Abstract
The expression of most genes is regulated by multiple transcription factors. The interactions between transcription factors produce complex patterns of gene expression that are not always obvious from the arrangement of cis-regulatory elements in a promoter. One critical element of promoters is the TATA box, the docking site for the RNA polymerase holoenzyme. Using a synthetic promoter system coupled to a thermodynamic model of combinatorial regulation, we analyze the effects of different strength TATA boxes on various aspects of combinatorial cis-regulation. The thermodynamic model explains 75% of the variance in gene expression in synthetic promoter libraries with different strength TATA boxes, suggesting that many of the salient aspects of cis-regulation are captured by the model. Our results demonstrate that the effect of changing the TATA box on gene expression is the same for all synthetic promoters regardless of the arrangement of cis-regulatory sites we studied. Our analysis also showed that in our synthetic system the strength of the RNA polymerase–TATA interaction does not alter the combinatorial interactions between transcription factors, or between transcription factors and RNA polymerase. Finally, we show that although stronger TATA boxes increase expression in a predictable fashion, stronger TATA boxes have very little effect on noise in our synthetic promoters, regardless of the arrangement of cis-regulatory sites. Our results support a modular model of promoter function, where cis-regulatory elements can be mixed and matched (programmed) with outcomes on expression that are predictable based on the rules of simple protein–protein and protein–DNA interactions.
A major component of transcriptional regulation is the recruitment and interaction of sequence-specific DNA binding proteins known as transcription factors (TFs). Most genes have multiple regulators. Because of the high complexity of some promoters, and because of phenomena such as cooperativity and competition between TFs for similar cis-regulatory sites, the interactions between TFs often produce patterns of expression that are difficult to predict from the effects of individual TFs (Yuh et al. 2001; Istrail and Davidson 2005; Kulkarni and Arnosti 2005; Hsiau et al. 2007). Because of the centrality of transcriptional regulation in so many biological processes, unraveling the rules that govern combinatorial cis-regulation is an important and challenging task (Bintu et al. 2005). One recent approach to this problem focuses on synthetic promoters as models of transcriptional regulation (Ligr et al. 2006; Cox et al. 2007; Murphy et al. 2007; Gertz and Cohen 2009; Gertz et al. 2009). In previous work we studied combinatorial cis-regulation using a library of synthetic promoters in yeast coupled with a formal thermodynamic model that relates the promoter sequence to the expression of the downstream gene. We showed that a large fraction of the variability in gene expression can be explained by simple TF–TF and TF–DNA interactions. In addition, we used this system to investigate the effects of low-affinity TF binding sites (Gertz et al. 2009) and the effects of different growth environments on combinatorial cis-regulation (Gertz and Cohen 2009).
One important variable in gene expression is the interaction of RNA polymerase with DNA. A critical step in the regulation of eukaryotic genes is the recruitment of the TATA binding protein (TBP), which bends the DNA to allow binding of RNA polymerase II and transcription initiation (Struhl 1995). The canonical binding site for TBP is an A/T-rich sequence, known as the TATA box. In Saccharomyces cerevisiae only 17% of genes contain a conventional TATA box at their promoters (Iyer and Struhl 1995; Basehoar et al. 2004), but TBP is still required for the transcription of virtually all genes (Pugh and Tijan 1990; Cormack and Struhl 1992; Martinez et al. 1994). TATA and TATA-less promoters show structural and functional differences: In TATA-containing promoters TBP is delivered by the SAGA complex, while in TATA-less promoters TBP is delivered by TFIID (Lemon and Tijan 2000; Basehoar et al. 2004); a conventional TATA box is usually found at stress-related promoters as opposed to housekeeping genes (Basehoar et al. 2004). Keeping in mind these structural and functional differences, we sought to investigate whether the presence and strength of the TATA box affects combinatorial cis-regulation by TFs.
Several studies on individual promoters have shown how mutants of the TATA box affect both gene expression and promoter noise (Raser and O'Shea 2004; Blake et al. 2006). However, it is unclear whether these effects follow a general rule or if they are promoter-specific. It is well established that changes to the sequence of the TATA box affect the interaction between polymerase and DNA (Maicas and Friesen 1990; Stewart and Stargell 2001; Blake et al. 2006; Tirosh et al. 2006). However, it is still unclear whether these changes also affect the combinatorial interactions of TFs with polymerase and with each other. To address these questions we constructed synthetic promoter libraries with TATA boxes of different strengths and used a thermodynamic framework to model the interactions that occur on these promoters. The synthetic library allows us to study a wide variety of promoters in a controlled system. We found that TATA is a remarkably modular component of our promoters. Changes in the strength of the TATA box have similar effects on all promoters regardless of the arrangement of their components (TF binding sites). Changing the strength of the TATA box appears to affect the interaction between polymerase and DNA without affecting any of the combinatorial interactions between TFs, or between TFs and RNA polymerase. We also measured the variance in gene expression in each of our synthetic libraries and showed that while stronger TATA boxes increase gene expression, stronger TATA boxes do not contribute additional noise to gene expression. Our results suggest that TATA amplifies gene expression in the same way for all promoters without adding significant noise. Thus, the TATA box appears to be a modular component of promoters.
Results
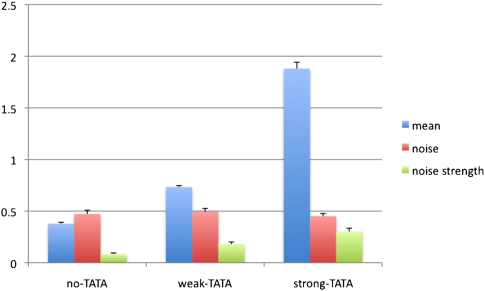
We built three different synthetic promoter libraries with random combinations of four TF binding sites following procedures previously described (Gertz et al. 2009). In each library we used a different basal promoter, one for a strong-TATA, one for a weak-TATA, and one for a no-TATA library (Blake et al. 2006). All three basal promoters were derived from the TSA1 promoter (see Methods for details). The strong-TATA basal promoter (here called S) contains one strong and one weak TATA box; the weak-TATA basal promoter (here called W), contains two weak TATA boxes; and the no-TATA basal promoter (here called N) contains no conventional TATA box (see Supplemental Table S2 for the exact sequence). These promoters were cloned upstream of the yellow fluorescent protein (YFP) gene, and each cassette was then integrated into the S. cerevisiae genome at the TRP1 locus as described in the Methods. Flow cytometry assays allowed us to obtain estimates of gene expression for each promoter, which we used as an estimate of promoter activity. As predicted, the three basal promoters showed different expression levels (Fig. 1).
Figure 1.
Average expression, noise, and noise strength in arbitrary units (AU) of the three basal promoters from four technical replicates. Error bars represent one standard deviation. Noise is CV (standard deviation divided by mean), and noise strength is VMR (variance divided by mean).
For our study of the relationship between the strength of the TATA box and the interactions among cis-regulatory sites we chose to study four TF binding sites. We picked binding sites for Gcr1, Reb1, Rap1 (activators), and Mig1 (repressor) as in Gertz et al. (2009). These sites are known to co-occur in promoters across the genome, and can be stimulated by glucose growth conditions (Scott and Baker 1993; Tornow et al. 1993; Pilpel et al. 2001; Harbison et al. 2004).
The vast majority of cis-regulatory sites occur evenly between TATA and non-TATA promoters (Supplemental Table S1). This suggests that most cis-regulatory sites function in both TATA and non-TATA promoters. We were therefore careful to choose sites that show no statistical enrichment in either TATA or non-TATA promoters as “typical” cis-regulatory sites function in both classes of promoters. All four sites we chose occur at expected frequencies in both TATA and non-TATA promoters (Supplemental Table S1). Among the known targets for each of these four transcription factors are both TATA and TATA-less promoters (Basehoar et al. 2004; Harbison et al. 2004). These four sites, Mig1, Reb1, Rap1, and Gcr1, are reasonable representative cis-regulatory sites, because like most cis-regulatory sites they occur evenly between both classes of promoters.
Binding sites for Gcr1, Reb1, Rap1, and Mig1 were randomly inserted upstream of each basal promoter in the S. cerevisiae genome. Each integrant was individually sequenced. We obtained 235 clones for the strong-TATA library (here called L-S), 298 for the weak-TATA library (here called L-W), and 241 for the no-TATA library (here called L-N). We arrayed our library clones in 96-well microplates. Each plate contained 92 individual clones and four replicates of the basal promoter as controls. We measured volume and YFP expression of single cells using flow cytometery. We then calculated the normalized fluorescence by dividing the fluorescence signal by the volume for every cell. The mean expression of every well was then normalized by the average of the mean expression of the four control wells to control for plate-to-plate variation, resulting in a measurement of fold change with respect to the basal promoter. Individual clone sequences and their associated expression values are available in Supplemental Table S2.
TATA is a simple scaling factor for all promoters
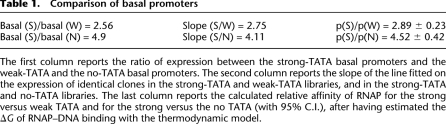
In each plate of libraries L-W and L-N we also added controls for the strong basal promoters (S). This allowed us to compare S and W, and S and N, in the same assay. We then estimated the strength of the strong-TATA with respect to the weak-TATA and to the no-TATA clones by calculating the ratio of expression of the basal promoters. The basal promoter with the strong TATA box was 4.9-fold stronger than the no-TATA basal promoter and 2.56-fold stronger than the weak-TATA basal promoter (Table 1).
Table 1.
Comparison of basal promoters
The first column reports the ratio of expression between the strong-TATA basal promoters and the weak-TATA and the no-TATA basal promoters. The second column reports the slope of the line fitted on the expression of identical clones in the strong-TATA and weak-TATA libraries, and in the strong-TATA and no-TATA libraries. The last column reports the calculated relative affinity of RNAP for the strong versus weak TATA and for the strong versus the no TATA (with 95% C.I.), after having estimated the ΔG of RNAP–DNA binding with the thermodynamic model.
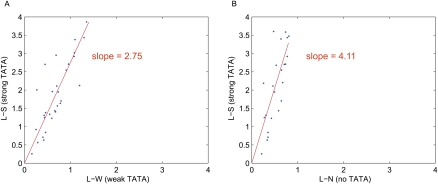
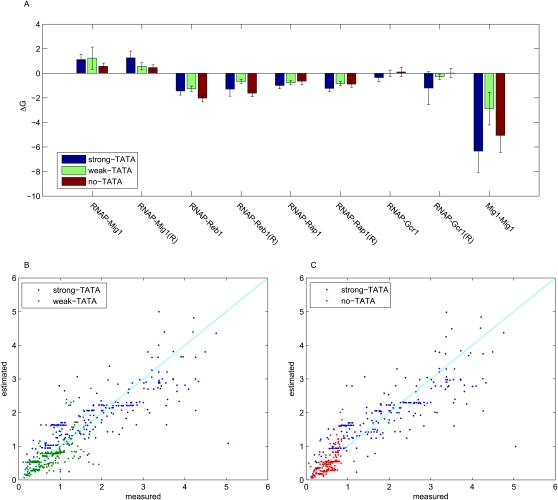
We next asked whether the effect of different strength TATAs depends on promoter context. To address this question we compared identical clones between the L-S and L-W libraries, and between the L-S and L-N libraries. Identical clones are promoters with the same sequence of TF binding sites but with different TATA boxes. We identified 30 unique clones identical between the L-S library and the L-W library. The scatter plot in Figure 2 shows the average expression of biological replicates of the identical clones. The slope of the best fit regression between identical clones in these libraries was 2.75, which is comparable to the ratio of expression from the strong and weak basal promoters (2.56). Similarly, the slope of the regression line between the 23 unique clones identical between the L-S and L-N libraries (4.11) is close to the ratio of expression of the strong- and no-TATA basal promoters (4.9). Furthermore, the residual variance in Figure 2, A and B, is almost entirely explained by the biological replicate variance. The biological replicate variance (Table 2) of our libraries explains 100% of the residual variance in Figure 2A, and 86% of the variance of the residuals in Figure 2B. This suggests that the small number of points that lie off of the regression lines in Figure 2, A and B, is due to experimental error, not to contextual differences caused by different strength TATAs. Moreover, the points that do lie off the regression line do not show any enrichment in size or composition of the promoter. Taken together, our results suggest that TATA acts as a simple scaling factor: Gene expression scales with the strength of the TATA box in a predictable way regardless of the upstream arrangement of cis-regulatory sites we studied.
Figure 2.
Comparison of the expression of identical library clones. (A) Scatter plot of identical clones between the L-S and L-W libraries. The slope of the best-fit regression line is 2.75. (B) Scatter plot of identical clones between the L-S and L-N libraries. The slope of the best-fit regression line is 4.11. The Pearson correlation is 0.82 for identical clones between L-S and L-W and between L-S and L-N.
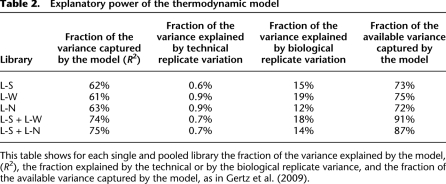
Table 2.
Explanatory power of the thermodynamic model
This table shows for each single and pooled library the fraction of the variance explained by the model, (R2), the fraction explained by the technical or by the biological replicate variance, and the fraction of the available variance captured by the model, as in Gertz et al. (2009).
The TATA box is a modular component of promoters
The interactions that occur on a promoter form a dense web of connections. We therefore sought to address whether the strength of the TATA box might affect the interactions between TFs and RNA polymerase (RNAP), or between TFs. We tested whether changing the strength of the TATA box had effects other than changing the affinity of RNAP for DNA (Stewart and Stargell 2001; van Werven et al. 2009). We applied a thermodynamic model previously developed to estimate the binding affinities between TF–RNAP and TF–TF given the expression level of each promoter (Gertz et al. 2009). The model's final outputs are proportional to the free energies (ΔG) of TF–RNAP and TF–TF interactions. We first fit each library independently to obtain estimates for the interactions between TFs and RNAP, and between TFs in the context of different strength TATA boxes. In each case a thermodynamic model specific for each library explained a high fraction of the variation in gene expression we observed in our data. Models that include only TF–RNAP interactions explained 62%, 61%, and 63% of the variance for L-S, L-W, and L-N, respectively (Table 2). Strikingly, the strengths of TF–RNAP and TF–TF interactions were not significantly different for the three libraries (Fig. 3; Supplemental Table S3). This result suggests that changes in the strength of the TATA box only affect RNAP binding, and that the change in RNAP binding does not alter TF–RNAP or TF–TF binding. Changes in the TATA box appear to be modular with respect to the web of interactions that occur on promoters.
Figure 3.
(A) Estimated ΔG for the interactions between TFs and RNAP. (R) Binding site in the reverse orientation. Each library was fit individually with the thermodynamic model. The figure shows the value of the estimated ΔG for TF–RNAP and TF–TF interactions with a 95% confidence interval (only the significant parameters are shown here); there are no significant differences in the energies of the interactions between RNAP and TF and in TF cooperativity in the three libraries. (B,C) Scatter plots of the pooled libraries. Each dot represents a library clone, with its measured expression on the x-axis, and its estimated expression on the y-axis (using the thermodynamic model). (Blue dots) Clones in the strong-TATA library; (green dots) clones in the weak-TATA library; (red dots) clones in the no-TATA library. (Cyan line) The perfect model, in which estimated values are exactly equal to the measured values.
To compare the effect of each TATA box, we pooled the data from two libraries together. We fit the pooled data using the thermodynamic framework by adding one extra parameter to the model that describes the relative affinity of RNAP for different strength TATAs. We obtained better fits to the pooled data than to data from individual libraries. The R2 for L-S + L-W was 74% and for L-S + L-N was 75% (Table 2; Fig. 3B,C), indicating that the thermodynamic model captured the added complexity introduced by different TATA boxes with the addition of only a single parameter that describes the relative affinity of RNAP for different TATA boxes. The extra variance in the pooled data, coupled with the improved model significantly increased our predictive power, allowing us to explain 15% more variance. The model performs significantly better with the extra DNA–RNAP parameter than without such a parameter, demonstrating that the improved fit is due to a better model of cis-regulation and not just to data with a larger dynamic range (see Methods for details). Using the fits from the pooled data we estimated the ΔG of the RNAP–TF interaction of the weak TATA and of the no TATA with respect to the strong TATA. Given these ΔG values we then calculated the probability of RNAP binding at each basal promoter. The calculated relative affinity of RNAP for the strong versus weak TATA was 2.89 ± 0.23, a number very similar to the ratio of expression of S and W (2.56), and to the slope of the line regressed for identical clones in L-S and L-W (2.75) (Table 1). Similarly, the calculated relative affinity of RNAP for the strong- versus no-TATA clones was 4.52 ± 0.42, which is similar to the ratio of expression of S and W (4.9), and to the slope of the line regressed for identical clones in L-S and L-W (4.11). Thus, in both comparisons (strong versus weak, and strong versus no TATA), three independent estimates suggest that the TATA box acts as a linear amplifier of gene expression, and that its effect is independent of the cis-regulatory content of the promoter.
TATA amplifies gene expression with little effect on noise
We investigated whether the strength of the TATA box affects the noise in transcriptional regulation in our system. Previous studies have extensively modeled the noise in protein expression as a result of stochastic processes that occur during TF binding, RNA polymerase binding, transcription, mRNA degradation, and translation (Paulsson 2004; Bar-Even et al. 2006; Kaufmann and van Oudenaarden 2007). Noise in specific S. cerevisiae promoters has been studied using a double color assay (Raser and O'Shea 2004), by introducing multiple copies of the same promoter into cells (Volfson et al. 2006), or by mutating the TATA box at a particular promoter (Raser and O'Shea 2004; Blake et al. 2006). Here, rather than measuring single promoters or single proteins, we extend these studies by analyzing noise in libraries of promoters with different strength TATA boxes and different cis-regulatory contents, but with the same translational efficiency.
Each flow cytometry run consisted of measurements of volume and fluorescence for 25,000 events (single cells). Cross-validation tests showed that 5000 events were sufficient to obtain robust estimations of the mean and variance of the distribution of expression measurements for each promoter (data not shown). To control for extrinsic sources of noise we normalized each fluorescence measurement by the volume of the cell from which it was derived (see Methods for details).
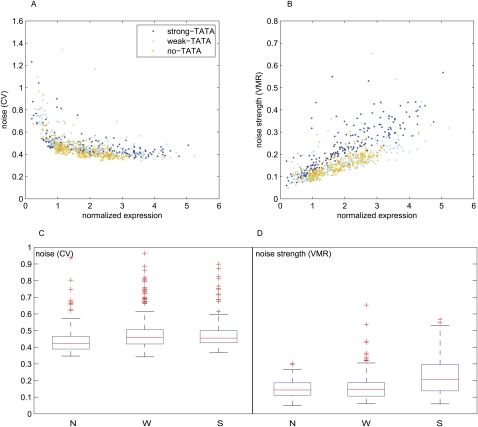
A simple comparison of the three basal promoters only (N, W, and S) showed that noise, as measured by the coefficient of variation (CV), which is the standard deviation to mean ratio, does not depend on the strength of the TATA box, while the noise strength, or variance to mean ratio (VMR), does (Fig. 1). Because the VMR depends on the mean expression level, as already described in Raser and O'Shea (2004), we sought to determine whether TATA has an effect on transcriptional noise that is independent of its effect on the mean expression of promoters.
We compared the variances in expression between clones from each of our libraries. To compare clones from different libraries we normalized their mean expression to the mean expression of their specific basal promoter (i.e., each clone is expressed in terms of fold changes with respect to its specific basal promoter). When CV is used as the measure of noise we find no differences between strong- and weak-TATA clones (P > 0.3, Students t-test), and only a small difference between strong- and weak-TATA and no-TATA clones (P = 10−6 and P = 10−7, respectively, Students t-test) (Fig. 4A,C). This result holds across the entire range of normalized expression levels and across all cis-regulatory combinations we studied. When VMR is used as the measure of noise (Fig. 4B,D) we find no significant difference between weak-TATA and no-TATA clones (P > 0.08, Students t-test). Clones that contain a strong TATA show a very small, but statistically significant, increase in noise strength (P = 10−4 and P = 10−7 with respect to no TATA and weak TATA, respectively, Students t-test). Although CV and VMR are the two most commonly used metrics of noise, our results suggest that using the CV (in Fig. 4A) skews the no-TATA data (the lowest expressing clones) slightly downward, while using the VMR (in Fig. 4B) skews the strong-TATA data (the highest expressing clones) slightly upward. If we use a different noise model (see Supplemental material) we do not find any significant difference in the three libraries.
Figure 4.
Transcriptional noise in different promoter libraries. Noise of each individual clone is plotted against the mean expression. Normalized expression means that the expression value (fluorescence/volume) for each clone was divided by the expression of its basal promoter (promoter with same TATA but no TF binding site), so that each clone is expressed in terms of fold changes with respect to their basal promoter. (A) Noise versus normalized expression for the three libraries. (B) Noise strength versus normalized expression. (C,D) Data of A and B, respectively, in a boxplot representation.
Taken together, our results suggest that the strength of the TATA box has very little effect on transcriptional noise. The TATA box thus acts as a clean amplifier: It amplifies the expression level, without changing the rules of combinatorial cis-regulation, and adding very little transcriptional noise, regardless of the arrangement of the cis-regulatory sites used in this study.
Discussion
We examined the effect of the strength of the TATA box on transcription in synthetic yeast promoters. We found that the TATA box acts as a simple scaling factor: Stronger TATA boxes result in predictably stronger expression levels. The increase in transcription was not promoter-specific as the same scaling factor applied to all our synthetic promoters. The TATA box amplifies gene expression in a predictable way regardless of the arrangement of the cis-regulatory sites used in this study. The strength of the TATA box does not affect the cis-regulatory interactions that occur on promoters, except by affecting the affinity of RNAP for the promoter DNA. Thus, the affinity of RNAP–TF and TF–TF are not affected by the affinity of RNAP–DNA. It was previously shown that the TATA box does not directly affect TBP binding but rather the stability of the TFIIA and TBP complex, which is crucial for both TBP recruitment (Stewart and Stargell 2001) and turnover rate (van Werven et al. 2009). We have shown here that this process does not affect TF binding to the promoters, allowing us to conclude that the TATA box, as well as transcription factor binding sites, is a modular component of synthetic promoters.
In our system TATA is a clean amplifier. TATA does not have a large effect on transcriptional noise, as it does on expression levels. Several previous studies showed correlations of CV and VMR with gene expression levels or protein abundance (Raser and O'Shea 2004; Blake et al. 2006; Newman et al. 2006; Volfson et al. 2006). In our study we separated the effects of TATA from the effects of mean expression level. By isolating the effect of TATA we showed that promoters with the same mean expression level, but different strength TATAs, have the same CV (Fig. 4A). This result suggests that TATA does not have a large effect on the CV that is independent of its effect on the mean expression level of promoters.
Although most cis-regulatory sites in yeast are distributed evenly between both TATA and non-TATA promoters, different cis-regulatory sites also show different levels of bias in their occurrence between these classes of promoters, with some sites showing significant skew toward one or the other promoter class. It is therefore possible that we might have obtained qualitatively different results with another set of cis-regulatory sites. Some cis-regulatory sites may function in very different modes when they occur in TATA versus non-TATA promoters. As additional sites are tested in synthetic promoter systems the likelihood of this hypothesis will be determined.
This work clarified the role of the basal promoter in our synthetic library system (Gertz et al. 2009). The thermodynamic model we developed explained ∼62% of the variability in gene expression by only accounting for TF–RNAP binding. Here, we varied another element in our system, the strength of the TATA box, and showed that the thermodynamic model still explains a large fraction of the variability in gene expression (∼75%), even with the added complexity introduced by different strength TATA boxes. These results support a modular model of transcriptional regulation, where promoter elements can be mixed and matched with outcomes on expression that are predictable based on the rules of simple protein–protein and protein–DNA interactions.
One striking result was the behavior of the promoter with no conventional TATA box (called N in the text) and the library we built using it. Despite the fact that TATA and TATA-less promoters have functional and structural differences, we did not find any differences in the rules of combinatorial transcriptional regulation in TATA versus TATA-less library clones. Our results suggest that the TATA box does not drive the recruitment of SAGA or TFIID, but rather only affects the transcription rate, regardless of the way TBP is delivered to the DNA. This hypothesis is supported by the fact that SAGA and TFIID are functionally redundant (Lee et al. 2000).
Methods
Strains and plasmids
We used the 160 nucleotides upstream of the start codon of the TSA1 gene of S. cerevisiae as the basal promoter, as already cloned into plasmid pJG102 (Gertz et al. 2009). The strong-TATA promoter was derived from the original TSA1 promoter by introducing a 3-bp substitution at positions −105 to −103, from GTC to TCG, to create a restriction site for XhoI (forming plasmid pIM102). Plasmids pIM104 and pIM103 carried the mutations for the weak-TATA and for the no-TATA basal promoters. The cassette containing the selection marker (URA3 gene), the basal promoters, and the YFP gene was then integrated into strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) at the TRP1 locus, as described in Gertz et al. (2009). Sequences are available in Supplemental Table S2.
Construction of the TATA libraries
Libraries L-S, L-W, and L-N were cloned first into plasmids pIM102, pIM104, and pIM103, respectively, and then integrated into S. cerevisiae BY4742 following the procedure described in Gertz et al. (2009). The oligonucleotide building blocks we used to construct library inserts contained binding sites for the transcription factors Mig1, Gcr1, Reb1, and Rap1 (see Gertz and Cohen 2009 for the exact sequence for these binding sites). Each individual clone was sequenced as in Gertz et al. (2009).
Flow cytometry assay
All library strains, including the basal promoter control strains, were arrayed into 96-well microplates. All cultures were grown in 500 μL of synthetic complete media lacking uracil with 2% glucose in 2-mL 96-well plates with shaking for 4 h at 30°C. They were then fixed with paraformaldehyde as described in Gertz et al. (2009). The fluorescence intensities and electronic volumes of 25,000 events from each well were measured on a Beckman Coulter Cell Lab Quanta SC with a multiplate loader. Fluorescence was then divided by volume to obtain a normalized fluorescence value for every cell. For each well, mean and variance were then calculated from the normalized fluorescence values for 25,000 events.
The purpose of normalizing by cell volume was to control for extrinsic components of noise. The major source of extrinsic noise is cell volume (Di Talia et al. 2007; Nachman et al. 2007; Skotheim et al. 2008); thus, dividing fluorescence by volume for each cell is a way of focusing on intrinsic sources of noise. In previous studies, investigators have attempted to gate on cells in the G1 phase of the cell cycle using forward scatter and side scatter (Newman et al. 2006). Because G1 cells are relatively uniform in size, this results in a lower noise population of cells to study. However, gating by forward scatter and side scatter is not an accepted method of determining the cell cycle state of yeast cells, so there is no guarantee that this method gates on a uniform population of G1 cells. For example, after gating, Newman et al. (2006) retained only 1% of total cells, when it is known that 30%–35% of yeast cells from an asynchronous population are in the G1 phase. While this method does reduce noise in the measurements, the analysis necessarily focuses on a biased subset of the data. Simply dividing fluorescence by volume corrects for the largest source of extrinsic noise and yields a similar relationship between CV and mean expression, as Newman et al. (2006) observed (Supplemental Fig. S3). Thus, we reduce the extrinsic noise, as Newman et al. (2006) did, without resorting to gating on a very small subpopulation of the data.
Thermodynamic model
To model gene expression, we implemented a thermodynamic model of polymerase occupancy that was originally proposed by Shea and Ackers (1985). The model and implementation were described previously in Gertz et al. (2009). For each promoter, the probability of RNAP binding is calculated as the sum of Boltzmann weights for the states with RNAP bound by the sum of Boltzmann weights for all states. Boltzmann weights are calculated for each possible state of every promoter. Each state includes parameters for ΔG changes for RNAP–DNA, TF–DNA, RNAP–TF, and TF–TF interactions. The parameters representing the ΔG are then estimated with a nonlinear fit. Goodness-of-fit was estimated with R2 and RSS. Akaike Information Criterion (Akaike 1974), which introduces a penalty term for the number of parameters, was used to determine whether the model applied to the pooled libraries with the extra parameter for RNAP–DNA interaction was significantly more accurate than the simpler model applied to the single libraries. All calculations were performed using the Matlab package from The Mathworks, Inc..
Acknowledgments
We thank members of the Cohen lab for helpful discussions and critical readings of the manuscript. This work was supported by a grant from the National Institutes of Health, R01 GM078222-01A1.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106732.110.
References
- Akaike H 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19: 716–723 [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N 2006. Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R 2005. Transcriptional regulation by the numbers: Models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Cormack BP, Struhl K 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69: 685–696 [DOI] [PubMed] [Google Scholar]
- Cox RS III, Surette MG, Elowitz MB 2007. Programming gene expression with combinatorial promoters. Mol Syst Biol 3: 145 doi: 10.1038/msb4100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448: 947–951 [DOI] [PubMed] [Google Scholar]
- Gertz J, Cohen BA 2009. Environment-specific combinatorial cis-regulation in synthetic promoters. Mol Syst Biol 5: 244 doi: 10.1038/msb.2009.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA 2009. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457: 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC 2007. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE 2: e643 doi: 10.1371/journal.pone.0000643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Davidson EH 2005. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci 102: 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann BB, van Oudenaarden A 2007. Stochastic gene expression: From single molecules to the proteome. Curr Opin Genet Dev 17: 107–112 [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN 2005. cis-regulatory logic of short-range transcriptional repression in Drosophila melanogaster. Mol Cell Biol 25: 3411–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704 [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev 14: 2551–2569 [DOI] [PubMed] [Google Scholar]
- Ligr M, Siddharthan R, Cross FR, Siggia ED 2006. Gene expression from random libraries of yeast promoters. Genetics 172: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas E, Friesen JD 1990. A sequence pattern that occurs at the transcription initiation region of yeast RNA polymerase II promoters. Nucleic Acids Res 18: 3387–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Lagna G, Roeder RG 1994. Overlapping transcription by RNA polymerases II and III of the Xenopus TFIIIA gene in somatic cells. J Biol Chem 269: 25692–25698 [PubMed] [Google Scholar]
- Murphy KF, Balazsi G, Collins JJ 2007. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci 104: 12726–12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman I, Regev A, Ramanathan S 2007. Dissecting timing variability in yeast meiosis. Cell 131: 544–556 [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Paulsson J 2004. Summing up the noise in gene networks. Nature 427: 415–418 [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R 1990. Mechanism of transcriptional activation by Sp1: Evidence for coactivators. Cell 61: 1187–1197 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Baker HV 1993. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol 13: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MA, Ackers GK 1985. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol 181: 211–230 [DOI] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR 2008. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JJ, Stargell LA 2001. The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J Biol Chem 276: 30078–30084 [DOI] [PubMed] [Google Scholar]
- Struhl K 1995. Yeast transcriptional regulatory mechanisms. Annu Rev Genet 29: 651–674 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N 2006. A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Tornow J, Zeng X, Gao W, Santangelo GM 1993. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J 12: 2431–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J 2006. Origins of extrinsic variability in eukaryotic gene expression. Nature 439: 861–864 [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH 2001. Cis-regulatory logic in the endo16 gene: Switching from a specification to a differentiation mode of control. Development 128: 617–629 [DOI] [PubMed] [Google Scholar]