Abstract
The intestinal microbiota consists of over 1000 species, which play key roles in gut physiology and homeostasis. Imbalances in the composition of this bacterial community can lead to transient intestinal dysfunctions and chronic disease states. Understanding how to manipulate this ecosystem is thus essential for treating many disorders. In this study, we took advantage of recently developed tools for deep sequencing and phylogenetic clustering to examine the long-term effects of exogenous microbiota transplantation combined with and without an antibiotic pretreatment. In our rat model, deep sequencing revealed an intestinal bacterial diversity exceeding that of the human gut by a factor of two to three. The transplantation produced a marked increase in the microbial diversity of the recipients, which stemmed from both capture of new phylotypes and increase in abundance of others. However, when transplantation was performed after antibiotic intake, the resulting state simply combined the reshaping effects of the individual treatments (including the reduced diversity from antibiotic treatment alone). Therefore, lowering the recipient bacterial load by antibiotic intake prior to transplantation did not increase establishment of the donor phylotypes, although some dominant lineages still transferred successfully. Remarkably, all of these effects were observed after 1 mo of treatment and persisted after 3 mo. Overall, our results indicate that the indigenous gut microbial composition is more plastic that previously anticipated. However, since antibiotic pretreatment counterintuitively interferes with the establishment of an exogenous community, such plasticity is likely conditioned more by the altered microbiome gut homeostasis caused by antibiotics than by the primary bacterial loss.
The human intestinal tract harbors the most abundant, and among the most diverse, microbial community of all body sites (Ley et al. 2008; Costello et al. 2009). As in most mammals, the gut microbiome is dominated by four bacterial phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (Ley et al. 2008), which represent more than 1000 different molecular species or phylotypes (Dethlefsen et al. 2008; Claesson et al. 2009). Remarkably, this phylotype composition can be specific and stable for each individual. Repeated sampling of the same individuals indicates that samples from the same subject are more similar than samples from different subjects (Costello et al. 2009; Turnbaugh et al. 2009), and in a 2-yr interval an individual conserves over 60% of phylotypes of the gut microbiome (Manichanh et al. 2008).
The gut is considered the primary site for cross-talk between the host immune system and microorganisms, in part because of the size and complexity of its microbiota and the presence of specialized lymphoid structures in the mucosa (Guarner et al. 2006). This close relationship is important for maintaining an adequate homeostasis between the individual and the external environment (Backhed et al. 2005; Guarner et al. 2006). Imbalances of the intestinal microbial composition, named dysbiosis, may disturb homeostasis, and therefore lead to a dysfunction or disease state. For instance, specific changes of this microbial ecosystem were recently associated with two of the major inflammatory bowel diseases (IBD) (Ott et al. 2004; Manichanh et al. 2006; Frank et al. 2007; Dicksved et al. 2008). A large reduction of microbial diversity was found in patients with Crohn's disease (Manichanh et al. 2006; Dicksved et al. 2008), and a selective reduction of Faecalibacterium prausnitzii, a member of the Firmicutes phylum, was reported in patients with ulcerative colitis (Sokol et al. 2009). Remarkably, in both inflammatory bowel diseases, most bacteria that decrease in abundance relative to healthy controls are producers of butyrate, which has strong anti-inflammatory effects (Nancey et al. 2002; Hamer et al. 2009). Therefore, although the mechanisms underlying these disorders are yet unclear, it is now well accepted that intestinal microorganisms play a key role in the initiation and maintenance of IBD (Round and Mazmanian 2009).
Experimental manipulation has great potential to go beyond observational studies and allow us to decode the physiological roles of the gut bacterial community, and also define new therapeutic strategies based on altering this microbiome. In principle, the stability of the gut microbiome could be disrupted by the use of prebiotics, probiotics, and antibiotics. Intake of prebiotics (i.e., specific nondigestible food ingredients) is expected to stimulate the growth and/or bacterial activity in the gut. So far, however, no prebiotic has been shown to have a persistent effect in modifying the gut microbial composition. Similarly, intake of probiotics (i.e., live microorganisms) confers only transient effects on digestive physiology, and long-term persistent alteration of the indigenous gut microbial composition remains controversial. Attempts to manipulate the composition of the intestinal microbiome by fecal bacteriotherapy have now become the focus of an extensive body of clinical case reports with promising results (Borody et al. 2003; You et al. 2008; Khoruts et al. 2009; Shanahan 2009). For instance, it has recently been shown that fecal transplantation from a healthy donor restored both gut microbiota composition and function in a human patient that suffered from recurrent Clostridium difficile–associated diarrhea (Khoruts et al. 2009). Finally, in contrast to prebiotic and probiotic intake, antibiotics have been shown to produce drastic short- and long-term alterations of the human indigenous microbiota. In these studies, microbial compositions were examined using DNA fingerprint techniques (Lofmark et al. 2006; Jernberg et al. 2007), microarrays (Palmer et al. 2007), and, more recently, by taking advantage of DNA pyrosequencing (Dethlefsen et al. 2008; Antonopoulos et al. 2009). All of the above studies indicated that after antibiotic intake there is a drastic disruption of the intestinal microbiota, resulting in a long-term decrease of its overall diversity.
The above observations clearly anticipate that experimental manipulation of the gut bacterial community should be feasible to some extent, for example, in the well-established transplantation of exogenous microbiota into germ-free animals. The result of this procedure is a stable colonization by the transplanted community that keeps most of its original diversity (Rawls et al. 2006; Alpert et al. 2008). Therefore, although host factors probably have a major effect in broadly shaping the intestinal microbial ecosystem, long-term alterations of an indigenous consortium might also be induced, especially at the phylotype level. Such changes can now be uncovered due to the rapid development of genomic approaches and computational methods, which permit more detailed comparisons of the compositions of microbial ecosystems. In the present study, we used recently developed tools for deep sequencing and phylogenetic clustering to examine the degree to which the gut ecosystem could be intentionally manipulated. Using rats as a model system, we compared the long-term effects of exogenous microbiota transplantation combined with and without an antibiotic pretreatment. We tested the hypothesis that antibiotics, by reducing bacterial load, would promote establishment of the transferred microbiota, this outcome would have important implications for clinical practice in situations where the goal is to colonize the gut with a new microbiota. The results were surprising, and indicated that the indigenous gut microbial composition could be reshaped to an extent not anticipated in previous studies.
Results and Discussion
Experimental strategy
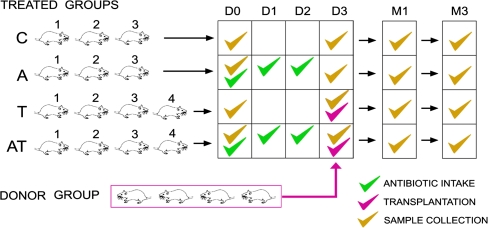
A total of 18 rats were included in this study. Four rats were used as cecal content donors, and 14 were used as recipients of the different treatments (transplantation, antibiotics, transplantation following antibiotics, and controls). Antibiotic treatment was conducted for 3 d, and transplantation of exogenous microbiota was performed at day four (Fig. 1).
Figure 1.
Experimental design. Four groups of rats were used as recipients of different treatments: (C) controls; (A) ATB intake during 3 d; (T) transplantation; (AT) 3-d ATB intake followed by transplantation. The cecal content of four donor rats was pooled and transplanted to a recipient rat once by gavage. Fecal samples of all rats were collected at different time points, day 0 (D0), day 3 (D3), month 1 (M1), and month 3 (M3).
All recipient rats were from the same strain (Lewis), but the donor rats belonged to different strains (Sprague Dawley and Wistar), and their cecal content was pooled in a single sample before administration to the recipients. This strategy ensured that the exogenous microbiota being transplanted would be highly diverse and different from the recipients, thus increasing the effect size and facilitating the discovery of changes following treatment.
The effects of the various treatments were analyzed by determining the bacterial density and bacterial diversity in the fecal samples of the recipient and control rats collected at different time points: at day 0 (before any treatment), at day 3 (after antibiotic intake but before transplantation), then at month 1 and month 3 (after antibiotic intake and/or transplantation). Two additional samples were collected at week 2 for one rat in each of the control and antibiotic groups.
As summarized in Figure 1, a total of 58 fecal samples were collected. Genomic DNA was extracted from these 58 samples and from the donor pool sample. The V4 hypervariable region of the bacterial 16S gene was amplified by PCR and used to determine the bacterial density and bacterial diversity in each sample. Bacterial load was estimated by means of quantitative real-time PCR, and was calculated from the number of copies of the 16S gene found per weight of stool. In order to analyze bacterial composition, unidirectional reads of the PCR-amplified V4 region were used for the analysis, so that any biases introduced during amplification would be common among all samples (allowing similarities and differences among samples to be interpreted). The 16S rRNA V4 amplicons were subsequently pyrosequenced on a 454 Life Sciences (Roche) Genome Sequencer FLX. The sequence reads were normalized and processed by the QIIME pipeline as described in the Methods section. Briefly, we first filter by quality and denoise the raw sequencing reads. Next, we define the operational taxonomic units (OTUs) or phylotypes by choosing a 97% identity threshold. Finally, in order to cluster the bacterial populations regarding the received treatment, we used the UniFrac metric, which compares microbial communities using phylogenetic information (Lozupone and Knight 2005).
Composition of the rat intestinal microbiome
Pyrosequencing of the samples described above produced, in total, 546,230 reads of raw data, which were submitted to trimming and denoising steps (see Methods). Of these reads, around 20% corresponded to a single fecal sample of one of the control rats (C1), which had been submitted to a very deep sequencing in order to capture bacteria present at very low abundance.
Analysis of the sequence reads from sample C1 allowed us to identify 926 phylotypes by using a 97% similarity cut-off. The final bacterial richness of this sample was then estimated to be 2621 phylotypes with the Chao1 estimates: This result was supported by rarefaction curves, which did not saturate with the number of sequences obtained (many previous studies have shown that saturation of each sample is not required to reveal biological patterns, e.g., Ley et al. 2008; Costello et al. 2009; Turnbaugh et al. 2009). The identified phylotypes indicated that this microbial community harbors at least eight bacterial divisions dominated by two major phyla: Firmicutes (74%) and Bacteroidetes (23%) (Supplemental Fig. S1). These data allowed us to examine how different the rat and human intestinal microbiome are. The human samples were obtained from fecal samples of two healthy female individuals (Turnbaugh et al. 2009). In order to properly compare the rat and the two human datasets, we first randomly sampled even numbers of sequences from all (30,100 sequences per sample), and calculated the alpha metrics. Unexpectedly, our results, also illustrated in the Supplemental Figure S2, showed that the number of observed species of the rat sample (621) was two to three times higher than the two human samples (271 and 277), with chao1 estimators of 1168 versus 426 and 483, respectively. The phylogenetic classification suggested that the rat and human microbiome are similar at the phylum level, but different at the genus level (Supplemental Fig. S3). Faecalibacterium and Bacteroides genera appear to be human specific, whereas Lactobacillus, Turibacter, and an uncharacterized member of the Porphyromonadaceae family were restricted to the rat microbiome. These taxonomic proportions are similar whether or not singletons are included: The singleton sequences are expected to contain any incorrect reads that escape denoising, and any chimeras.
Because of cost considerations, deep sequencing was performed only for sample C1. However, the smaller number of reads obtained in each of the other samples still allowed us to identify more than 200 species-level phylotypes in most of them (using a 97% similarity cut-off). As detailed below, these spectra of bacterial richness proved to be enough to observe significant reshaping of the gut microbiome following antibiotic and transplantation treatments.
Reshaping the gut microbiome by transplantation of an exogenous cecal content
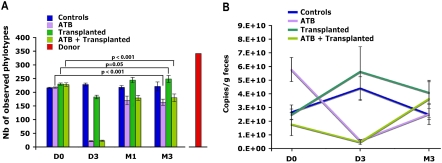
We determined whether exogenous bacterial phylotypes could reshape the microbial composition of the gastrointestinal tract by transplanting the gut microbiota from donor to recipient rats. To better differentiate microbial composition between recipient and donor samples, we used different strains of rats (Lewis for the recipient and Sprague Dawley and Wistar for the donors) coming from different farms. We surgically removed the cecal content from four donor rats and pooled them together. By using cecal contents, the bacterial composition of which is expected to be different than in fecal samples, and by pooling them we aimed to obtain a different diversity and a greater richness between the exogenous transplant and the endogenous microbiota of the recipients. These assumptions were validated by evaluating the bacterial richness in the donor sample, which was greater than in any of the recipient stools analyzed prior to inoculation (341 phylotypes in the donor and an average of 229 [SD = 11] in the recipients [P < 0.001; one sample t-test]; Fig. 2A); and by the UniFrac PCoA analyses, which showed that the community structure of donor sample (red dot in Fig. 3A) clustered separately from the recipient baseline and control samples (Cluster I, see the projection on plane PC2-PC3).
Figure 2.
Variation of bacterial load and richness. (A) Number of observed phylotypes as defined at 97% sequence identity. For both figures, mean value (n = 3 for controls and ATB; n = 4 for Transplanted and ATB + Transplanted) ±SD are plotted. (B) Bacterial quantification assessed by real-time PCR of the 16S gene at three time points: baseline (D0), day 3 (D3), and month 3 (M3).
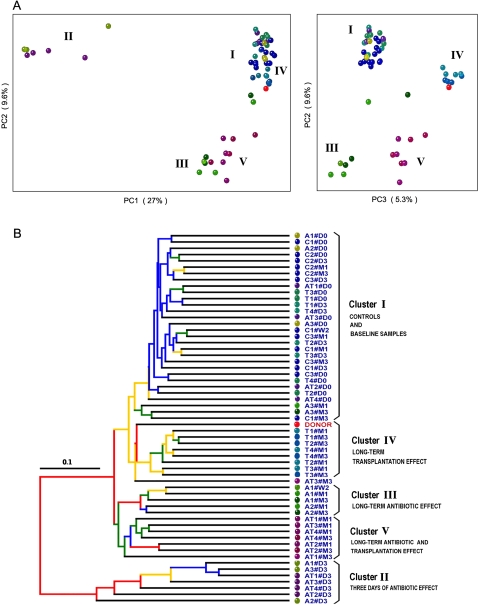
Figure 3.
16S gene surveys show clustering of bacterial communities by treatments. (A) Principal coordinates analysis (PCoA) performed on pairwise unweighted UniFrac distances shows a 3-d antibiotic effect (PC1 and PC2) and a long-term effect for all treated groups (PC2 and PC3). (B) Hierarchical cluster tree built using UPGMA (unweighted pair group method with arithmetic mean) from the same UniFrac distance matrix that was used for the PCoA. Each dot represents a sample codified by either C (controls), A (ATB), T (Transplanted), or AT (ATB and Transplanted), followed by the number of the animals (from one to three or to four) in each group and by a date of sample collection (#D0, #D3, #W2 [week 2], #M1, and #M3). The effect of each treatment leads to five clusters of samples (I to V). Branches in the UPGMA tree are colored according to their jackknife support: red, 75%–100%; yellow, 50%–75%; green, 25%–50%; blue, <25% support.
Transplantation was conducted by a single gavage of the pooled cecal content to recipient rats. UniFrac analyses revealed that 1 mo after transplantation, the fecal bacterial diversity of the recipient was modified to highly resemble that of the donor sample (red dot in Fig. 3A,B), and that this clustering persisted to a remarkable extent 3 mo after transplantation (Cluster IV in Fig. 3). The variation 3 mo apart in the bacterial structure of the recipients was likely due to the combination of an increase in the number of phylotypes (P = 0.05; Fig. 2) and a significant change in the proportion of Firmicutes (P = 0.05; Fig. 4). These results thus indicate that a single inoculation of a very complex microbial community by gavage can be sufficient to initiate a long-term reshaping of the recipient gut microbiome.
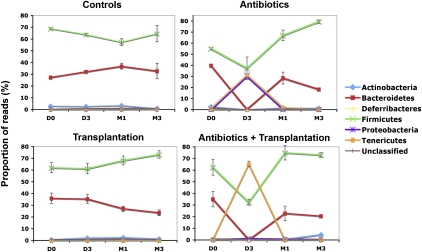
Figure 4.
Variation of the diversity of the microbial communities. Phylotypes were assigned a taxonomy using the RDP classifier at the phylum level and at different time points. Mean value (n = 3 for controls and ATB; n = 4 for Transplanted and ATB+Transplanted) ±SD are plotted.
Reshaping the gut microbiome by transplantation of an exogenous cecal content combined with antibiotic pretreatment of the recipients
Before analyzing the effects of transplantation after antibiotic intake, we first needed to determine the reshaping effects produced by the antibiotic treatment alone. We administered vancomycin and imipenem to the rats in drinking water for 3 d. This mixture of antibiotics has a broad-spectrum activity, acting against Gram-positive and Gram-negative bacteria, respectively, and is known to have an antimicrobial effect in the rat intestinal microbiome (Videla et al. 1994). Our analyses corroborated this effect. After 3 d of intake, we observed a 10-fold decrease in bacterial load (Fig. 2B) and reduced bacterial phylotype richness (from 217 to 21 OTUs, on average; Fig. 2A). UniFrac principal coordinates analyses (PCoA) showed that the microbiome of all treated rats clustered far from the controls (Cluster II and Cluster I, respectively, in Fig. 3A). This change in composition was mainly due to the near-extermination of Bacteroidetes and a significant decrease in Firmicutes (P < 0.001; Fig. 4). Strikingly, in all of these samples, there was a large increase in the Proteobacteria and Tenericutes phyla (from 1% to 31% of the reads). Therefore, although the antibiotics clearly affected a large proportion of the two major phyla (Bacteroidetes and Firmicutes), two minor ones (Proteobacteria and Tenericutes) either presented a higher proportion due to the depletion of the other microbes or took advantage of the empty niche to overgrow. One month after discontinuation of the antibiotics, the fecal samples regained a similar bacterial load to the controls. Bacteroidetes and Firmicutes recovered as the two major phyla, and Proteobacteria and Tenericutes returned to their initial proportions (Fig. 4). However, as shown by the PCoA analyses, bacterial diversity (Cluster III in Fig. 3A) was not resilient (i.e., did not return to its original structure). Phylotype richness was still decreased from its original values (from an average of 217 to 164 OTUs; P < 0.001). This loss of diversity was mainly due to phylotypes of the Bacteroidetes phylum (Supplemental Fig. S4). Three months after drug discontinuation, these changes still persisted, leading to both a global diversity loss and a reshaping of the two main bacterial phyla: The proportion of Firmicutes increased and of Bacteroidetes decreased (P < 0.01) in comparison to the control samples. One of the three rats showed a partial resilience clustering with the rats of Cluster I as pictured (green dots in Fig. 3A,B). Such behavior from a microbial community denotes some specificity in drug response at the individual level, as previously observed for ciprofloxacin (Dethlefsen et al. 2008) and acetaminophen (Clayton et al. 2009).
Our antibiotic mixture produced a marked, persistent change in the microbiota. We therefore anticipated that decreasing the bacterial load of the recipient prior to transplantation would produce different patterns of gut recolonization compared with transplantation alone. We thus treated a subgroup of rats with vancomycin and imipenem for 3 d, as described above, prior to transplantation. As expected, immediately before transplantation, fecal samples from these rats had a twofold decrease in microbial load and had radically altered bacterial communities compared with control rats (Figs. 2B, 3A). One month after transplantation, the bacterial load in the recipient fecal samples increased markedly and slightly surpassed the pretreatment values (Fig. 2B). UniFrac analyses revealed that these fecal bacterial communities (Cluster V in Fig. 3) clustered far away from the baseline and the control samples (Cluster I) and, surprisingly, were also distant from both the donor sample and the long-term antibiotic-effect cluster (Cluster III). Bacterial diversity was considerably altered, with a significant decrease of Bacteroidetes (P < 0.01) and increase of Firmicutes (P = 0.05) (Fig. 4). Three months after both treatments, all of these effects persisted (Cluster V). These results did not support our hypothesis that antibiotic intake to clear out the original gut communities would significantly enhance the reshaping effect of transplantation. Rather, combining antibiotic and transplantation treatments appeared to result in a combination of their effects, yet, producing a larger reshaping of the gut community than did each of the two treatments separately.
Analysis of phylotype capture and conservation following transplantation with and without antibiotic pretreatment
To further compare the alterations of the microbial diversity by the treatments described above, we examined the phylotype conservation of the original microbial composition in all individual rats 3 mo after treatment using two methods.
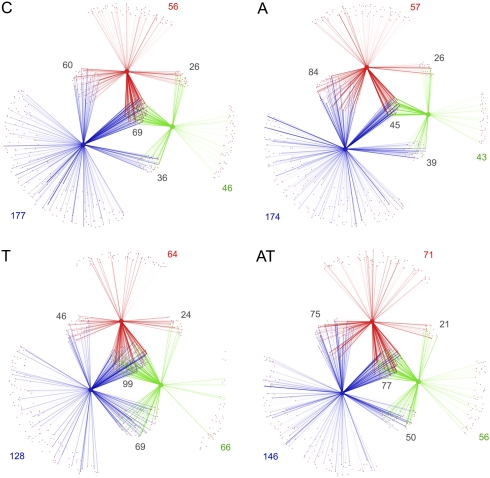
First, we identified shared phylotypes between the initial and final time points of each treatment, and the donor. These analyses were performed using bipartite networks showing the relationships between phylotypes and samples (Ley et al. 2008), such as those illustrated in Figure 5. Intensity of the network lines reflects the relative abundance of each detected phylotype in a given sample. The different network connections depict the fractions of vanished and conserved phylotypes, as well as the contribution of the donor to the gain in diversity in recipient animals. Taking as reference the network of an untreated animal (Fig. 5, panel C), the loss of diversity and extent of reshaping produced by antibiotic treatment (Fig. 5, panel A) can be appreciated by the decreased number of observed phylotypes and the marked reduction of shared phylotypes between the initial and final time points (red and green spectra, respectively). In the case of transplantation (Fig. 5, panel T), the network reveals that the clear gain of diversity is mainly contributed by phylotypes of the donor (compare the corresponding intersections between panels C and T). When transplantation was done after antibiotic intake (Fig. 5, panel AT), it is observed that antibiotics minimized the transplantation effect. The network reveals that transplantation contributes to recover the loss of diversity produced by the antibiotics (Fig. 5, cf. panels A and AT), although the surfacing of new phylotypes not present at the initial time point is not as prominent as that observed with the transplantation alone.
Figure 5.
Network plots of shared microbial diversity. The relationships between phylotypes and samples are represented as a bipartite graph in which nodes are either phylotypes (small) or samples (large), and connecting lines between small and large nodes mean that the phylotype was found in the given sample. Colors of lines and large nodes indicate the donor sample (blue), the sample before treatment (red), and the sample obtained 3 mo after treatment (green). The intensity (opacity) of each line reflects the relative abundance of each detected phylotype in a given sample; the groups of phylotypes that join any given pair of samples indicate the proportion of shared phylotypes, and the phylotypes connected to only a single sample are unique. The number of shared phylotypes between samples and phylotypes uniquely found in each sample is indicated. The panels represent the controls (C), effects of antibiotics only (A), transplantation only (T), or transplantation with antibiotics (AT). Intersections of red, blue, and green lines show common phylotypes between donor and recipient rats at any time-point.
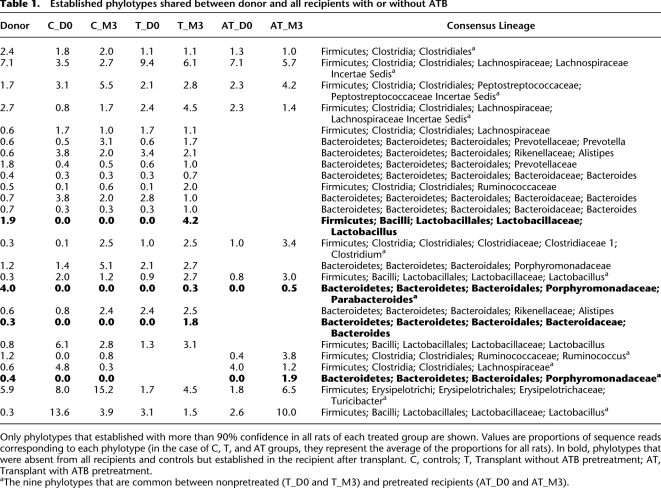
Second, we examined whether or not some phylotypes were captured from the donor and established by all of the treated rats, and whether such common phylotypes were different or not in the antibiotic pretreated recipients. Our analyses revealed that 3 mo after transplantation 22 phylotypes shared with the donor were commonly established (≥90% confidence) in all of the recipient rats nontreated with antibiotics (Table 1, columns T_D0 and T_M3). Among these 22, three phylotypes were not detected in all baseline samples of the recipients. Therefore, these species phylotypes had to be acquired through the donor sample. Conversely, when the recipient rats were pretreated with antibiotics, only 12 phylotypes (columns AT_D0 and AT_M3) shared with the donor were commonly established in all them (≥90% confidence). From these 12 phylotypes, nine overlapped (footnote a in Table 1) with the core of 22 observed in the nonpretreated recipients, and two were likely acquired through the donor since they were not detected in any of the recipient baseline samples. These results, therefore, support the idea that antibiotic pretreatment interferes with the reshaping effect of the exogenous transplantation.
Table 1.
Established phylotypes shared between donor and all recipients with or without ATB
Only phylotypes that established with more than 90% confidence in all rats of each treated group are shown. Values are proportions of sequence reads corresponding to each phylotype (in the case of C, T, and AT groups, they represent the average of the proportions for all rats). In bold, phylotypes that were absent from all recipients and controls but established in the recipient after transplant. C, controls; T, Transplant without ATB pretreatment; AT, Transplant with ATB pretreatment.
aThe nine phylotypes that are common between nonpretreated (T_D0 and T_M3) and pretreated recipients (AT_D0 and AT_M3).
Finally, the fact that the above analyses also show that the overlap of phylotypes, established in all recipient animals regardless of the antibiotic pretreatment, is highly significant (P < 10−13). This core represents less than 20% of the most abundant phylotypes found in the donor sample. Therefore, the relative abundance of phylotypes at the time of transplantation is not the only determinant for phylotype establishment. Clearly, host factors also play a role in shaping the intestinal microbial ecosystem, and those factors might be also altered by antibiotic intake.
Conclusion
In this study, we took advantage of recently developed tools for deep sequencing and phylogenetic clustering to investigate the manipulation of the rat intestinal microbial community in a nongerm-free condition. Our findings are summarized as follows. First, the diversity of the rat intestinal microbiome seems to surpass by two to three times the diversity of the human gut microbiome, suggesting that these laboratory-raised mammals may establish a higher complexity in their gut bacterial ecosystem to harvest more nutrients from their basic diet. Second, a short intake of the antibiotic cocktail (vancomicyn and imipenem) produces profound long-term effects on the rat intestinal microbiome. Although there is some individual variability in this long-term response, gut microbial diversity is notably reduced along with a reshaping of its two major phyla (Firmicutes and Bacteriodetes). Third, transplantation of a rich pool of exogenous bacteria by a single gavage leads to an increase in bacterial diversity. Phylogenetic clustering shows that the microbiome of the recipients changed to resemble that of the donor and, remarkably, that this reshape does not present any resilience after 3 mo of transplantation. These results indicate that an indigenous gut microbial community can be reshaped to an extent not previously anticipated. Finally, we show that minimizing the recipient microbiota with antibiotic intake prior transplantation does not facilitate the establishment of the exogenous microbiota. Phylogenetic clustering indicates that combination of both treatments simply produces a combination of their reshaping effects. Although this finding makes ecological sense, that antibiotics might be almost as deleterious to the input community as to the endogenous community, it is a highly counterintuitive result that should be taken into account in designing future bacteriotherapy protocols.
Methods
Rat husbandry
All experiments with rats were conducted using protocols approved by the Institut de Recerca de l'Hospital Universitary Vall d'Hebron Studies Committee. A total of 18 conventionally raised rats were included in the study (14 males Lewis, one female, and one male Sprague Dawley, and one female and one male Wistar). Each animal was isolated in a sterilized cage in order to avoid the transmission of microbiota between individuals. Cages were changed once a week and also before starting any treatment.
All rats drank sterilized water and were fed an autoclaved chow diet with no prevention of coprophagia. At the end of the study, all the rats were sacrificed by CO2 asphyxiation.
Experimental design (Fig. 1)
Six rats (Lewis male) were used for studying the antibiotic effect alone. Three rats were treated with 50 mg/kg/day of vancomycin and 50 mg/kg/d of imipenem in drinking water during 3 d; the remaining three, without any treatment, were used as controls. Twelve rats were used for studying the effect of gut microbiota transplantation alone and the combined effect of both antibiotics and transplantation. These included four donor rats (Sprague Dawley female and male, Wistar female and male) and eight recipient rats (Lewis male). Before transplantation, the eight recipient rats were given an oral gavage of omeprazol for 3 d at the concentration of 50 mg/kg/d. Omeprazol reduces the gastric acid secretion by the inhibition of proton pumps, thus allowing the survival of microorganisms through the stomach. In this period, all recipient rats also received oral gavages of water daily in order to avoid the stress of gavage manipulation. During this 3-d period, four of the recipient rats received the same antibiotic treatment described above. Microbiota transplantation was conducted at day 4. Four rats (Sprague Dawley and Wistar, one female and one male from each strain) were used as donors. Their cecal content was surgically extracted, pooled, and immediately administrated to the eight recipient rats by one oral gavage.
Genomic DNA extraction from cecal and fecal sample collections
Just before transplantation to recipient rats, aliquots of the pooled cecal content of the four rat donors were kept at −80°C for bacterial composition analyses. From all of the remaining rats, fecal samples were collected at four time points: 3 d before antibiotic treatment (basal time point, D0); 3 d after antibiotic intake but just before transplantation (D3); and 1 and 3 mo after transplantation or/and antibiotic intake (M1 and M3). These 56 fecal samples were collected directly from the anus of each rat and were immediately stored at −80°C until analysis.
For genomic DNA extraction, 100 mg of each of the above samples were suspended in 1400 mL of ASL (lysis buffer) provided by the QIAamp DNA stool mini kit (Qiagen). The suspensions were transferred to a Lysing Matrix E tube that contained a mixture of ceramic and silica particles designed to efficiently lyse microorganisms (QBiogen). Tubes were shaken in a FastPrep Bio 101 Bead apparatus (QBiogen) at 6.5 m/sec for 30 sec. DNA was then extracted by using a QIAamp DNA stool minikit from Qiagen, as recommended by the manufacturer (protocol for isolation of DNA for pathogen detection), except that a supplemental mixture of enzymes (mutanolysin at 90 U and lysozyme at 9 mg/mL) was added at the proteinase K step.
Calculation of bacterial load by quantitative real-time PCR
In order to determine the bacterial load per volume of fecal sample, genomic DNA extracted was submitted to real-time PCR of the V4 hypervariable region of the bacterial 16S gene. V4 regions (289 bp) were amplified by using universal primers, V4F_517_17 (5′-AGGCAGCAGTGGGGAAT-3′) and V4R_805_19 (5′-GCCAGCAGCCGCGGTAA-3′). We selected these primers among several pairs commonly found in the literature because they matched the most bacterial sequences deposited in the Ribosomal Database Project (RDP). To calibrate the Q–PCR reactions, calculated amounts of a linearized plasmid, in which the V4 region from one of the control rats had been inserted, were used. Plasmid concentration was measured using a NanoDrop ND-1000 Spectrophotometer (Nucliber), and the number of plasmid copies was calculated from the plasmid's molecular weight. To extrapolate the bacterial number in each sample, serial dilutions of the plasmid were amplified (copy number ranging from 182 to 1.82 × 107).
Amplification and detection of DNA by real-time PCR were performed with the 7500 Fast Real-Time PCR System (Applied Biosystems) using optical-grade 96-well plates. The PCR reaction was performed in a total volume of 25 μL using the Power SYBR Green PCR Master Mix (Applied Biosystems), containing 100 nM of each of the universal forward and reverse primers. The reaction conditions for amplification of DNA were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. All reactions were conducted in triplicate and mean values calculated. Data analysis made use of Sequence Detection Software version 1.4 supplied by Applied Biosystems.
Tag-pyrosequencing
In order to analyze bacterial composition, the V4 hypervariable region of the 16S gene was amplified from the DNA extracted from the 57 collected samples using the two universal primers described above. Each of the multiplex identifiers (MIDs) has been added upstream of the forward primer sequence (V4F_517_17). Standard PCR was run in a Mastercycler gradient (Eppendorf) at 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 56°C for 20 sec, 72°C for 40 sec, and a final cycle of 72°C for 7 min. Ten of 12 MIDs proposed by Roche resulted in a good amplification of the PCR product. The 16S rRNA V4 amplicons were subsequently sequenced on a 454 Life Sciences (Roche) Genome Sequencer FLX platform (Center for Genomic Regulation) according to 454 platform protocols.
Bioinformatics analyses
The sequences were analyzed using the QIIME pipeline (http://qiime.sourceforge.net/). Of the 546,230 reads recovered from two runs of the 454 GSFLX sequencing machine, ∼25% were removed by initial quality filters. These filters checked for the correct primer sequence, the proper barcode sequence, a read length of 200–300 nucleotides (nt), and an average quality score of 25, following the recommendations of Huse et al. 2007. Also, sequences containing ambiguous nucleotides (“N”) or long homopolymers >6 nt were removed. Strict quality filters ensure high quality in the downstream analysis. The remaining 415,785 reads were denoised using a modified version of PyroNoise (Quince et al. 2009). Denoising removes most of the common sequencing errors on the 454 platform by clustering reads that were most likely derived from the same sequence, and greatly reduces the number of incorrectly inferred OTUs or phylotypes.
After denoising, 3004 clusters were passed to the QIIME pipeline. Here, cd-hit (Li and Godzik 2006) was used to define OTUs at 97% sequence identity, which were assigned a taxonomy using the RDP classifier (Wang et al. 2007). Representative sequences for each OTU were aligned with PyNast (Caporaso et al. 2010) and columns uninformative for phylogeny building were filtered out using the Lanemask_PH file from Greengenes (DeSantis et al. 2006), either because they are too variable (hypervariable) or too conserved. The resulting alignments were used to build a phylogeny using FastTree (Price et al. 2009).
Rarefaction analysis was done for all samples with 10 repetitions using a step size of 100 from 100 to 2000 sequences per sample. For beta diversity analysis all samples were subsampled to 2000 sequences per sample to remove all possible side effects of sample size. The principal coordinates analysis (PCoA) was performed on pairwise unweighted UniFrac distances (Lozupone and Knight 2005). The hierarchical cluster tree was built using UPGMA (unweighted pair group method with arithmetic mean) on the UniFrac distance matrix derived from a subsample with up to 3000 sequences per sample. Jackknife support was based on 20 additional subsamples with 2000 sequences per sample.
The two human gut samples from the V2 region (Turnbaugh et al. 2009) were analyzed and integrated using the same protocol.
Network analyses, which allows the visualization of shared phylotypes between samples, were performed as previously described (Ley et al. 2008): Shared phylotypes were extracted from a table of sample by phylotype and annotated for use in Cytoscape (http://www.cytoscape.org/). Briefly, nodes represent either phylotypes or samples; an edge indicates that a given phylotype was found in a given sample; and the opacity of each edge is proportional to the count of phylotypes found in that sample. A fixed number of sequences per sample was used to ensure that effects were due to intrinsic diversity rather than sampling effort, and a spring-embedded layout was used so that samples that share more phylotypes cluster together naturally.
Shared phylotypes were identified by clustering all sequences in all samples at the 97% OTU level, then identifying which of these groups contained sequences that originated in multiple samples from the table linking OTUs to samples. Confidence values for the acquisition of shared phylotypes were established by 100 repetitions of subsampling with equal sequence numbers per sample.
Unless indicated in the text, all P-values were obtained after statistic tests using the Poisson model.
Acknowledgments
We thank M. Casellas, M. Gallart, C. Alastrue, D. Datta, M. Zehnsdorf, and M. Hummel for their technical assistance. We also thank J. Roca for helpful comments and critical reading of the manuscript. This work was supported in part by grant SAF 2007-64411 (Ministerio de Ciencia e Innovacion, Spain), and by the National Institutes of Health, and the Howard Hughes Medical Institute. Ciberehd is funded by the Instituto de Salud Carlos III (Spain).
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA020673.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.107987.110.
References
- Alpert C, Sczesny S, Gruhl B, Blaut M 2008. Long-term stability of the human gut microbiota in two different rat strains. Curr Issues Mol Biol 10: 17–24 [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77: 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI 2005. Host-bacterial mutualism in the human intestine. Science 307: 1915–1920 [DOI] [PubMed] [Google Scholar]
- Borody TJ, Warren EF, Leis S, Surace R, Ashman O 2003. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol 37: 42–47 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R 2010. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4: e6669 doi: 10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK 2009. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci 106: 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280 doi: 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK 2008. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2: 716–727 [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci 104: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Bourdet-Sicard R, Brandtzaeg P, Gill HS, McGuirk P, van Eden W, Versalovic J, Weinstock JV, Rook GA 2006. Mechanisms of disease: The hygiene hypothesis revisited. Nat Clin Pract Gastroenterol Hepatol 3: 275–284 [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers DM, Bast A, Vanhoutvin SA, Fischer MA, Kodde A, Troost FJ, Venema K, Brummer RJ 2009. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr 28: 88–93 [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1: 56–66 [DOI] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, and Sadowsky MJ 2009. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent clostridium difficile-associated diarrhea. J Clin Gastroenterol. 44: 354–360 [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. 2008. Evolution of mammals and their gut microbes. Science 320: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A 2006. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Lofmark S, Jernberg C, Jansson JK, Edlund C 2006. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother 58: 1160–1167 [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R 2005. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Varela E, Martinez C, Antolin M, Llopis M, Dore J, Giralt J, Guarner F, Malagelada JR 2008. The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. Am J Gastroenterol 103: 1754–1761 [DOI] [PubMed] [Google Scholar]
- Nancey S, Bienvenu J, Coffin B, Andre F, Descos L, Flourie B 2002. Butyrate strongly inhibits in vitro stimulated release of cytokines in blood. Dig Dis Sci 47: 921–928 [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO 2007. Development of the human infant intestinal microbiota. PLoS Biol 5: e177 doi: 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP 2009. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6: 639–641 [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127: 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F 2009. Therapeutic implications of manipulating and mining the microbiota. J Physiol 587: 4175–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J 2009. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videla S, Vilaseca J, Guarner F, Salas A, Treserra F, Crespo E, Antolin M, Malagelada JR 1994. Role of intestinal microflora in chronic inflammation and ulceration of the rat colon. Gut 35: 1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You DM, Franzos MA, Holman RP 2008. Successful treatment of fulminant Clostridium difficile infection with fecal bacteriotherapy. Ann Intern Med 148: 632–633 [DOI] [PubMed] [Google Scholar]