Abstract
The histone variant H2A.Z has been implicated in the regulation of gene expression, and in plants antagonizes DNA methylation. Here, we ask whether a similar relationship exists in mammals, using a mouse B-cell lymphoma model, where chromatin states can be monitored during tumorigenesis. Using native chromatin immunoprecipitation with microarray hybridization (ChIP-chip), we found a progressive depletion of H2A.Z around transcriptional start sites (TSSs) during MYC-induced transformation of pre-B cells and, subsequently, during lymphomagenesis. In addition, we found that H2A.Z and DNA methylation are generally anticorrelated around TSSs in both wild-type and MYC-transformed cells, as expected for the opposite effects of these chromatin features on promoter competence. Depletion of H2A.Z over TSSs both in cells that are induced to proliferate and in cells that are developing into a tumor suggests that progressive loss of H2A.Z during tumorigenesis results from the advancing disease state. These changes were accompanied by increases in chromatin salt solubility. Surprisingly, ∼30% of all genes showed a redistribution of H2A.Z from around TSSs to bodies of active genes during the transition from MYC-transformed to tumor cells, with DNA methylation lost from gene bodies where H2A.Z levels increased. No such redistributions were observed during MYC-induced transformation of wild-type pre-B cells. The documented role of H2A.Z in regulating transcription suggests that 30% of genes have the potential to be aberrantly expressed during tumorigenesis. Our results imply that antagonism between H2A.Z deposition and DNA methylation is a conserved feature of eukaryotic genes, and that transcription-coupled H2A.Z changes may play a role in cancer initiation and progression.
Histones are small basic proteins that package the genome and play important regulatory roles by modulating processes that require access to DNA (Wolffe 1992). Histone octamers wrap DNA to form nucleosomes, and both the modification of histones and the incorporation of nonallelic histone variants into nucleosomes have been shown to correlate with different chromatin states (Campos and Reinberg 2009). One of these histone variants, H2A.Z, is a universal member of the H2A family, and is required for viability in metazoans (Zlatanova and Thakar 2008). H2A.Z levels have been found to correlate with both active and repressed states of gene expression. For example, H2A.Z is enriched at active gene promoters, as well as at enhancer and insulator elements in terminally differentiated cells from chicken, mouse, and human, suggesting a positive role in regulating gene expression (Bruce et al. 2005; Thambirajah et al. 2006; Barski et al. 2007). Indeed, activation of a gene often includes promoter chromatin remodeling and incorporation of H2A.Z into nucleosomes surrounding the transcriptional start site (TSS) (Gevry et al. 2009; Hardy et al. 2009). H2A.Z is also localized to Polycomb protein binding sites in murine embryonic stem (ES) cells, suggesting a role in potentiating gene expression (Creyghton et al. 2008). In addition, loss of H2A.Z in Drosophila suppresses heterochromatin formation, suggesting a role in gene repression (Swaminathan et al. 2005).
In contrast to H2A.Z localization, DNA methylation is generally absent from the promoters of active genes, and methylation of a promoter can cause gene silencing (Baylin and Ohm 2006). One mechanism of gene silencing via DNA methylation is thought to be the recruitment of methyl binding proteins (Jones et al. 1998; Nan et al. 1998). These proteins may repress transcription, either by inhibiting the binding of RNA polymerase or associated factors at the transcriptional start site, or by recruiting chromatin modifiers, such as histone deactylases. Alternatively, gene silencing by DNA methylation might result from exclusion of chromatin factors that facilitate gene expression, and H2A.Z is one such candidate.
Recent work in the model plant, Arabidopsis thaliana, demonstrated an active antagonism between DNA methylation and deposition of H2A.Z (Zilberman et al. 2008). In that study, patterns of H2A.Z and DNA methylation were found to be quantitatively anticorrelated both at genes and at transposable elements. In DNA methyltransferase mutant plants, losses and gains of DNA methylation resulted in opposite changes in H2A.Z occupancy. Moreover, mutating the Swr1 nucleosome remodeler homolog, which replaces H2A with H2A.Z, led to hypermethylation of gene bodies. Very recently, H2A.Z occupancy and DNA methylation were shown to be strongly anti-correlated in the puffer fish, T. nigroviridis (Zemach et al. 2010). We wondered whether a similar reciprocal relationship exists between H2A.Z and DNA methylation in mammals, especially during cancer progression, where transcriptional programs have become misregulated. Cancer cells have been shown to undergo global hypomethylation, particularly in pericentric heterochromatin, which can contribute to genome instability associated with oncogenesis (Feinberg et al. 1988; Howard et al. 2007; Zilberman et al. 2008). Also, aberrant hypermethylation in the promoters of tumor suppressor genes has been observed in many cancers, and some of these changes occur early in transformation to the malignant state (Fraga et al. 2004). If these changes in DNA methylation are mirrored by changes in H2A.Z occupancy in cancer, this might suggest that antagonism between H2A.Z and DNA methylation contributes to aberrant silencing of promoters, including those of tumor suppressor genes, during oncogenic transformation.
To explore the possibility of antagonism between H2A.Z and DNA methylation in mammals, we used a classic mouse B-cell lymphoma system to ask whether changes in H2A.Z occupancy and DNA methylation levels are coordinated during progression from wild-type B cells to B-cell lymphomas. Indeed, we found that H2A.Z and DNA methylation levels are generally anticorrelated in wild-type mouse pre-B cells. Surprisingly, tumors showed elevated levels of H2A.Z in bodies of active genes. These gains in H2A.Z occupancy were seen with increasing transcription levels and corresponded to reductions in DNA methylation in gene bodies.
Results
H2AZ occupancy decreases around transcriptional start sites during oncogenesis
To ascertain whether H2A.Z occupancy changes during tumorigenesis, we used a mouse model system for B-cell lymphoma (Adams et al. 1985). These mice contain an IgH-MYC fusion transgene (Eμ-MYC), which mimics classical translocations between IgH cis-acting regulatory sequences and the MYC oncogene seen in human Burkitt's lymphomas. The IgH-MYC transgene results in overexpression of MYC in pre-B cells, which induces uncontrolled proliferation of these cells. This polyclonal population represents an intermediate step in the progression to lymphoma. A small subset of these hyperproliferative cells accumulates sufficient additional mutations to develop into independent monoclonal large B-cell lymphomas. By analyzing chromatin states in wild-type, Eμ-MYC pre-B, and lymphoma cells, we can identify changes that occur during tumorigenesis.
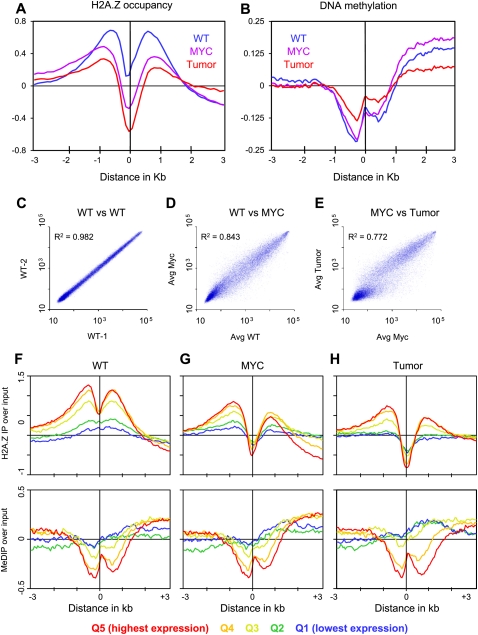
To determine H2A.Z occupancy, chromatin immunoprecipitations (ChIPs) from wild-type pre-B, Eμ-MYC pre-B, and Eμ-MYC induced lymphoma cells were used to prepare labeled DNA and were hybridized together with alternatively labeled input DNA to NimbleGen high-density mouse promoter arrays (Supplemental Fig. S1). We first asked how H2A.Z is distributed relative to TSSs genome-wide. To do this, we lined up genes at their annotated 5′ ends and then averaged H2A.Z occupancy in 50-bp intervals out to 3 kb upstream and 3 kb downstream. In agreement with other studies comparing ChIP with input, we found average H2A.Z levels to be high on either side of the TSS, with a relative depletion over TSSs (Fig. 1A; Supplemental Fig. S2; Raisner et al. 2005; Barski et al. 2007). When we compared the average H2A.Z occupancy around the TSS from each of the three cell types, we observed a substantial decrease at and around TSSs in both cycling Eμ-MYC pre-B and lymphoma cells as compared with wild-type pre-B cell controls. Interestingly, Eμ-MYC cells displayed an intermediate level of loss between wild-type pre-B and lymphoma cells, suggesting progressive loss of H2A.Z around TSSs with cellular transformation.
Figure 1.
H2A.Z and DNA methylation occupancy around TSSs. Genes were aligned at the TSS and the normalized probe signals were averaged in 50-bp bins for H2A.Z occupancy (A) and DNA methylation (B). (C–E) Comparisons of microarray expression data for each cell type. (F–H) Genes are aligned at the TSS and divided into quintiles (Q1–Q5) by decreasing expression level. The ChIP with microarray hybridization (ChIP-chip) data were normalized by computing standard deviates for each probe.
We also examined the relationship between H2A.Z occupancy and gene expression. We measured gene expression levels on microarrays and observed that changes in cell state are accompanied by large-scale differences in expression level (Fig. 1C–E). We then separated genes into five classes based on expression levels and displayed H2A.Z levels around TSSs for each quintile averaged over 50-bp intervals (Fig. 1F–H, top panels). This revealed that, for all three tissues, H2A.Z occupancy flanking TSSs decreases with decreasing transcription (Fig. 1F–H; Supplemental Fig. S3).
The 5′ ends of most mammalian genes are embedded in CpG islands, which are conspicuous in their lack of DNA methylation despite the fact that these islands are heavily enriched in DNA methylation substrates (Bird 1980). We examined H2A.Z occupancy at annotated CpG islands, and found that levels of H2A.Z showed depletion in MYC-transformed cells relative to wild-type and further depletion in lymphomas, both within the CpG islands and on CpG-island shores, defined as 2 kb on either side of the island boundaries (Supplemental Fig. S4A; Doi et al. 2009).
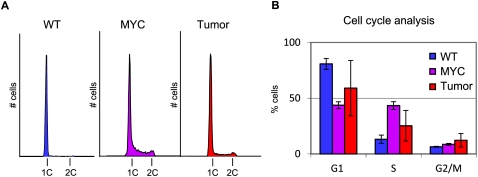
H2A.Z is deposited independently of replication. Consequently, an increase in the percentage of Eμ-MYC pre-B and lymphoma cells in S phase could potentially account for the reduced levels of H2A.Z occupancy we observed. To test this possibility, we asked whether the loss of H2A.Z-containing nucleosomes in the Eμ-MYC pre-B and lymphoma cells is associated with changes in cell cycle distribution. For this experiment, we stained cells with DAPI and then quantified the number of cells based on DNA content by fluorescence-activated cell sorting. We found that on average, 13% of the wild-type pre-B cells were in S phase, compared with 43% of the Eμ-MYC pre-B cells and 25% of the lymphoma cells (Fig. 2). Because Eμ-MYC pre-B displayed increased cell cycle rates as compared with wild-type pre-B cells, the decrease in H2A.Z that we observed could be either an active loss or a passive dilution. However, H2A.Z levels continued to decline over the TSS in the tumor population, despite having a smaller percentage of cells in S phase as compared with the Eμ-MYC cells. Therefore the observed loss of H2A.Z-containing nucleosomes around transcriptional start sites is unlikely to result from passive dilution, but rather appears to be an active feature of lymphomagenesis.
Figure 2.
Cell cycle analysis of DNA content during cell state changes. (A) Examples of FACS plots. (B) Five samples of each cell type were analyzed and averaged. DAPI staining was used for cell sorting.
Redistribution of H2A.Z over active gene bodies during B-cell lymphomagenesis
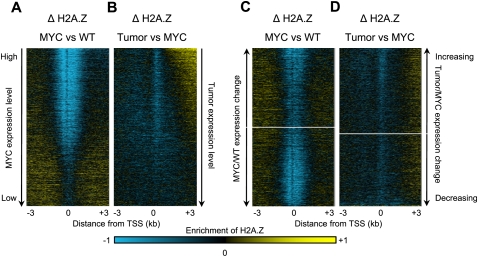
To ask whether cell state changes affect H2A.Z occupancy, we displayed differences in H2A.Z occupancy between cell types as heat maps ordered by decreasing gene expression (Fig. 3A,B). Comparisons between wild-type and Eμ-MYC pre-B cells showed a substantial loss of H2A.Z around the TSSs of active genes (Fig. 3A). As transcription levels decreased, the loss of H2A.Z over TSSs became less pronounced, whereas a gain of H2A.Z was observed over promoter regions and gene bodies of weakly expressed genes. In the transition from polyclonal Eμ-MYC pre-B cells to lymphoma, we observed a continued depletion of H2A.Z over the TSS. Strikingly, we found that, relative to Eμ-MYC pre-B cells, lymphoma cells also showed gains of H2A.Z downstream of TSSs of highly transcribed genes. This H2A.Z gain progressively decreased with decreasing expression (Fig. 3B). These same genes also showed the most dramatic loss of H2A.Z nucleosomes at the TSS, suggesting redistribution from the TSS to active gene bodies in the more advanced disease state. In contrast to the observation in lymphoma cells, we did not see a gain of H2A.Z over highly expressed gene bodies in the transition from wild-type pre-B cells to Eμ-MYC pre-B cells (Fig. 3A). Therefore, redistribution of H2A.Z to active gene bodies is a feature specific to lymphoma cells and is not a result of increased proliferation.
Figure 3.
Redistribution of H2A.Z from TSSs to gene bodies. (A) Changes in H2A.Z occupancy from wild-type (WT) to MYC-induced pre-B cells. (B) Changes in H2A.Z occupancy from MYC-induced pre-B to lymphoma cells. Heat maps were ordered by decreasing gene expression levels in the MYC (A) or lymphoma (B) cells. (C,D) Changes in H2A.Z occupancy from WT to MYC pre-B cells (C) and from MYC to lymphoma (D). Genes were ordered by the change in expression between cell types. Standard deviate normalized probe signals are represented as heat maps: (yellow) an enrichment of ChIP over input signal for H2A.Z, (blue) depletion. The genes with the largest gains in expression in the tumor are at the top, and the genes with the largest drops in expression level are at the bottom.
Because the lymphoma-specific gain of H2A.Z was seen in the bodies of expressed genes, and decreased with decreasing expression levels, we reasoned that redistribution of H2A.Z might be coupled to transcription. One possibility is that changes in gene expression during oncogenesis drive the redistribution of H2A.Z. To test this, we sorted the genes by expression differences between Eμ-MYC pre-B cells and lymphoma cells (Fig. 3C,D; Supplemental Fig. S5), rather than by absolute expression levels. When sorted in this manner, genes featuring a redistribution of H2A.Z to the gene body in lymphoma cells showed no trend over gene bodies, regardless of the method used to compute expression changes. This indicates that, although the gain in H2A.Z occupancy correlates with expression, the redistribution seen over active genes is not specific for genes that gain expression in lymphoma cells.
Chromatin solubility increases during MYC-induced oncogenesis
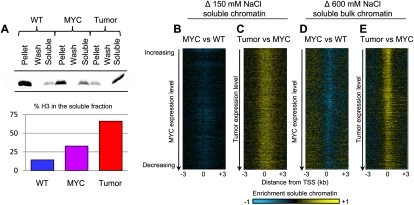
A potential explanation for the redistribution of H2A.Z over the bodies of expressed genes is that lymphoma cell chromatin is more dynamic over active genes (Henikoff et al. 2009). Consequently, if the lymphoma cells have a more dynamic chromatin configuration over active gene bodies, then the gain in H2A.Z occupancy could reflect in part a greater portion of active chromatin extracted from these cells. To test this, we digested chromatin in nuclei with MNase and washed with 150 mM NaCl to extract the low-salt soluble nucleosomes representing classical active chromatin. We then used quantitative Western blot analysis to measure H3 protein levels in the active chromatin fraction as a percentage of the total H3 (Fig. 4A). Intriguingly, we found that the percentage of H3 in this fraction is increased in both the Eμ-MYC pre-B and the lymphoma cells relative to wild-type cells. This suggests that a profound global change in chromatin solubility occurs upon overexpression of MYC, which is consistent with the global condensation that occurs with loss of N-MYC from neural progenitor cells (Knoepfler et al. 2006). In addition, because the amount of soluble chromatin recovered from lymphoma cells increased relative to that from Eμ-MYC pre-B cells, increases in chromatin solubility are not simply consequences of MYC-driven cellular proliferation.
Figure 4.
Chromatin solubility increases during oncogenesis. (A) Western blot of histones from chromatin fractions probed with an anti-H3 antibody. The fraction of H3 in the soluble input material is represented as a percentage of total histone H3 for each cell type (average of two samples for each cell type). (B,C) The ratio of 150 mM salt-soluble input chromatin between cell types displayed as heat maps. We obtained similar results by profiling individual 150 mM salt fractions versus MNased nuclei (Supplemental Fig. S6). (D,E) Changes in 600 mM salt-soluble chromatin between cell types displayed as heat maps: (yellow) an enrichment of soluble chromatin, (blue) depletion of soluble chromatin. Heat maps are ordered by decreasing gene expression levels in the MYC (B,D) or tumor (C,E) cells.
We next tested whether an increase in chromatin solubility could account for the pattern of H2A.Z gain over gene bodies seen in the lymphomas. To do this, we directly compared the 150 mM salt-soluble fractions, which were used as input for profiling H2A.Z from the Eμ-MYC pre-B and lymphoma cells. We aligned all the genes at the TSS and constructed heat maps with the genes arranged by decreasing expression level (Fig. 4B,C). We observed that at least part of the additional chromatin solubilized in the lymphoma cells corresponds to the region surrounding the TSS (Fig. 4C), which is in accordance with the dynamic histone turnover near promoters observed in other studies (Henikoff 2008). However, in contrast to the pattern seen for H2A.Z, we did not observe a gain in total nucleosomes over gene bodies in a transcription-dependent manner (Fig. 4C). As a control, we tested for total nucleosome occupancy by comparing bulk chromatin extracted in high salt (600 mM) between the cell types. We found the changes in total nucleosome occupancy and solubility were very similar to those seen in 150 mM salt (Fig. 4D,E; Supplemental Fig. S7). Thus, although chromatin from lymphoma cells is more soluble than chromatin from Eμ-MYC pre-B cells, this difference does not seem to be responsible for the gain of H2A.Z over active gene bodies seen in lymphoma cells. We conclude that the redistribution of H2A.Z seen over active gene bodies in lymphoma cells is specific to H2A.Z and is not a function of total nucleosome density in these regions.
H2A.Z levels do not increase in Eμ-MYC pre-B or lymphoma cells
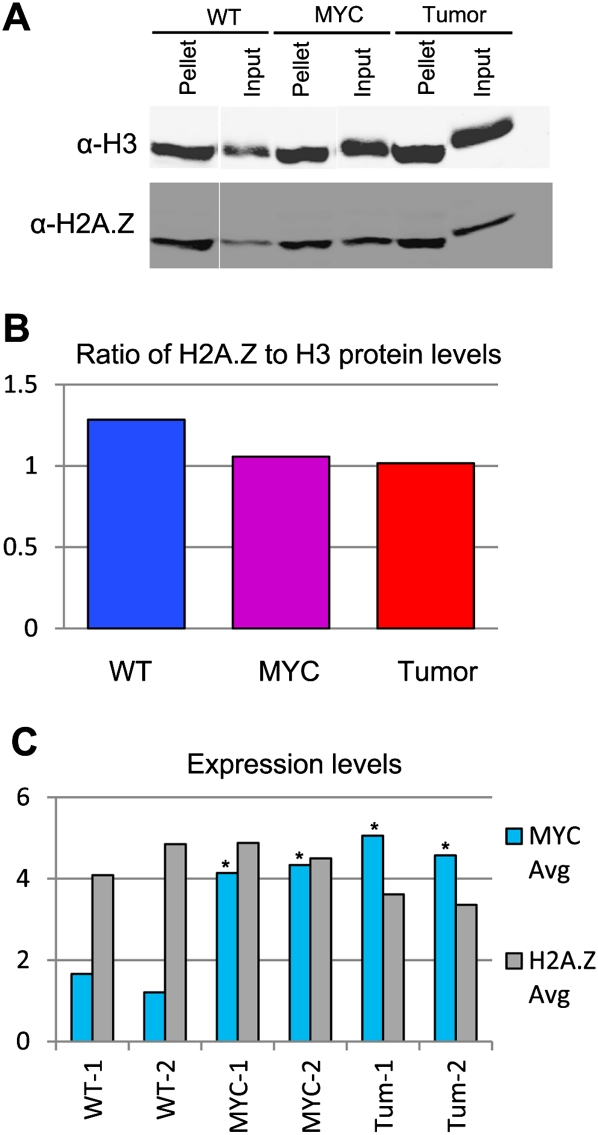
H2A.Z protein levels were reported to be globally increased in breast cancer patient samples by staining tissue microarrays with an antibody against H2A.Z (Hua et al. 2008). In addition, the investigators showed that in an estrogen-sensitive breast cancer cell line, H2A.Z protein levels were increased as a consequence of MYC binding to the H2A.Z promoter in response to estrogen signaling. Because increased deposition of H2A.Z during transcription could reflect an increase in total H2A.Z concentration, we asked if the increase in active gene body-associated H2A.Z levels seen in the lymphoma cells might result from the greater availability of this histone substrate. To test this, we compared H2A.Z protein levels from wild-type pre-B, Eμ-MYC pre-B, and lymphoma cells by quantitative Western blot analysis (Fig. 5A,B). We did not observe an increase in the total H2A.Z protein levels versus histone H3 (used as a standard) in either the Eμ-MYC pre-B cells or lymphoma cells. In contrast, we saw a small decrease in both as compared with wild-type. We also ascertained H2A.Z mRNA levels from our expression microarrays and observed no significant change between wild-type pre-B, Eμ-MYC pre-B, and lymphoma cells, despite a threefold increase of MYC mRNA in both the Eμ-MYC pre-B cells and lymphoma cells (Fig. 5C). Thus, the gain of H2A.Z over gene bodies in tumor cells is not caused by an increase in H2A.Z protein concentration.
Figure 5.
H2A.Z abundance in wild-type (WT), MYC-transformed, and tumor cells. (A) Western blots for H3 (above) and H2A.Z (below) for the soluble extracted (Input) material and the insoluble pellet. The antibody against histone H3 recognizes H3.1, H3.2, and H3.3 and therefore detects essentially all nucleosomes, regardless of the percentage of cells in S phase. (B) The ratio of total H2A.Z protein over H3, which includes both soluble and pellet fractions. (C) Expression of H2A.Z and MYC in the three cell types by microarray analysis (two replicates of each). (*) Expression changes that were significant at the P < 0.0001 level using Cyber-T.
An inverse relationship between H2A.Z and DNA methylation
To test whether there is an inverse relationship between H2A.Z and DNA methylation, as was observed in Arabidopsis, we used methyl DNA immunoprecipitation (MeDIP) on promoter methylation arrays to measure DNA methylation levels over the same regions for which we have determined H2A.Z profiles. We first asked whether MeDIP followed by a methylation promoter tiling array readout provides an accurate map of DNA methylation by comparing these profiles with profiles calculated in the same way but using an unrelated technique and readout platform. One such data set comes from DNA methylation profiling of human B cells by deep sequencing of HpaII-generated fragments (Ball et al. 2009). To compare the methylation data sets, we separated genes into five classes based on expression levels and displayed DNA methylation profiles around TSSs for each quintile averaged over 50-bp intervals. The profiles are remarkably similar, showing depletions on either side of the TSS, more for active than for inactive genes (Supplemental Fig. S8). We infer that MeDIP and methylation tiling arrays provide an accurate representation of the promoter methylome, and that average methylation around promoters is generally conserved between mouse and human lymphocytes.
To further validate that DNA methylation differences we observe on methylation promoter microarrays correspond to differences measured by other means, we chose two loci, Ism1 and Stat4, for bisulfite sequencing. Both loci were among those that showed the largest increases in DNA methylation near promoters during lymphomagenesis. For both loci, we confirmed that increases in DNA methylation observed on microarrays corresponded to increases in methylation determined by bisulfite sequencing, from 0.5% to 42% for Ism1 (Supplemental Fig. S9) and from 5% to 42% for Stat4 (Supplemental Fig. S10).
We next compared the average patterns of H2A.Z around TSSs with the corresponding patterns of DNA methylation in the three cell types. As expected, peaks of H2A.Z corresponded to dips of DNA methylation on either side of the TSS (Fig. 1A,B; Supplemental Fig. S8). Likewise, the increasing gradient of H2A.Z with increasing expression was reflected in a decreasing gradient of DNA methylation by 5′ ends analysis of expression quintiles (Fig. 1F–H). Thus, H2A.Z and DNA methylation show an approximate inverse relationship in mammalian cells. We observed a similar inverse relationship for CpG islands and shores in the transition to lymphoma, where methylation increased in both the CpG islands and in the shores (Supplemental Fig. S4). This is in agreement with findings in colon cancer, where 17% of CpG islands and 13% of the shores (based on the weighted average over the 2-kb shores) were hypermethylated (Irizarry et al. 2009).
DNA methylation decreases where H2A.Z increases in gene bodies
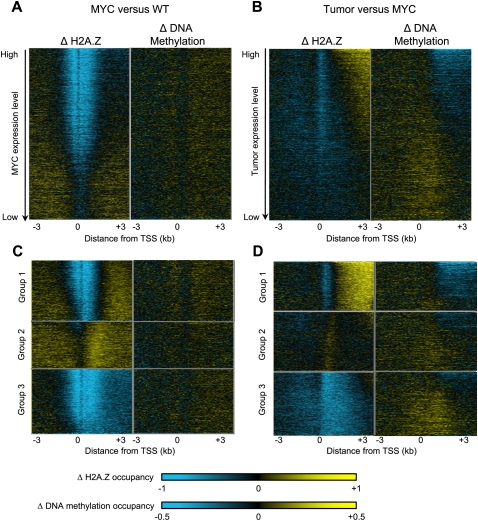
Another test of whether there is an inverse relationship between H2A.Z and DNA methylation during lymphomagenesis is to examine whether H2A.Z changes between cell states correspond to opposite changes in DNA methylation, as had been observed for Arabidopsis (Fig. 6A,B). In Arabidopsis, loss of the Swr1 complex that deposits H2A.Z had almost no effect on DNA methylation of promoters. Similarly, almost no change in methylation was seen over active gene promoters in the transition from wild-type to Eμ-MYC cells, despite a loss of H2A.Z. This may be due to protection of promoter elements and CpG islands by remodelers and transcription factors in the Eμ-MYC cells. In contrast, the gene body gains in H2A.Z during tumorigenesis were reflected by losses of DNA methylation, which showed a similar expression-dependent pattern in the lymphoma cells as compared with the Eμ-MYC cells (Fig. 6B). Likewise, in Arabidopsis, loss of Swr1 resulted in DNA methylation gains over gene bodies. No such gene body changes were seen in DNA methylation between wild-type and MYC-transformed cells, consistent with the observation that redistribution of H2A.Z to active gene bodies is specific to tumorigenesis (Fig. 6A). Taken together with the inverse patterns of H2A.Z and DNA methylation around promoters, we infer that changes in H2A.Z during transformation and oncogenesis correspond in general to opposite changes in DNA methylation. The striking appearance of expression-dependent gradients of H2A.Z and DNA methylation over gene bodies in the heat map display suggests that a large fraction of expressed genes undergo redistribution. To identify the genes undergoing redistribution in an unbiased manner, we used unsupervised k-means clustering to separate genes that show redistribution of H2A.Z from those that do not. We asked for three clusters in order to separate genes that have undergone redistribution of H2A.Z from genes that have either gained or lost H2A.Z over promoters without overall redistribution (Fig. 6C,D). In one group (Group 1), H2A.Z was strongly depleted from a tight region over the TSS and ∼500 bp downstream in the transition from MYC-transformed cells to B-cell lymphomas (Fig. 6D). This group comprised 30% of all annotated genes and contained nearly all of the genes in which H2A.Z was gained over the gene bodies in lymphomas. In this group, DNA methylation decreased over gene bodies, as expected if H2A.Z and DNA methylation are antagonistic as they have been shown to be in Arabidopsis. Another 35% of annotated genes fell into Group 2, which showed a mild gain of H2A.Z over TSSs and inconsistent changes in DNA methylation. Group 3 genes (35% of the total) displayed an overall loss of H2A.Z over the entire 6-kb region around the TSS. These genes also displayed modest gains in DNA methylation over TSSs, in the 5′ ends of gene bodies, and to some extent in upstream regions as well. We conclude that most genes undergo major changes in H2A.Z distribution during tumorigenesis, and that these changes are accompanied by extensive opposite changes in DNA methylation. These corresponding changes are specific for tumorigenesis, because in the transition from wild-type to Eμ-MYC cells, almost no DNA methylation changes were seen despite large changes in H2A.Z (Fig. 6C).
Figure 6.
Changes in H2A.Z and DNA methylation in the transition from overproliferation to tumor. Changes in H2A.Z and DNA methylation were compared as heat maps for MYC versus WT (A,C) and tumor versus MYC (B,D). (A,B) Genes were aligned at their TSSs and ranked by decreasing expression level. Data are represented as heat maps: (yellow) an enrichment of ChIP or MeDIP over input signals, (blue) depletion. (C,D) Genes were sorted into three groups using k-means clustering by weighting bins from the TSS to +3 kb downstream only in the H2A.Z sample. Each gene is in the same horizontal position for H2A.Z and DNA methylation.
Discussion
We have shown that cell state transitions during B-cell lymphomagenesis in the mouse involve large-scale changes in the distribution of the universal histone variant H2A.Z in genic regions. These changes include strong reductions at and around promoters of active genes, and a redistribution of H2A.Z from around promoters to gene bodies in a transcription-coupled manner. H2A.Z has been implicated in regulation of gene expression in numerous organisms, including animals, plants, and fungi, and therefore the changes that we have described might well play a role in gene expression changes in cancer. For example, the increases in H2A.Z abundance with increasing expression over the bodies of active genes during tumor progression are accompanied by a reciprocal decrease in DNA methylation, and either change has the potential of affecting gene expression levels. Our demonstration of large-scale changes in H2A.Z and nucleosome solubility during B-cell lymphomagenesis suggests that nucleosome dynamics also contributes to cancer progression.
One possible explanation for the gain in H2A.Z over active gene bodies is suggested by a recent study showing that MYC globally up-regulates transcriptional pause release (Rahl et al. 2010). The increased number of polymerase transits could result in greater H2A.Z deposition and less DNA methylation over gene bodies. However, this model does not explain why gene-body changes were seen only during tumorigenesis, and not in Eμ-MYC-transformed cells. An alternative possibility, not mutually exclusive, is that the global increase in nucleosome solubility that we observed in tumors relative to MYC-expressing cells synergizes with the increased pause release, causing increased transcriptional elongation, increased H2A.Z deposition and decreased DNA methylation.
H2A.Z is conspicuously enriched at sites of Polycomb complex binding in mouse embryonic stem cells, where it is required to maintain pleuripotency (Creyghton et al. 2008), which indicates that many of the genes that require H2A.Z regulate epigenetic changes. It has been proposed that cancer begins with a polyclonal epigenetic disruption in a field of cells, perhaps triggered by the fluctuating cellular microenvironment, that would predispose them to progress to the oncogenic state (Feinberg et al. 2006). The migration of Eμ-MYC pre-B cells from the bone marrow to the spleen and lymph nodes represents a dramatic microenvironment change that might be responsible for the chromatin changes that we see in the lymphoma. The redistribution of H2A.Z over bodies of 30% of all mouse genes makes this chromatin component a candidate for mediating such an epigenetic disruption. Furthermore, recent evidence that H2A.Z acts as an ambient temperature sensor in plants and fungi (Kumar and Wigge 2010) suggests that this unique histone variant has evolved to respond to environmental changes. Our findings point the way to future studies aimed at determining whether carcinogens alter H2A.Z occupancy and thus promote oncogenesis.
Despite the large-scale changes in the distribution of H2A.Z over promoters and genes, we were unable to detect changes in H2A.Z protein levels during B-cell lymphogenesis. In contrast, H2A.Z has been reported to be up-regulated in human breast cancer tissues (Hua et al. 2008). One potential explanation for this apparent discrepancy is that without induction of the estrogen pathway, MYC does not act on the H2A.Z locus, despite the presence of two E-boxes in the H2A.Z promoter. Indeed, MYC and the estrogen receptor have been shown to synergize in transcriptional activation of a subset of MYC target promoters in a ligand-dependent manner (Cheng et al. 2006). Thus, we find that global up-regulation of H2A.Z expression is not a hallmark of this lymphoma system and does not account for the lymphoma-specific gain of H2A.Z over active gene bodies.
Our finding of opposite patterns for H2A.Z and DNA methylation around TSSs was not unexpected based on the known distributions of these two epigenetic marks; however, their redistribution in gene bodies for such a large fraction of genes was surprising. Nevertheless, there are parallels between our observation of redistribution in lymphomas and previous findings in Arabidopsis, where loss of H2A.Z led primarily to gene body hypermethylation (Zilberman et al. 2008). Although mere anticorrelation between these marks does not necessarily imply a causal relationship between H2A.Z occupancy and DNA methylation, the corresponding pattern changes in the transition to lymphoma indicates interdependence. Taken together with the evidence for antagonism between these two marks in plants, our findings suggest that this interdependence is an ancient feature of eukaryotic genes that represents a new paradigm for epigenetic changes in cancer.
Methods
Mice
Pre-B cells with and without the Eμ-MYC transgene were harvested from the bone marrows of 6-wk-old mice (prior to any clonal expansion in potential tumor cells). Tumors were harvested from axillary or inguinal lymph nodes when mice exhibited late-stage disease symptoms and tumor masses exceeded 1 cm. C57BL/6J (wild-type) and B6.Cg-Tg(IghMYC)22Bri/J (Eμ-MYC) mice were purchased from The Jackson Laboratory. The Eμ-MYC strain was maintained by mating hemizygous males to wild-type C57BL/6J females. Animals were genotyped by PCR according to instructions from the supplier. Eμ-MYC transgenic animals were inspected twice weekly for overall signs of distress and for lymph node enlargement. Animals were euthanized when tumor mass reached 1 cm in diameter. All mouse procedures were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee.
B-cell purification
Single-cell suspensions were prepared from bone marrows or tumors following standard procedures. Mononuclear cells were initially separated from erythrocytes, granulocytes, platelets and dead cells by centrifugation on a density gradient (Histopaque 1083, Sigma Aldrich). B cells were subsequently recovered by using CD45 (B220) magnetic microbeads and a positive selection protocol on an automated magnetic sorter (Miltenyi Biotec). The purity of the samples, measured as percentage of B220+ cells, was verified by flow cytometry by using an FITC anti-mouse CD45R/B220 antibody (clone RA3-62B, BD Bioscience Pharmingen). Samples consistently contained >95% B220 positive cells. Using antibodies against CD43, IgM, and IgD, we determined that 50%–80% of the CD45-positive cells isolated from the bone marrows of 6-wk-old mice were immature B cells (Supplemental Fig. S11A). Of this immature B-cell population, ∼80% were pre-B cells based on CD43 staining (Supplemental Fig. S11B). Thus, we have called the population of cells assayed for H2A.Z and DNA methylation “pre-B cells” throughout the manuscript.
Cell cycle analysis
Purified B cells from bone marrow and tumor were washed in 1× PBS and fixed in 70% ethanol. After fixation, cells were washed in 1× PBS and resuspended in DAPI staining buffer (1× PBS, 0.1% Triton X-100, 1 mg/mL DAPI, 100 mg/mL DNase-free RNase A) and subjected to flow cytometry analysis on a BDT LSR II analyzer (Becton Dickinson). Cell cycle analysis was performed with FlowJO software (Tree Star Inc.).
Chromatin immunoprecipitation
Nuclei were isolated by lysing purified 10 × 106 B220+ cells in 10 mM Tris-HCl, 2 mM MgCl2, protease inhibitors (Roche, EDTA-free), and 0.08% NP-40. CaCl2 to a final concentration of 2 mM, and 0.2 units of micrococcal nuclease (MNase) (Sigma) were added to the nuclei and allowed to incubate for 1 min at 37oC. The MNase reaction was stopped by adding EGTA to a final concentration of 5 mM on ice. The nuclei were washed then resuspended in the ChIP buffer: 16 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.25 mM EDTA, and protease inhibitors. The nuclei were incubated for 10 min on ice then passed through a 30-gauge needle 10 times. Soluble chromatin was separated from the debris by centrifugation and was used as the input for ChIP experiments. Prior to the ChIP, the input fraction was precleared for 30 min with protein-A agarose beads. ChIPs were performed overnight with 10 μg of anti-H2A.Z antibody (Active Motif, #39113). Protein A agarose beads were used to isolate the antibody–chromatin complexes, and the DNA was recovered by proteinase-K digestion followed by phenol–chloroform extraction and desalting on a Qiagen mini-elute column. Protein and DNA recovery from ChIPs was determined by gel electrophoretic analysis (Supplemental Fig. S12). Input and ChIP DNAs were amplified using a whole-genome amplification kit (Sigma WGA2) then labeled with Cy3 and Cy5, respectively, using random 9-mers according to the NimbleGen ChIP with microarray hybridization (ChIP-chip) protocol.
MeDIP
Genomic DNA was isolated from cells using the DNeasy Blood and Tissue kit from Qiagen. Six micrograms of purified genomic DNA was sonicated to produce fragments of 100–1500 bp, with an average size of ∼600 bp (by ethidium bromide staining). An aliquot of 1.5 μg of the sonicated DNA was removed and set aside as an “input” fraction. The remaining 4.5 μg of DNA was denatured for 10 min at 95°C. After denaturation, 475 μL of MeDIP Buffer and 20 μg of anti-5-methyl C antibody (Eurogentec) were added to the sample, which was incubated for 15 h at 4°C (with end-over-end rotation). After the primary incubation, 50 μL of sheep anti-mouse magnetic dynabeads was added and the sample was rotated for an additional 2 h at 4°C. A magnet was used to separate methylated DNA bound to the dynabeads from unbound DNA. The input and MeDIP fractions were amplified using the WGA-2 kit then labeled with Cy3 and Cy5, respectively, using random 9-mer primers according to the NimbleGen ChIP-chip protocol.
RNA isolation and expression analysis
Total RNA was isolated using the Qiagen RNeasy kit, then amplified and labeled according to the NimbleGen protocol for expression arrays. Labeled RNA was hybridized to NimbleGen single-plex mouse MM8 expression arrays according to the manufacturer's directions. Two biological replicates were done for each cell type, all samples were normalized together using quantile normalization, and gene calls were made using the Robust Multichip Average protocol (Bolstad et al. 2003; Irizarry et al. 2003a,b). Cyber-T was used to compute confidence scores for changes in gene expression (Long et al. 2001).
Microarrays
ChIP and MeDIP samples were both hybridized to NimbleGen high-density 2.1 million probe tiled promoter arrays. H2A.Z ChIP samples were hybridized to arrays constructed using the mouse MM8 genome build, while MeDIP samples were hybridized to methylation-specific promoter arrays constructed using the mm9 genome build. Data were analyzed using NimbleGen software and protocols.
Data processing and analysis
To standardize for dynamic range differences, each data set was transformed to a normalized scale by converting the individual probe log2(IP/input) values to standard deviates; z = (x − μ)/σ, where x = the log2 ratio of the probe, μ = the mean of the data set, and σ = the standard deviation of the data set. Ends analysis was performed by lining up all Ref-seq genes (minus olfactory receptors and large overlapping genes) at their transcriptional start sites and averaging the normalized probe values as a function of distance. To map changes in either H2A.Z or DNA methylation, we first normalized the dynamic range of the data sets by converting scores to standard deviates of the mean. Data sets were then subtracted on a probe-by-probe basis to yield differences. To determine the relative levels of chromatin extracted from different cell types, we divided the signal from the input of the lymphoma cells by the signal of the input of the Eμ-MYC pre-B cells for each probe on the array. The input fraction represents the total chromatin digested to mostly mononucleosomes with micrococcal nuclease and extracted into the soluble phase.
Acknowledgments
We thank Jorja Henikoff for invaluable assistance with data processing and analysis; Andy Marty, Jeff Delrow, and the Hutch Genomics Facility for processing arrays and for technical advice; and Peter Cheung for the gift of an H2A.Z antibody. This work was supported by the Howard Hughes Medical Institute (S.H.) and NIH Grant R01CA57138 (R.N.E.).
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE19884.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106542.110.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538 [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE 2006. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116 [DOI] [PubMed] [Google Scholar]
- Bird AP 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C 2005. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res 33: 5633–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D 2009. Histones: Annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. 2006. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell 21: 393–404 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA 2008. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M 1988. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res 48: 1159–1161 [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S 2006. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Herranz M, Espada J, Ballestar E, Paz MF, Ropero S, Erkek E, Bozdogan O, Peinado H, Niveleau A, et al. 2004. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res 64: 5527–5534 [DOI] [PubMed] [Google Scholar]
- Gévry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L 2009. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev 23: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S, Jacques PE, Gévry N, Forest A, Fortin ME, Laflamme L, Gaudreau L, Robert F 2009. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS Genet 5: e1000687 doi: 10.1371/journal.pgen.1000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet R, Jaenisch R, Eden A 2007. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27: 404–408 [DOI] [PubMed] [Google Scholar]
- Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, et al. 2008. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol 4: 188 doi: 10.1038/msb.2008.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP 2003a. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003b. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA 2010. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem 276: 19937–19944 [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev 19: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J 2006. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J Biol Chem 281: 20036–20044 [DOI] [PubMed] [Google Scholar]
- Wolffe AP 1992. Chromatin: Structure and function. Academic Press, San Diego [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A 2008. H2A.Z: View from the top. Structure 16: 166–179 [DOI] [PubMed] [Google Scholar]