measured binding in the basal condition across all sites is indicated. Unbound and Bound are as in A; Intermediate corresponds to the remaining 50% of sites, defined as possessing intermediate p53 binding. (D) When cloned in a plasmid in front of a luciferase reporter gene, bound and unbound sites are equally effective in conferring p53 responsiveness. H1299 cells were transfected separately with luciferase plasmids carrying either bound or unbound sites as indicated, or no site (empty), together with increasing amounts of a TP53 expression plasmid (x-axis). Shown are averages and standard deviations (three biological replicates) of Firefly luciferase values, normalized by cotransfected Renilla luciferase reporter values. Full site details are shown in Supplemental Table 1. (E) When randomly integrated into the human genome, bound and unbound sites are equally bound by p53. HCT116 cells were stably transfected separately with luciferase plasmids carrying either bound or unbound sites or no site (empty), as indicated. ChIP was performed with antibodies against p53, followed by quantification by qPCR of ChIP vs. input-sonicated genomic DNA. Values for each site were divided by the values of p53 binding obtained from the empty plasmid. Shown are the average and standard deviation (calculated from duplicate qPCR reactions) of relative p53-binding values at each site.
To identify features that discriminate bound sites from weakly bound sites, we arbitrarily defined the 25% of the sites with the highest ChIP with microarray hybridization (ChIP-chip) binding signals in the basal condition as “p53-bound” sites and the lowest 25% of the sites as “p53-unbound” (Fig. 1A). Using ChIP-qPCR, we tested five sites from the bound group, and indeed found that they exhibit significant binding under basal condition and increased binding upon p53 activation, whereas five sites selected from the unbound group did not display detectable p53 binding in either condition (Fig. 1B; Supplemental Table 1). As expected, all sites tested by ChIP-qPCR displayed no binding in MCF7 cells in which TP53 had been stably knocked down by shRNA (Fig. 1B; Supplemental Fig. 1). Equivalent results for these sites were obtained in HCT116 cells under basal conditions (Supplemental Fig. 2). We further compared our data with a previous sequencing-based ChIP-paired-end diTag (ChIP-PET) measurement for p53 binding (Wei et al. 2006), and found that over the approximately 300 sites that overlap between the two studies there is a general agreement between our binding measurements and their corresponding measured PET density (Supplemental Fig. 3), further supporting the validity and sensitivity of our measurements.
To test whether the affinity of p53 discriminates bound from unbound sites in our data, we compared the distribution of predicted affinities of the group of bound and unbound sites using a model of p53-binding specificities (Wei et al. 2006). We found that the score distributions of the bound and unbound groups were similar, and that there was no correlation between site score and binding levels (Fig. 1C). Equivalent conclusions were obtained using another model of p53-binding specificities (Perez et al. 2007), which was derived from in vitro SELEX measurements (data not shown). Notably, when cloned in a plasmid in front of a luciferase gene, sites of the unbound group were as effective as bound sites in conferring transcriptional responsiveness to p53, indicating that these sites can interact productively with p53 when removed from their native chromatin context (Fig. 1D). Moreover, sites of both groups displayed comparable p53 binding when stably integrated into the genome at random locations and subjected to p53 ChIP (Fig. 1E). Thus, our results suggest that the affinity of p53 to a site is not a sufficient predictor of p53-binding strength to that site in vivo, consistent with previous studies that reached similar conclusions for the TP53 family member TP63 (Yang et al. 2006) and the MYC (Chen et al. 2007) transcription factors (TFs).
p53 binds preferentially to regions with high predicted intrinsic nucleosome occupancy
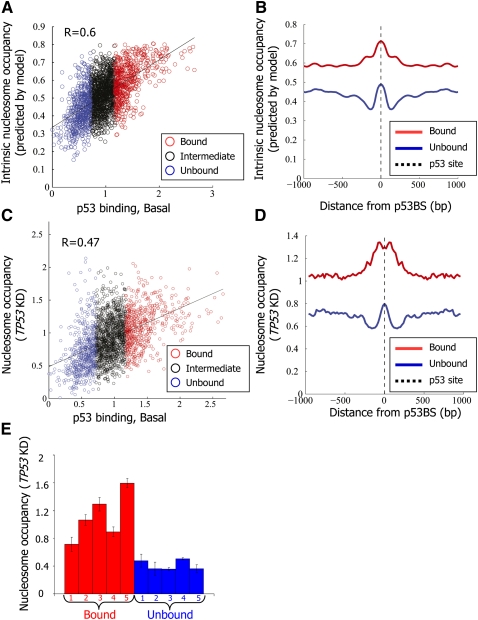
These findings suggested that information beyond the affinity of p53 to the site itself is critical for in vivo site choice by p53. We hypothesized that as in yeast, p53-bound sites may exhibit low intrinsic nucleosome occupancy, partly explaining how p53 selects its target sites in the genome. To test this hypothesis, we compared the intrinsic nucleosome occupancy around the p53-bound and unbound sites using a computational model of nucleosome sequence preferences that we previously derived from an in vitro nucleosome reconstitution map in yeast, in which nucleosome occupancy was governed only by the intrinsic sequence preferences of nucleosomes (Kaplan et al. 2009). Notably, we indeed found significant differences in predicted intrinsic nucleosome occupancy between the p53-bound and unbound sites. However, in contrast to yeast, we found that the bound sites have much higher predicted nucleosome occupancy than the unbound sites in a broad region surrounding the sites, and that there is generally a significant positive correlation between the predicted occupancy around an individual site and its measured level of p53 binding (Fig. 2A,B, R = 0.6). Consistent with the higher intrinsic nucleosome occupancy and the strong positive correlation between G/C content and intrinsic nucleosome occupancy (Lee et al. 2007; Peckham et al. 2007; Field et al. 2008; Kaplan et al. 2009; Tillo and Hughes 2009), genomic regions encompassing the group of p53-bound sites also exhibit much higher G/C content, on average, than unbound sites (Supplemental Fig. 4). These results suggest that high intrinsic nucleosome occupancy is predictive of p53-binding strength in vivo.
Figure 2.
p53 binds preferentially regions with high intrinsic nucleosome occupancy. (A) For each of the putative p53-binding sites around which p53 binding was measured, shown is the log-ratio between p53 ChIP in nonstressed MCF7 cells and input-sonicated genomic DNA in the 500 bp surrounding the p53BS (p53 binding, Basal, x-axis; the same as in Fig. 1A), and the intrinsic nucleosome occupancy predicted by a model of nucleosome sequence preferences (Kaplan et al. 2009) (y-axis). Model predictions are shown as the average nucleosome occupancy per base pair across the 2000-bp region centered on each site. Sites are colored blue, black, or red, according to their classification from Figure 1A into unbound, intermediate-binding, or bound sites, respectively. The correlation between the nucleosome model predictions and measured p53 binding in the basal condition is indicated. (B) Model-predicted nucleosome occupancy per base pair, averaged across all p53-bound sites (red) and p53-unbound sites (blue), and shown along the 2000-bp region centered on each site. (C,D) Same as A and B, respectively, except that the y-axis corresponds to experimental measurements of nucleosome occupancy using the same tiling array as that used for measuring p53 binding in MCF7 cells with shRNA-mediated stable TP53 knockdown (TP53KD). Nucleosome measurements are shown as log-ratio between the nucleosome sample and sonicated genomic DNA. (E) Small-scale validation of the nucleosome occupancy measurements from C. Mononucleosomes were prepared from MCF7 cells with shRNA-mediated stable TP53 knockdown (TP53KD) by limited micrococcal nuclease digestion. Mononucleosomal DNA was subjected to quantification by qPCR for the five bound and five unbound sites listed in Supplemental Table 1, and values were normalized for qPCR readings of input-sonicated genomic DNA. Shown are the average and standard deviation (calculated from duplicate qPCR reactions).
p53 binds preferentially regions with high nucleosome occupancy in vivo
As the model for nucleosome sequence preferences used above was derived from yeast data, we sought to experimentally validate its predicted association between high intrinsic nucleosome occupancy and in vivo p53 binding by measuring nucleosome occupancy in vivo prior to p53 activation. To this end, we isolated mononucleosomes from micrococcal nuclease-digested chromatin of MCF7 cells in which TP53 had been stably knocked down (TP53KD), and hybridized the mononucleosomal DNA to the above high-resolution custom array, in comparison to input-sonicated genomic DNA. We found, on average, a strong correlation between the measured nucleosome occupancy and the model predictions (R = 0.82; Supplemental Fig. 5). Indeed, the measured nucleosome occupancy was much higher at the p53-bound sites compared with the unbound sites, and more generally, was positively correlated with the level of p53 binding (Fig. 2C,D, R = 0.47). Analysis of a recent nucleosome map, determined for “resting” T cells using high-throughput sequencing (Schones et al. 2008) yielded similar results, wherein regions surrounding the MCF7 p53-bound sites had significantly more nucleosome reads than the genome average and those regions surrounding the unbound sites (Supplemental Fig. 6).
A more detailed positional correlation analysis implied that the observed positive correlation between p53-binding strength and G/C content, predicted intrinsic nucleosome occupancy, and in vivo measured nucleosome occupancy (in both the TP53 knocked-down cells and the “resting” T cells), is insensitive to the particular thresholds by which we defined the bound and unbound sites, and holds throughout the spectrum of p53-binding signals (Supplemental Figs. 7,8A). We found equivalent results when repeating the analysis with p53 binding defined according to our measurements in the activated condition (NCS treatment) rather than the basal condition (Supplemental Fig. 8B). Similarly, equivalent results were obtained when we restricted our analysis to only those sites that are predicted to have high affinity according to two independent models of p53 sequence specificities (Supplemental Fig. 8C; Wei et al. 2006; Perez et al. 2007), and also when we considered separately sites according to their distance from the nearest known transcription start site (TSS) (Supplemental Fig. 9). We note that the positional correlation analyses (Supplemental Figs. 7–9) suggest the existence of a weak trend of local nucleosome depletion directly over the binding site, compared with its surrounding region, which is associated with p53 binding; however, the primary (and much stronger) association with p53 binding that we observed is high nucleosome occupancy over the wider surrounding region.
As further validation, we quantified by qPCR the nucleosome occupancy around the center of each binding site for the aforementioned representative five p53-bound sites and five unbound sites, using mononucleosomal DNA obtained as above versus input-sonicated genomic DNA. Indeed, all five p53-bound sites displayed higher nucleosome occupancy than all five p53-unbound sites (Fig. 2E). A similar trend was also found in TP53-null H1299 cells (Supplemental Fig. 10). Furthermore, we compared the measured nucleosome occupancy over the CDKN1A and MDM2 binding sites to a previously reported nucleosome-depleted region at the BRCA1 promoter and another region within the same promoter reported to be highly nucleosome occupied (Ozsolak et al. 2007), and found that both CDKN1A and MDM2 p53-binding sites possess nucleosome levels comparable to the BRCA1 nucleosome-occupied region. We note, however, that repetitive measurements of nucleosome occupancy over the MDM2 site showed substantial variability, suggesting high responsiveness to mild changes in experimental conditions (Supplemental Fig. 11).
Together, and in agreement with the model predictions, these experimental measurements of nucleosome occupancy confirm that strongly in vivo-bound p53 sites typically reside within regions that have relatively high nucleosome occupancy in the absence of p53, whereas sites that are only weakly bound by activated p53 tend to be embedded within regions of low nucleosome occupancy.
p53 binding is also associated with proximity to genes and specific histone modifications
To test whether the p53-bound and unbound sites also differ by the composition of their nucleosome modification marks, we compared our data with available genome-wide measurements of various histone modifications and variants (Barski et al. 2007; Wang et al. 2008), although we note that those measurements were obtained in a different experimental system from ours, namely, resting T cells. Nevertheless, as discussed above (Supplemental Figs. 6–8), these cells also exhibit a general trend of high nucleosome occupancy at p53-bound sites, supporting the relevance of the histone modification data obtained in these cells for our study. We found an enrichment at p53-bound sites for specific sets of euchromatin marks that are characteristic of enhancer elements (Barski et al. 2007; Wang et al. 2008): H3K4me1,2,3, H3K9me1, H3K4ac, H3K9ac, and H2A.Z (Supplemental Fig. 12); some of these marks were previously reported to be associated with p53 binding (Gevry et al. 2007; Kaneshiro et al. 2007; Heintzman et al. 2009). In contrast, we found that regions encompassing unbound sites were enriched with silent heterochromatin marks (H3K9me2, H3K9me3). Some histone modifications considered to be representative of euchromatin (H3K14ac) or heterochromatin (H3K27me3) were not differentially associated with the two groups. As these data were obtained in different cell types from ours, we further directly measured the state of one euchromatic (H3K4me1) and one heterochromatic (H3K9me3) modification for our aforementioned representative five bound and five unbound sites (see Supplemental Table. 1) in MCF7 cells, in which TP53 had been stably knocked down (Supplemental Fig. 13). Indeed, with only one exception (“unbound4”), all of the sites behaved similarly to the data analysis above. We also found that an additional differential characteristic of the bound sites is that they tend to reside closer to known TSSs (Supplemental Fig. 14). Thus, binding sites preferred by p53 in vivo are further characterized by proximity to genes and by the presence of euchromatin-associated specific histone modifications and variants.
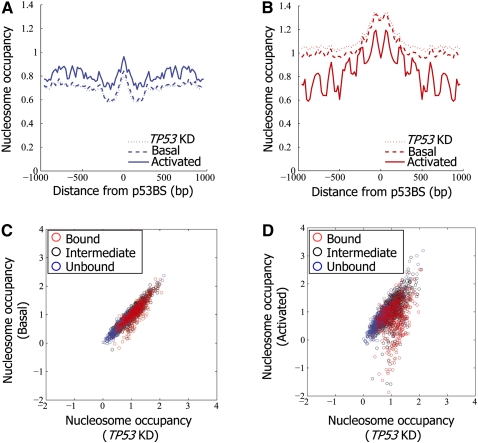
p53 binding leads to rapid and reversible reduction in regional nucleosome occupancy
Our finding that p53 binding is preferred in regions with high nucleosome occupancy prompted us to examine whether the nucleosome organization over the binding site is altered in response to p53 binding. We thus measured nucleosome occupancy using the custom array as above, except that mononucleosomes were prepared from MCF7 cells in basal condition and, separately, following p53 activation by NCS treatment. As expected, average nucleosome occupancy around sites that are not bound by p53, upon p53 activation, did not detectably change when p53 was activated (Fig. 3A,C,D). In contrast, following p53 activation, we found a marked reduction in average nucleosome occupancy over the bound sites (Fig. 3B–D). More sensitive analysis indicated that in both basal and activated conditions, the stronger the binding of p53, the higher the measured reduction in nucleosome occupancy; moreover, the reduction is in general much stronger upon p53 activation (Supplemental Figs. 7–9). On average, the reduction in nucleosome occupancy over the entire region surrounding the binding site does not imply that nucleosomes from the entire region are evicted in every locus. Instead, each locus showed a distinct pattern of nucleosome reorganization with an overall tendency to reduced occupancy, but not at consistent specific locations relative to the binding site (Supplemental Figs. 15,16).
Figure 3.
Nucleosomes are depleted from p53-bound sites upon activation of p53. (A) Nucleosome occupancy measurements per base pair, averaged across the p53-unbound sites (as defined in Fig. 1A), and shown along the 2000-bp region centered on each site. Results are shown for nonstressed MCF7 cells (Basal), MCF7 cells in which p53 was activated by Neocarzinostatin (Activated), and MCF7 cells with shRNA-mediated stable TP53 knockdown (TP53KD). Nucleosome measurements are shown as log-ratio between nucleosome sample and sonicated genomic DNA. (B) Same as in A, for the p53-bound sites. (C) Shown are nucleosome occupancy measurements (log-ratio between nucleosomal sample and sonicated genomic DNA) across the unbound (blue), intermediate-binding (black), and bound (red) p53 sites as defined in Figure 1A, averaged across all of the array probes that tile the 2000-bp region centered around the site. Results are shown for nonstressed MCF7 cells expressing endogenous wild-type p53 (Basal, y-axis) as well as cells with shRNA-mediated stable TP53 knockdown (TP53KD, x-axis). (D) Same as C, except that the y-axis corresponds to nucleosome measurements in MCF7 cells in which p53 was activated by Neocarzinostatin (Activated). Note the marked reduction of average nucleosome occupancy at many p53-bound sites (red group).
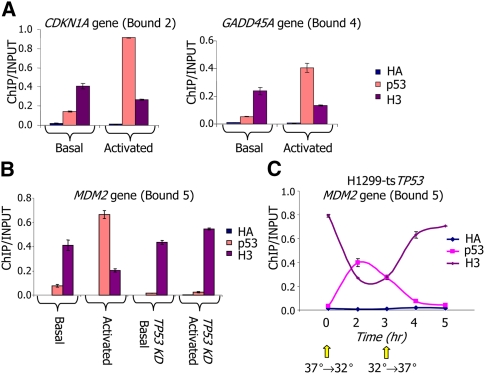
As further validation, using an alternative nucleosome readout we performed ChIP for histone H3 around three p53-bound sites. Indeed, for all three sites, the increase in p53 binding upon p53 activation was accompanied by a marked reduction in nucleosome occupancy over the sites (Fig. 4A,B). This effect was p53 dependent, as it was not observed in MCF7 cells depleted of TP53 (Fig. 4B). Similar results were found when the experiments were performed in another cell line, HCT116, and when the endogenous wild-type p53 was activated by 5-FU (Supplemental Fig. 17).
Figure 4.
Nucleosome depletion from p53-bound sites is transient and rapidly reversible. (A) ChIP was performed with antibodies against p53, histone H3, or HA epitope tag (HA) as control, followed by quantification by qPCR of ChIP and input-sonicated genomic DNA for two bound sites, as defined in Figure 1A. Shown are the average and standard deviation (calculated from duplicate qPCR reactions) in nonstressed MCF7 cells (basal), and in MCF7 cells in which p53 was activated by Neocarzinostatin. Full details of selected sites are shown in Supplemental Table 1. (B) Similar to A but for a different p53-bound site, and where in addition to the cellular conditions measured in A, also shown are measurements in MCF7 cells with shRNA-mediated stable TP53 knockdown (TP53KD, Basal), and MCF7 cells with stable TP53 knockdown and treatment with Neocarzinostatin (TP53KD, Activated). (C) Similar to B, but measured throughout a time-course in H1299-tsTP53 cells harboring a temperature-sensitive TP53 mutant. At time zero, cultures were shifted from 37°C to the permissive temperature (32°C), resulting in p53 activation. After 3 h from the initial shift, cultures were shifted back to 37°C to turn off p53 transcriptional activity.
Binding of p53 to the CDKN1A, MDM2, and GADD45A sites promotes transcriptional activation of the corresponding genes, raising the question of a link between nucleosome occupancy changes and transcriptional activity. Notably, the trend of reduced nucleosome occupancy was also observed for sites residing far away from any known TSS (Supplemental Fig. 9A). This is consistent with the notion that such reduction is not due to adjacent transcriptional activity. However, the possibility that transcription of noncoding RNAs occurs in the vicinity of those sites and affects their chromatin organization cannot be ruled out. To further investigate the relationship between nucleosome reduction and transcriptional changes, we subjected RNA from TP53-depleted MCF7 cells and from NCS-treated wild-type MCF7 cells to microarray expression analysis. Notably, this analysis revealed a significant, albeit not very strong, positive correlation between the relative change in transcript levels and the extent of nucleosome occupancy reduction over adjacent p53-bound sites (Supplemental Fig. 18). Thus, on average, sites displaying a stronger decrease in nucleosome occupancy are likely to reside preferentially in the vicinity of genes that are more strongly activated following p53 activation. However, there are also numerous sites where a strong reduction in nucleosome occupancy is not accompanied by transcriptional changes as measured in this analysis. It is presently unclear whether these sites modulate transcription of distant genes or of nearby noncoding RNAs or whether their reduced nucleosome occupancy is transcription independent. We note, however, that there are also sites in which a substantial increase in adjacent transcripts occurs without much change in nucleosome occupancy, suggesting that in those cases p53 binding may augment transcription via mechanisms that do not call for nucleosome reorganization.
Finally, we studied the dynamics of the nucleosome reorganization induced by p53 using H1299ts cells, which harbor a temperature-sensitive TP53 mutant that is active at 32°C but inactive at 37°C. As with the activation of p53 by NCS in MCF7 cells, inducing p53 activity in H1299ts cells by shifting from 37°C to 32°C resulted in increased p53 binding and rapid nucleosome displacement from the tandem p53-binding sites in the MDM2 gene (Fig. 4C). Notably, within 1 h of inactivating p53 by transferring the cells back to 37°C, we observed reduced p53 binding and nearly full restoration of nucleosome occupancy around the p53 site, attesting to the highly dynamic nature of the process. Taken together, our results demonstrate that p53 activation leads to a selective reduction in nucleosome occupancy in the regions spanning many p53-bound sites.
Discussion
To explain why some high-affinity sites are bound much stronger by p53 upon p53 activation while others are only weakly bound, we studied the determinants of p53 binding in vivo through computational and experimental analyses of approximately 2000 known and predicted high-affinity sites genome-wide. In yeast, most regulatory regions, including binding sites, core promoters, and origins of replication, exhibit low intrinsic nucleosome occupancy (Field et al. 2008; Kaplan et al. 2009). Since access to nucleosome-associated DNA is relatively occluded for most DNA-binding proteins, this low intrinsic occupancy likely enhances the accessibility of these sites, providing a partial explanation for how yeast TFs find their targets. In principle, a similar mechanism may have explained p53 site selection in humans. However, unexpectedly, using both a model of nucleosome sequence preferences and experimental measurements of nucleosome occupancy in the absence of p53, we found that p53 binds preferentially to sites that have high nucleosome occupancy, over themselves and over their surrounding regions. Moreover, there is a marked reduction in nucleosome occupancy over p53-bound sites and their surrounding regions upon p53 activation, suggesting a more complex mechanism than a simple competition between p53 and nucleosome occupancy over the binding site itself. Furthermore, the rapid repositioning of nucleosomes following p53 inactivation suggests that these changes are reversible and do not have any long-term effects on the bound region.
Our findings thus point to clear differences in the utilization of nucleosome positioning signals between yeast and humans, where the yeast genome encodes low intrinsic occupancy around most bound TF sites, while, at least for p53, the human genome encodes high intrinsic occupancy over bound sites. Since many regulatory regions in the human genome are known to be associated with high G/C content, one may predict that regulatory regions in human DNA would tend, in general, to have relatively high intrinsic nucleosome occupancy (Tillo et al. 2010). Our experimental data for p53 support this prediction.
Our work raises the question of whether nucleosomes interfere with or assist p53 binding. One possibility is that p53 preferentially binds nucleosome-associated DNA. This model is supported by earlier work on one p53 site in the CDKN1A gene, which showed that p53 binds with higher affinity to chromatin than to naked DNA (Espinosa and Emerson 2001). Alternatively, a nucleosome occupying the binding site may compete with p53 binding due to occlusion by steric hindrance. In that case, we would have expected the observed nucleosome displacement from bound sites upon p53 activation. This model would be more consistent with most studies in both yeast (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Field et al. 2008; Kaplan et al. 2009) and humans (Ozsolak et al. 2007; Fu et al. 2008; Cuddapah et al. 2009; Zheng et al. 2009), which reported nucleosome depletion from regulatory regions including binding sites and core promoters, upon protein binding. However, this same model of competition between p53 and nucleosomes cannot easily explain the stronger p53 binding that we observed at sites that have high intrinsic nucleosome occupancy.
Since p53, like many human TFs, acts in a context-specific manner, one may propose that high intrinsic nucleosome occupancy over functional p53-binding sites may in fact be advantageous, as it may provide a mechanism by which access to sites could be restricted in most conditions and cell types. However, this proposal does not explain why high-affinity p53 sites that are embedded in regions with low intrinsic nucleosome occupancy are generally only weakly bound upon p53 activation. One possible explanation may be related to the observed differential association of strongly and weakly bound sites, respectively, with specific euchromatic and heterochromatic histone variants and modifications. Another possible explanation may be differential clustering of binding sites for p53, as well as for additional TFs, which may lead to different binding strengths through cooperative interactions.
In sum, the striking differences in intrinsic nucleosome occupancy that we found between strongly and weakly bound p53 sites, and the dynamic reorganization of nucleosomes at bound sites upon p53 activation, strongly suggest that in vivo site selection by p53 depends on modulation of the chromatin context surrounding the binding site. Moreover, and contrary to expectation, our results also suggest that high intrinsic nucleosome occupancy is a good predictor of p53 binding in vivo.
Methods
Cell culture and NCS treatment
Cells were grown as previously described (Raver-Shapira et al. 2007). MCF7EcoR and MCF7sip53 were generously provided by R. Agami (Brummelkamp et al. 2002). MCF7EcoR cells are referred to in the main text as MCF7 cells expressing endogenous wild-type p53. For p53 activation, MCF7EcoR cells were treated with Neocarzinostatin (NCS, 200 ng/mL, 4 h).
Chromatin immunoprecipitation and microarray hybridization
For p53 ChIP, 2.5 × 106 MCF7EcoR were plated in 15-cm plates a night before. Cells were either nontreated or treated with NCS. The next day the experiment was performed as previously described (Shema et al. 2008) using antibodies against p53 (CM-1). Sonication was performed in a Bioruptor sonication device (Diagenode) using 15 rounds of 30-sec sonication and 1-min off-sonication. A total of 1 μg of immunoprecipitated DNA or input DNA was labeled and hybridized to NimbleGen custom arrays (performed by NimbleGen Systems). This was done in two biological replicates. ChIP-qPCR was performed with antibodies against human p53 (CM-1), mouse p53 (CM-5) (in H1299ts), histone H3 (Abcam1791), and HA epitope (sc-805); immunoprecipitated and input DNA served as qPCR templates as described (Raver-Shapira et al. 2007).
Mononucleosome preparation
To prepare mononucleosomes, 2.5 × 106 MCF7sip53 and MCF7EcoR were plated in a 15-cm plate a night before. The next day, MCF7EcoR were either nontreated or treated with NCS. Nuclei were isolated by incubation in lysis buffer (10 mM Tris at pH7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM Spermine, 0.5 mM Spermidine) for 10 min on ice. Nuclei were washed once (10 mM Tris at pH7.4, 15 mM NaCl, 60 mM KCl, 0.15 mM Spermine, 0.5 mM Spermidine) and resuspended in 500 μL of MNase buffer (Wash buffer + 1 mM CaCl2) followed by MNase digestion (1 U/12 million cells, 10 min at 37°C; MNase was obtained from Sigma). Reaction was stopped by addition of EDTA to 0.01 M. SDS and NaCl were added to a final concentration of 2% and 0.2 M, respectively. RNase (0.1 mg) was added for 1 h at 37°C. DNA was extracted using the Qiagen PCR purification kit, separated on a 1.5% agarose gel, and fragments of ∼150 bp were excised and extracted using the Qiagen Gel extraction kit. In parallel, input DNA was isolated with a Qiagen DNeasy Blood and Tissue Kit and sonicated in TE to ∼500-bp fragments. Sonication was performed as above. A total of 1 μg of nucleosomal DNA and input DNA were labeled and hybridized to NimbleGen custom arrays as above. This was done in two biological replicates. For qPCR, mononucleosomal DNA and sonicated input DNA served as templates.
Luciferase plasmid construction and Luciferase assays
Luciferase reporter plasmids were produced by inserting annealed oligonucleotides corresponding to individual p53-binding sites (20 bp) with flanking tags of SacI/XhoI into pGL3 promoter digested with SacI/XhoI. Luciferase assays were performed in H1299 cells. A total of 40,000 cells were plated a night before in 12-well plates. The next day cells were transfected using the JetPEI transfection reagent (according to manufacturer's instructions). On the following day, luciferase assay was performed as previously described (Raver-Shapira et al. 2007).
Stable transfection
To obtain pools of stable transfectants, 1.3 × 106 HCT116 cells were plated in a 10-cm plate, The next day cells were transfected with the different luciferase plasmids, together with a plasmid conferring puromycin resistance, using the JetPEI reagent according to the manufacturer's instructions. Cells were grown under puromycin selection for an additional 10 d and then subjected to p53 ChIP, as described above.
RNA preparation and expression microarray analysis
RNA purification and qRT–PCR quantification were as described (Shema et al. 2008). For oligonucleotide microarray hybridization, RNA was extracted using a Qiagen RNeasy Mini Kit according to the manufacturer's protocol. Ten micrograms of RNA were labeled and hybridized to an Affymetrix Human U133 plus 2.0 oligonucleotide array. Expression value for each gene was calculated using Affymetrix Microarray software 5.0. Average intensity difference values were normalized across the sample set. Microarray expression data presented in this study is part of a larger experiment (E Lidor Nili and M Oren, unpubl.) and has been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession no. GSE22783).
Tiling microarray design
Predicted p53 sequence preferences were taken from and computed according to Wei et al. (2006) as the log-ratio between the position-specific distribution according to the reported specificities and a background zero-order Markov model based on single-nucleotide frequencies in the human genome. Predictions were taken as the maximum score when allowing zero or one-nucleotide spacers between the two p53 half-sites. Sites were selected for the array design from multiple sources: high predicted scores using the computation above; exact matches to sites previously reported (Wei et al. 2006; Kaneshiro et al. 2007; Riley et al. 2008), and bound sites according to a previous ChIP-chip experiment for p53 binding that we performed with a lower resolution custom array design (data not shown). Sites were only selected if their surrounding region could be mapped in high resolution when repeated regions were removed by RepeatMasker. Together, 2204 sites were analyzed, of which 1299 sites have predicted binding log-ratio scores above 7, 730 sites have scores in the range of from 0 to 7, and 175 sites have scores lower than zero. Coordinates are reported relative to build hg18 of the human genome. The custom tiling microarray was manufactured by NimbleGen Systems and included a total of 385,000 probes of 50-bp length, confirmed to meet the manufacturer's specifications (regarding melting temperature, self complementarity, etc.) and to not contain any contiguous 40 bp with a perfect match to other genomic locations (according to build hg17 of the human genome). The data has been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession no. GSE22783).
Microarray analysis of nucleosome measurements
Log2 ratios of sample to input DNA were standardized in each array across all probes, and replicates were averaged following quantile normalization. The nucleosome arrays were further scaled based on analysis of all binding sites with respect to the average nucleosome occupancy in the 2000-bp centered around each site (as in Fig. 3C) to minimize differences between samples with consecutively increasing p53 concentration. The nucleosome arrays with stable knockdown of TP53 were shifted to have a minimum value of zero, the nucleosome arrays measured in nonstressed MCF7 cells (basal state) were scaled and translated to minimize the mean square difference relative to the normalized TP53 knockdown arrays, and the nucleosome arrays that were measured after p53 activation were similarly transformed to minimize the differences with the normalized p53-basal arrays. For all profile analyses (predicted and measured nucleosome occupancies and G/C content) the value in each point was taken as the centered 30-bp window average over the corresponding data, with a resolution of 20 bp between points.
Microarray analysis of p53 ChIP-chip array
Log2 ratios of sample to input DNA were standardized in each array across all probes, and replicates were averaged following quantile normalization. The p53-binding values were taken as the average of all probes in the 500 bp centered on each binding site. The p53 arrays measured in nonstressed MCF7 cells (basal state), and following p53 activation (activated state) were calibrated according to the qPCR measurements of the five p53-bound and five p53-unbound sites (Fig. 1B) by scaling and translating the probe values of the p53-activated array to match the average fold enrichment for these two groups relative to the p53-basal array.
Acknowledgments
We thank Sylvie Wilder for excellent technical assistance; Yael Aylon, Reut Shalgi, and Efrat Shema for helpful discussions and inspiring ideas; and Shirley Horn-Saban for Affymetrix expression array hybridization. This work was supported by grant R37 CA40099 from the NCI and the Yad Abraham Center for Cancer Diagnosis and Therapy (M.O.), grant U54 CA143869-01 from the NCI PSOC (J.W and E.S.), grant 5R01CA119176-03 from the NIH (E.S.), and by an ERC grant (E.S.). M.O. is the incumbent of the Andre Lwoff Chair in Molecular Biology. E.S. is the incumbent of the Soretta and Henry Shapiro career development chair.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE22783.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103945.109.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M 2007. Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Chen Y, Blackwell TW, Chen J, Gao J, Lee AW, States DJ 2007. Integration of genome and chromatin structure with gene expression profiles to predict c-MYC recognition site binding and function. PLoS Comput Biol 3: e63 doi: 10.1371/journal.pcbi.0030063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 8: 57–69 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z 2008. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138 doi: 10.1371/journal.pgen.1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev 21: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Iggo RD 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci 99: 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneshiro K, Tsutsumi S, Tsuji S, Shirahige K, Aburatani H 2007. An integrated map of p53-binding sites and histone modification in the human ENCODE regions. Genomics 89: 178–188 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AJ, Hammond EM, Giaccia AJ 2006. Functional analysis of p53 binding under differential stresses. Mol Cell Biol 26: 7030–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE 2007. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25: 244–248 [DOI] [PubMed] [Google Scholar]
- Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z 2007. Nucleosome positioning signals in genomic DNA. Genome Res 17: 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CA, Ott J, Mays DJ, Pietenpol JA 2007. p63 consensus DNA-binding site: Identification, analysis and application into a p63MH algorithm. Oncogene 26: 7363–7370 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA 2003. The structure of DNA in the nucleosome core. Nature 423: 145–150 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. What controls nucleosome positions? Trends Genet 25: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I 2008. Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res 68: 9671–9677 [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M 2008. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res 36: 3639–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442 doi: 10.1186/1471-2105-10-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE 5: e9129 doi: 10.1371/journal.pone.0009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593–602 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zheng D, Zhao K, Mehler MF 2009. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol 10: r9 doi: 10.1186/gb-2009-10-1-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]