Abstract
Transcriptome analysis has important applications in many biological fields. However, assembling a transcriptome without a known reference remains a challenging task requiring algorithmic improvements. We present two methods for substantially improving transcriptome de novo assembly. The first method relies on the observation that the use of a single k-mer length by current de novo assemblers is suboptimal to assemble transcriptomes where the sequence coverage of transcripts is highly heterogeneous. We present the Multiple-k method in which various k-mer lengths are used for de novo transcriptome assembly. We demonstrate its good performance by assembling de novo a published next-generation transcriptome sequence data set of Aedes aegypti, using the existing genome to check the accuracy of our method. The second method relies on the use of a reference proteome to improve the de novo assembly. We developed the Scaffolding using Translation Mapping (STM) method that uses mapping against the closest available reference proteome for scaffolding contigs that map onto the same protein. In a controlled experiment using simulated data, we show that the STM method considerably improves the assembly, with few errors. We applied these two methods to assemble the transcriptome of the non-model catfish Loricaria gr. cataphracta. Using the Multiple-k and STM methods, the assembly increases in contiguity and in gene identification, showing that our methods clearly improve quality and can be widely used. The new methods were used to assemble successfully the transcripts of the core set of genes regulating tooth development in vertebrates, while classic de novo assembly failed.
Transcriptomic information is used in a wide range of biological studies and provides fundamental insights into biological processes and applications such as levels of gene expression (Torres et al. 2008), gene expression profiles after experimental treatments or infection (Hegedus et al. 2009), discovery of tissue biomarkers (Disset et al. 2009), cancer gene expression (Morrissy et al. 2009), gene discovery (Hahn et al. 2009), gene content (Reinhardt et al. 2009), and isolation of conserved ortholog genes for phylogenomic purposes (Hughes et al. 2006; Dunn et al. 2008), among others.
However, transcriptomic information is generally abundant only for model organisms on which international research effort and funding is concentrated, setting aside non-model organisms. This situation is drastically changing with the emergence and generalization of next-generation DNA sequencing technologies that tremendously reduce cost, labor, and time, providing the opportunity to conduct large-scale genomic projects at lower cost for non-model organisms.
The most economical next-generation sequencing technologies are those that generate short sequence reads, typically in the range of 30–100 bp, and are the method of choice for “re-sequencing” model organisms (e.g., the Illumina technology) (Porreca et al. 2007). In this case, the analysis is performed by mapping the short-reads onto the reference genome or transcriptome. This approach has recently been used for transcriptome profiling in a method called RNA-seq that is expected to allow major breakthroughs in transcriptome analysis (Mortazavi et al. 2008; Nagalakshmi et al. 2008; Wilhelm et al 2008; Wang et al. 2009; Montgomery et al. 2010).
However, de novo assemblies of sequences without a known reference using short reads have been considered difficult (Schuster 2008), and researchers working on non-model organisms have often turned to the more expensive longer sequence reads (250–450 bp) obtained by the 454 Life Sciences (Roche) technology (Margulies et al. 2005; e.g., Vera et al. 2008; Hale et al. 2009). However, the applicability of short-reads methods as an appropriate choice for de novo transcriptome assembly has recently received attention. By reassembling the transcriptome of a species with a known genome using a de novo assembler, Gibbons et al. (2009) have shown that short-reads can be of considerable utility for assembling transcriptomes of non-model organisms.
Despite the fast development of assemblers able to efficiently handle more and more reads (Zerbino and Birney 2008; Simpson et al. 2009), transcriptome assembly is still difficult. For instance, elongation of contigs is not only impeded by repeats or allelic variations but also by alternatively spliced transcripts. Moreover, while genomic sequencing coverage is generally uniform across the genome, transcriptome coverage is highly variable, depending on gene expression level, excluding the use of coverage information to resolve repeated motifs (Zerbino and Birney 2008). Therefore, the quality of a de novo transcriptome assembly is highly dependent on the user-defined sequence overlap length between two reads required to consider them as contiguous (referred as k-mer length). The best k-mer value for a given assembly depends on the sequencing depth, the read error rate, and the complexity of the genome/transcriptome to be assembled (Simpson et al. 2009). For transcriptome assembly, in which coverage is not uniform, using higher k-mer length will theoretically result in a more contiguous assembly of highly expressed transcripts. On the contrary, poorly expressed transcripts will be better assembled if lower k-mer lengths are used (Zerbino and Birney 2008). These theoretical expectations have been experimentally supported in a controlled de novo transcriptome assembly of a model organism (Gibbons et al. 2009). The choice of the k-mer length is then a subjective decision of whether to emphasize on transcript diversity by using a short k-mer length (that will lead to the assembly of numerous and highly fragmented transcript fragments), or to emphasize on contiguity by using a longer k-mer length (that will allow the recovery of longer transcript fragments but at the cost of a lower transcript diversity). Hence, in most cases, an intermediate k-mer length is chosen to reach a compromise between these two extremes. Therefore, an approach for de novo transcriptome assembly that takes advantage of the assembly performances of various k-mer lengths is highly desirable.
The analysis of genomes or transcriptomes of non-model organisms can be enhanced by performing comparisons with the genome of closely related model organisms. For instance, algorithms have been proposed for boosting the assembly of bacterial genomes using available genomes of related species (Salzberg et al. 2008). In eukaryotes, the transcriptome of a non-model plant (Pachycladon enysii)—which has recently diverged from the reference Arabidopsis thaliana (7–10 million yr ago, Mya)—was analyzed using a combination of classic read mapping against the reference transcriptome, de novo assembly, and contig mapping against the reference genome using BLAST (Collins et al. 2008). Likewise, the brain EST data set of the social wasp Polistes metricus, which was generated by next-generation 454 sequencing, was successfully analyzed by comparing it to the complete genome of the honey bee, from which it diverged 100 to 150 Mya (Toth et al. 2007). An unexplored extension of these comparisons is the use of a closely related model organism to serve as template for improving the assembly of the transcriptome of a non-model organism. However, at the nucleotide level this approach is limited to those non-model organisms that possess a very close relative with a complete genome. This limitation is due to the increasing amount of nucleotide differences between ortholog genes with increasing evolutionary distance, which will rapidly lead to the absence of good-quality matches between the two species. However, differences in amino acid sequence accumulate more slowly than nucleotide differences with increasing evolutionary distance, so comparing sequence translations against a reference proteome might be a promising approach to improve the assembly of the coding fraction of the transcriptome of non-model organisms, even if the reference model is distantly related.
Here we present two methods for improving de novo transcriptome assembly, which answer the expectations presented above. The principle of the first method is to perform multiple assemblies with various k-mer lengths and to retain the best part of each one to form the final assembly. In the second method, we assemble the coding contigs into scaffolds by mapping their translation on a distant reference proteome. The pipeline implementing this method can be applied to the results of any de novo transcriptome assembly as long as a reference genome or transcriptome of an evolutionarily linked species is available. We then validated the efficiency and the accuracy of our two methods by using simulated and real data from species with a known genome. To demonstrate the efficiency of both methods on real data from non-model organisms, we applied them in assembling reads from a next-generation short-read sequencing experiment that we performed on the transcriptome of the Neotropical catfish Loricaria gr. cataphracta. We demonstrate the practical efficiency of these methods by their success in recovering the full set of transcripts belonging to the gene network regulating dental development, while classic methods failed.
Results
De novo transcriptome assembly with multiple k-mer values
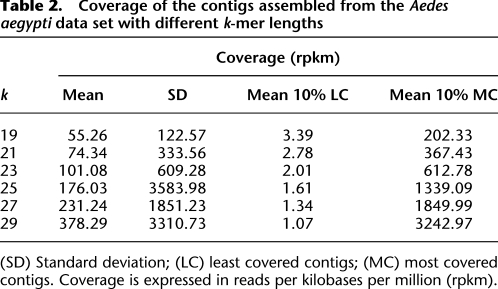
The basic assumption of this new method is that different k-mers will allow the assembly of transcripts with different abundances. To verify this assumption, we first assembled the recently published next-generation transcriptome sequence data set of the yellow fever mosquito, Aedes aegypti (Gibbons et al. 2009), with different k-mer values (Table 1). We then estimated the abundance of the transcripts assembled with the different k-mer values based on their read coverage (digital gene expression). This analysis was also conducted on a simulated 35-bp RNA-seq data set based on the set of zebrafish cDNA from Ensembl (Supplemental Table S1). As expected, the average coverage of assembled contigs increases with increasing k-mer values on both the real (Table 2) and the simulated (Supplemental Table S2) data sets. However, it is worth noting that the standard deviation of transcript abundances also increases with higher k-mer values. Hence, low k-mer values allow the assembly of numerous transcripts with relatively low abundance, while larger values allow the assembly of a lower number of transcripts but with a much larger range of abundances. Given the different characteristics of the transcripts assembled with different k-mer lengths, combining the results obtained with various k-mer lengths into a final assembly seems to be a promising way of improving de novo assembly of sequences with very variable coverage levels as is the case for nonstandardized transcriptomes.
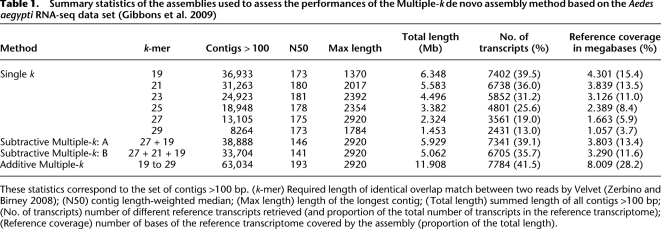
Table 1.
Summary statistics of the assemblies used to assess the performances of the Multiple-k de novo assembly method based on the Aedes aegypti RNA-seq data set (Gibbons et al. 2009)
These statistics correspond to the set of contigs >100 bp. (k-mer) Required length of identical overlap match between two reads by Velvet (Zerbino and Birney 2008); (N50) contig length-weighted median; (Max length) length of the longest contig; (Total length) summed length of all contigs >100 bp; (No. of transcripts) number of different reference transcripts retrieved (and proportion of the total number of transcripts in the reference transcriptome); (Reference coverage) number of bases of the reference transcriptome covered by the assembly (proportion of the total length).
Table 2.
Coverage of the contigs assembled from the Aedes aegypti data set with different k-mer lengths
(SD) Standard deviation; (LC) least covered contigs; (MC) most covered contigs. Coverage is expressed in reads per kilobases per million (rpkm).
To take advantage of the assembling properties of different k-mer lengths, we have designed two alternative methods of de novo assembly that use multiple k-values. In the first place, we designed the “subtractive Multiple-k” method that starts the assembly with a high k-mer length and then uses the nonassembled reads of this assembly to perform another assembly with a smaller k-mer value. This procedure can be reiterated. The second method, which we called the “additive Multiple-k” method, pools the contigs obtained with different k-mer lengths and subsequently removes redundant contigs (see Methods). We investigated the performances of the two alternative methods using the transcriptome of Ae. aegypti (Gibbons et al. 2009), for which the complete genome is known (Nene et al. 2007). Hence, it is possible to evaluate the number of transcripts recovered by the different assembly methods, as well as the proportion of the reference transcriptome covered by the assembled contigs. We also compared the results obtained with these new approaches to the optimum assembly obtained with a single-k. We first carried out Velvet assemblies using k-mer lengths from 19 to 29 and selected the assembly obtained with k = 21 since it gives a good compromise between the number of contigs and their length (as was already determined by Gibbons et al. 2009).
We tested two assembly variants (A and B) of the subtractive Multiple-k method: assembly A with two k-values (k = 27 followed by k = 19, which are the two most extreme k-values still displaying interesting statistics; see Table 1), and assembly B with three k-values (k = 27, then k = 21, and finally k = 19). As can be seen from Table 1, the subtractive Multiple-k method does not provide a clear improvement to the Velvet de novo assembly (the assembly shows the lowest N50 and a lower number of transcripts recovered than the single-k method) and will not be discussed further.
The additive Multiple-k method was performed with all the k-values between 19 and 29. The final assembly statistics indicate that this approach outperforms all others (Table 1). The number of contigs >100 bp and total length are both doubled as compared to the single-k Velvet assembly. Interestingly, this marked increase is accompanied by a higher N50 (median length-weighted contig length) (Zerbino and Birney 2008), indicating a substantial improvement in contiguity. Furthermore, the reference transcripts recovered are more numerous, reaching almost 40% of the reference transcriptome, and base coverage of reference transcripts is doubled as compared to the single-k Velvet assembly.
It can be noted that the number of transcripts identified with the Multiple-k method is similar to the number of transcripts identified with k = 19, but that the number of contigs is much higher in the former. This is due to the fact that the Multiple-k method pools contigs assembled with various k-mer lengths and covering different parts of each transcript. Hence, for a given transcript, more sequence information is available in the Multiple-k assembly. This situation may be explained by splice variants with different abundances resulting in a heterogeneous number of exons, which are therefore assembled with different k-mer lengths. Variation in the coverage within a transcript may also be due to regions more difficult to reverse-transcribe, amplify, or sequence (like repeated motifs or strong secondary structures), to genes with alternative transcription start or stop sites or to stochasticity.
When comparing the set of transcripts identified with the single-k Velvet assembly and the additive Multiple-k method, we identified 6697 transcripts in common between the two methods, while 1090 new transcripts were found only with the latter. Furthermore, the contigs belonging to this common set (30,749 contigs) were longer when they were assembled with the Multiple-k method (N50 = 218 vs. 180; maximum length = 2920 vs. 2017; total length = 6.3 vs. 5.5 Mb for the additive Multiple-k method and single-k Velvet assembly, respectively). Hence, the additive Multiple-k method does not only improve the transcript diversity of the assembly but also increases contiguity.
Scaffolding using translation mapping (STM)
De novo transcriptome assemblies may be substantially improved by the addition of a scaffolding step where the contigs belonging to a single transcript are ordered, orientated, and assembled. This scaffolding step is generally performed using paired-ends libraries, but the generation of such libraries doubles the cost of a sequencing experiment. An innovative way of scaffolding without incorporating additional sequence is to use the proteome of a related species as a reference to assemble contigs belonging to a same coding sequence. We have designed a method called “Scaffolding using Translation Mapping” (STM) that exploits the fact that, by translating contigs into amino acid sequences, it is possible to search for orthologous regions in a reference proteome, even when it belongs to a distantly related organism. In this way, all translated contigs matching a same reference protein can be assembled into a scaffold, provided that they pass some accuracy checks (a diagrammatic representation of the pipeline is presented in Fig. 1). In case reads are long enough (typically longer than 70 bp), we developed two flavors for this method: with or without the incorporation of the orphan reads not included in the initial assembly, named STM+ and STM−, respectively.
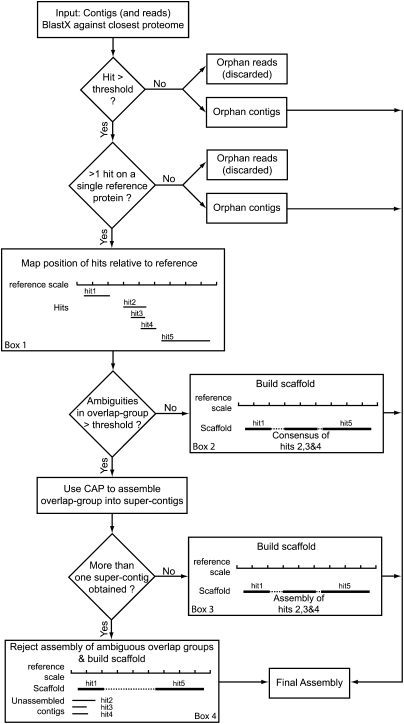
Figure 1.
Diagrammatic representation of the STM method. This pipeline can either use only contigs (STM- method) or, if reads are long enough, contigs plus unassembled reads (STM+ method). These contigs/reads are mapped on the reference proteome using BLASTX. When a contig has no significant hit or is the only one to map on a given reference protein, it cannot be further assembled and is directed into the final assembly. When there are several hits on a same reference protein (Box 1: an example with 5 hits) their relative positions are recorded on the reference scale. If there is an overlap in the positioning of several hits (here hits 2, 3, and 4 form an overlap group), their consensus sequence is computed, and when the number of ambiguities is below a user-defined threshold, the consensus is accepted and a scaffold is constructed (Box 2: dashed line represents N's added to join the contigs). Else, the consensus is rejected and the contigs of the overlap group are assembled using CAP. If the result of this assembly step is a single “super-contig,” it is accepted and a scaffold is constructed (Box 3). If more than one super-contig is obtained (Box 4), the overlap group assembly is rejected and the contigs are placed as independent transcripts in the final assembly. If present, the other nonoverlapping hits (or nonambiguous overlap groups) are joined into a scaffold, which is incorporated into the final assembly.
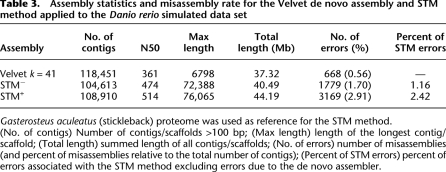
In order to assess the accuracy of our method, we have tested it using a simulated de novo transcriptome assembly of the zebrafish (Danio rerio), a model organism with a highly studied and richly annotated genome (for simulation details, see Methods). In this way, we could estimate the number of misassemblies by comparing the scaffolds obtained to the original transcriptome using BLASTN (Altschul et al. 1997). We considered as mis-assemblies all scaffolds that did not match perfectly an existing transcript of the original transcriptome. To identify the error rate that can be attributed solely to the STM method, we first determined the amount of misassembled contigs due to the Velvet de novo assembler via BLASTN against the initial transcriptome, which resulted in 668 erroneous contigs (0.56% of the total) (Table 3). We then performed the STM method on the zebrafish de novo assembly using the proteome of the stickleback (Gasterosteus aculeatus) as a reference. The results of the STM method show a clear improvement of the transcriptome assembly, either with STM+ or STM− (Table 3). The number of contigs >100 bp was decreased by ∼10% coupled to a marked increase in N50 of 31% and 42% for STM− and STM+, respectively. The STM method also leads to a much longer maximum scaffold length and a greater total length, especially for STM+, which globally shows better assembly statistics than STM− (Table 3). Nevertheless, the assembly error rate specific to STM− is 1.16% (1.70% when including the error rate of the de novo assembly), while it is 2.42% for the STM+ (2.91% when including the de novo assembly error rate). This test indicates that STM+ performs the best yet with a slightly higher error rate than STM−, which also enhances substantially the assembly with minor error risk.
Table 3.
Assembly statistics and misassembly rate for the Velvet de novo assembly and STM method applied to the Danio rerio simulated data set
Gasterosteus aculeatus (stickleback) proteome was used as reference for the STM method.
(No. of contigs) Number of contigs/scaffolds >100 bp; (Max length) length of the longest contig/scaffold; (Total length) summed length of all contigs/scaffolds; (No. of errors) number of misassemblies (and percent of misassemblies relative to the total number of contigs); (Percent of STM errors) percent of errors associated with the STM method excluding errors due to the de novo assembler.
We then investigated whether the efficiency of this scaffolding method varied depending on the characteristics of the transcripts being assembled. First, we classified the contigs that were most efficiently scaffolded with our new method (a set of 38 transcripts showing a 20-fold increase or more of their length after scaffolding) according to their Gene Ontology, showing no particular bias in GO categories (Supplemental Table S3). Then, to check whether the assembly efficiency depended on the known transcript length or on its abundance, we measured the correlation between the contigs' length increase after scaffolding and the real transcript length (as given in the reference transcriptome), and its read coverage. Both these correlations were quite low (Spearman rank correlation coefficient of 0.1218 and −0.1336, respectively), suggesting a lack of strong effect of transcript length or abundance on the STM method's efficiency.
Optimized de novo transcriptome assembly of the catfish Loricaria
Having demonstrated in controlled conditions the accuracy and high performance of the two new methods for de novo transcriptome assembly, we then used them to assemble the transcriptome of a non-model organism: the Neotropical catfish L. gr. cataphracta. The genus Loricaria belongs to the catfish family Loricariidae, the most species-rich family of freshwater fishes in the Neotropics. All loricariids share the presence of extra-oral and post-cranial denticles. These denticles develop in the same way as do teeth, and similar morphogenetic mechanisms underlying their formation may be inferred (Sire 2001).
Deciphering the genetic control of the development of loricariids' ectopic teeth may contribute to the understanding of teeth formation and regeneration in vertebrates and will certainly shed light on the evolutionary implication resulting from bearing such denticles, especially on the great species diversification of loricariids. To this aim, we sequenced and assembled the transcriptome of L. gr. cataphracta embryos to reconstruct the sequences of the transcripts known to control tooth development. Recently, the genes forming the core dental regulatory network have been identified and represent a conserved set of 14 genes that provides the molecular machinery and developmental constraints for all teeth, either jaw or pharyngeal teeth (Fraser et al. 2009). Out of these 14 genes, five are duplicated in teleosts, resulting in a set of 19 genes.
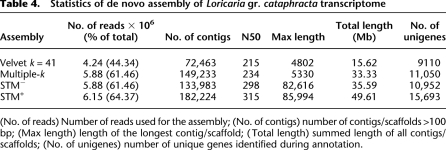
The transcriptome of full embryos of L. gr. cataphracta—from stages ranging from end of gastrula until hatching—was sequenced with 71-bp single-end reads on the Illumina Genome Analyzer II platform. One sequencing lane was used and resulted in 9.56 million reads. Reads were first assembled using Velvet and a range of single k-mer lengths. For this step we kept the assembly obtained with k = 41 as it gave a good compromise between the number of contigs obtained, the N50, and the number of unigenes recovered (Supplemental Table S4). Next, the additive Multiple-k method was performed, pooling the assemblies obtained with values of k = 37, 41, 45, 49, 53, 57, and 61. Summary statistics (Table 4) show that the additive Multiple-k assembly makes use of 38.7% more reads than the single-k Velvet assembly. It also displays twice as many contigs >100 bp and a higher N50, indicative of an increased contiguity. All other assembly statistics are also markedly improved. In particular, the additive Multiple-k method allowed the identification of about 2000 additional genes, representing an increase of >20% as compared to the single-k Velvet assembly (Table 4).
Table 4.
Statistics of de novo assembly of Loricaria gr. cataphracta transcriptome
(No. of reads) Number of reads used for the assembly; (No. of contigs) number of contigs/scaffolds >100 bp; (Max length) length of the longest contig/scaffold; (Total length) summed length of all contigs/scaffolds; (No. of unigenes) number of unique genes identified during annotation.
We implemented the two flavors of the STM method to the additive Multiple-k de novo assembly. STM− was performed with a minimum contig length threshold of 73 bp. Its resulting summary statistics indicated a reduction in the number of contigs/scaffolds >100 bp due to the assembly of some of them into scaffolds. Out of the 166,490 contigs >73 bp, 23,675 (14.2%) were successfully incorporated into 6613 scaffolds. As expected, a substantial increase is observed for N50 (+27.4%), and particularly for the maximum contig/scaffold length, which reaches >82 kb (Table 4). However, the number of different transcripts has slightly decreased (−0.88%). This decrease is probably due to the few instances where two or more contigs belonging to different transcripts were erroneously joined into a single scaffold.
The STM+ method integrated 4.6% more reads and further improved the assembly as indicated by the summary statistics (Table 4). Notably, this led to a marked increase in the number of unigenes identified (+72.3% compared to the single-k Velvet de novo assembly, +42% compared to the Multiple-k method). Using the STM+ method, we recovered 246 transcripts longer than the longest transcript obtained without it. The longest transcript identified was a transcript coding for the titin b (ttnb), and the size distribution of these 246 long transcripts is presented in Supplemental Figure S1.
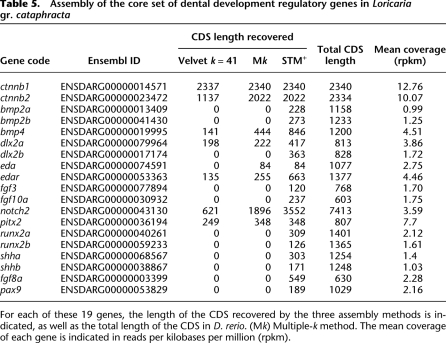
We then examined whether our new methods allowed a better assembly of the 19 genes representing the core set of dental development regulatory genes. Using classic de novo transcriptome assembly methods (Velvet with k = 41), we were able to retrieve transcript fragments of seven out of the 19 genes. By implementing the Multiple-k method, we identified an additional transcript belonging to the set of tooth development genes, and the sequence length of the seven transcript fragments already recovered was in most cases substantially increased (Table 5). Finally, the use of the Multiple-k together with the STM+ methods resulted in the assembly of transcript fragments of the full set of 19 genes, and with a marked increase in sequence length for those transcripts recovered earlier (Table 5).
Table 5.
Assembly of the core set of dental development regulatory genes in Loricaria gr. cataphracta
For each of these 19 genes, the length of the CDS recovered by the three assembly methods is indicated, as well as the total length of the CDS in D. rerio. (Mk) Multiple-k method. The mean coverage of each gene is indicated in reads per kilobases per million (rpkm).
Discussion
Improving de novo transcriptome assembly
The emergence of next-generation sequencing technologies has impressively enlarged the realm of transcriptomic analyses. For instance, these new technologies have been efficiently employed in the discovery of new genes (Hahn et al. 2009), the development of new tissue-specific or cancer biomarkers (Levin et al. 2009; Morrissy et al. 2009), the isolation of fast-evolving genes (Montoya-Burgos et al. 2010), the detection of new alternative splice variants (Carninci 2008; Gibbons et al. 2009; Tang et al. 2009), allele-specific gene expression (Main et al. 2009), SNP discovery in genes (Barbazuk et al. 2007), or epigenetic gene regulation (Elling and Deng 2009). These advances and future ones rely, however, on the size and quality of the transcriptome assembly.
In this study, we present methods to improve both the quantity and the quality of the information that can be extracted from a de novo transcriptome assembly. By taking advantage of the assembling properties of many different k-mer lengths, the Multiple-k method is able to incorporate the best parts (i.e., the more contiguous) of each assembly into the final assembly. We have demonstrated that this strategy leads to a considerable increase in both contig contiguity (by keeping long contigs of highly expressed genes assembled with high k-values) and in transcript diversity (by keeping contigs of poorly expressed genes that only assemble with low k-values). Furthermore, the use of this method avoids the subjective selection of a single k-mer length.
The second methodology we developed, the STM method, uses the information of a reference proteome to accurately join contigs into scaffolds. Simulated data demonstrated that this method efficiently joins multiple transcript fragments that are part of a single gene, providing new and valuable information on the order and the orientation of these fragments along the original transcript.
Importantly, the sequential application of these two methods to the new next-generation short-read data set of the catfish L. gr. cataphracta demonstrates their utility in improving the de novo assembly of a non-model organism transcriptome. First, the additive Multiple-k method makes use of more sequence information from the original data set than a single-k Velvet de novo assembly; the number of reads used is increased by 38.7%. This, together with an 8.8% increase in contiguity, leads to the identification of ∼21% more unigenes. A further increase in contiguity is observed when using the STM− method. The STM+ method, which includes orphan reads into the procedure (4.6% more reads used), leads to a remarkable increase in the number of unigenes identified in the Loricaria transcriptome (+72%, as compared to the single-k Velvet de novo assembly), which corresponds to >56% of the zebrafish gene set Zv8 (Ensembl GeneBuild). In the single-k Velvet de novo assembly, only 33% of these were recovered.
The number of unigenes identified using the different assembly methods is illustrated in a Venn diagram (Supplemental Fig. S2) and shows that 8986 unigenes were identified by all the methods tested here. However, an analysis of the contigs assigned to this set of shared unigenes (Supplemental Table S5) indicates that the new methods allow a more contiguous assembly of these contigs. Hence, these methods not only allow the identification of a higher number of unigenes, but also allow a better assembly of the transcripts belonging to the unigenes already identified using a single-k Velvet assembly.
The increase in the number of unigenes identified is probably not artifactual since the error rate of the STM method can likely not be higher in this experiment than the one determined in the assembly of the simulated zebrafish transcriptome data set using the proteome of the stickleback as reference; these two model fish species diverged ∼300 Mya, while Loricaria and the zebrafish have diverged more recently, ∼150 Mya (Steinke et al. 2006). Moreover, the expected small amount of misassemblies (scaffolding two or more contigs belonging to different transcripts) will only lead to an underestimate of the number of unigenes. Indeed, when using the best BLASTX hit to annotate a “chimeric” scaffold, a single gene identification will be obtained, while two or more would have been obtained with the unassembled contigs. Hence, when the primary goal of a de novo transcriptome assembly experiment is gene identification, the inclusion of reads (>70 bp) with the STM+ method is highly recommended.
The recovery of more diverse transcripts with higher contiguity is also demonstrated by the successful recovery of all the core set of dental development regulatory genes using a combination of the additive Multiple-k and STM+ methods, while less than half of them were recovered using single-k Velvet de novo assembly. Furthermore, this analysis demonstrates once again the interest of our methods to assemble transcripts with low abundance since the single-k method only assembled the transcripts with the highest sequence coverage (Table 5).
Application requirements
The Multiple-k method can be implemented in conjunction with any assembler that uses the k-mer length parameter, such as those based on de Bruijn graphs representation of sequence neighborhoods, as initially implemented in this field by Pevzner and coworkers (Pevzner and Tang 2001; Pevzner et al. 2001). Current assemblers using this graph-based approach include ALLPATHS (Butler et al. 2008) and ALLPATHS 2 (MacCallum et al. 2009), Edena (Hernandez et al. 2008), Velvet (Zerbino and Birney 2008), EULER-SR (Chaisson and Pevzner 2008), and ABySS (Simpson et al. 2009).
As to the STM method, it works using the output data set of the assembly and is therefore independent of the assembler used. This makes it of general use for de novo transcriptome assembly.
STM method limitations
The STM method relies on the assumption that the gene set of the reference proteome, which will serve as a template for joining contigs into scaffolds, is sufficiently similar in terms of gene composition, ortholog gene length, or multigene families, to the gene set of the transcriptome under assembly. Large differences may reduce the number of scaffolds or lead to an increase of misassemblies. The errors introduced by this method arise mainly when two contigs from different transcripts map on a single reference transcript. This can happen when recent paralogs or pseudogenes are present or when the reference proteome is not complete enough, particularly for multigene families for which not all members are present in the reference proteome. The error rate can be reduced by increasing the similarity cutoff in the STM procedure at the cost of a lower scaffolding efficiency. Hence, the choice of a similarity cutoff is a trade-off between accuracy and efficiency.
In this respect, a parallel can be drawn between the STM method and gene orthology prediction, for which substantial literature exists. EST databases are being used for predicting gene orthology among species, particularly for phylogenomic purposes (e.g., Burki et al. 2008; Dunn et al. 2008). However, it has been recently shown that ortholog prediction accuracy is significantly higher when at least one of the two transcriptomes compared is complete, and that comparing two partial transcriptomes results in many more false-positive predictions and in more unpredicted true orthologs (Gibbons et al. 2009). Interestingly, this same study showed that although the amount of predicted orthologs decreases with increasing evolutionary distance, the prediction accuracy remains the same. This observation is promising as it may well hold true in the context of the STM method for improving the assembly of coding parts of a non-model transcriptome, as suggested by the low error rate associated with this method, even when using the proteome of the stickleback to reconstruct the zebrafish transcriptome, two species that diverged 290–330 Mya (Steinke et al. 2006; Yamanoue et al. 2006).
In the future, the STM method will not only benefit from the promise of longer and more numerous reads resulting from next-generation sequencing technologies, but also from both the improvement of current model species transcriptomes/proteomes and the fast rate of development of new model organisms and their transcriptomes/proteomes.
Methods
Multiple-k method
As no optimal k-mer length exists for any de novo transcriptome assembly, we designed and investigated two procedures to combine the best assembly information obtained with different k-mer lengths into a final assembly. The first method consists in assembling the set of reads using a high k-value so that highly expressed genes are best assembled. The reads used in this initial assembly are then discarded, and a new assembly is performed with the remaining reads and using a lower k-value so that genes with lower expression levels are well assembled. These steps can be repeated one or more times using decreasing k-values. The contigs of the different assemblies are then pooled to form the final assembly. We called this approach the “subtractive Multiple-k” method. In the second method, the reads used in the assembly with a high k-value are not discarded before running the subsequent assembly with a lower k; each assembly uses the total set of reads. Some contigs will appear in two or more assemblies introducing redundancy. We used CD-HIT-EST (Li and Godzik 2006) to remove redundancy and retain the longest possible contigs; the full set of contigs is mapped against itself. The short-redundant contigs are removed, and the remaining contigs of the pool of assemblies compose the final assembly. We called this procedure the “additive Multiple-k” method.
Scaffolding using translation mapping (STM)
The bioinformatics pipeline for building scaffolds based on contig and read translations is diagrammed in Figure 1. Subsequent to a de novo transcriptome assembly, contigs and unassembled (orphan) reads longer than a given threshold are simultaneously translated and “blasted” against a reference proteome using BLASTX. The threshold size should be long enough to potentially result in sufficiently good BLASTX E-values (we used a threshold size of 71 bp, giving translations of 23 amino acids). BLASTX results are parsed to retain only good quality hits; the criteria we used are contig coverage >90%, identity >60%, and E-value ≤ 10−5. The contigs with no good BLASTX hit, or orphan contigs, are directly placed into the final assembly data set. If reads (longer than the threshold size) were included in the procedure, those that showed low-quality BLASTX hits are discarded.
The BLASTX results are parsed to retrieve the coding strand and mapping position of the contigs/reads on the reference protein. If only one contig/read maps on a given reference protein, it cannot be further assembled and is directly included in the final assembly (for contigs) or discarded (for reads). When multiple contigs/reads map on a same reference protein, their relative position is set according to their place along the sequence reference in nucleotide coordinates (termed “reference scale” in Fig. 1). The contigs/reads are then joined to form a scaffold, with N's filling the spaces between them. This way of proceeding ensures that the reading frame is maintained. Several contigs/reads belonging to the same scaffold may overlap, and sequence differences may exist in the overlapping regions. The overlapping contigs/reads, called overlap groups, are therefore checked for the presence of minor or major sequence differences at each position by computing a majority rule consensus sequence (here the majority rule parameter was set to 75%). Minor differences, which may represent allelic variations or sequencing errors, will be resolved in the consensus, and the scaffold is built by joining the various overlap groups. Ambiguous bases (N) will appear in the consensus when major sequence discrepancies exist at a given position. If ambiguous positions cover <1% of the consensus sequence length of the overlap group, they are still considered as allelic variations or sequencing errors. Else, when >1% of the consensus sequence length is composed by N, which may result from the misassembly of splice variants, or of transcripts displaying sequence affinities, or due to indels in the reference sequence relative to the transcript being assembled, then the overlap group is examined for discerning among the various cases. The assembler CAP (Huang 1992) is used to reassemble the sequences composing the overlap group, without using the positioning on the reference sequence. This realignment resolves instances where indels were the cause of the problem; the scaffold is thus assembled and included in the final assembly data set. If the problem persists, then the sequences composing the overlap group are separated and placed in the final assembly.
Validation of Multiple-k and STM methods
To investigate and test the performances of our two methods, we analyzed two independent data sets, one based on real data and one based on simulated data. To test the Multiple-k assembly method, we used the Ae. aegypti next-generation short-reads (36 bp) data set recently published by Gibbons et al. (2009), generated from the same strain as the one used to sequence the complete genome (Nene et al. 2007). This data set was subjected to de novo assembly using Velvet v0.7.59 (Zerbino and Birney 2008) and with k-mer lengths of 19 to 29. Unless otherwise specified, the assembly statistics were taken from the Velvet output file. We then applied the two versions of the Multiple-k method to this data set and evaluated their efficiency. As the Multiple-k method is not aimed at assembling reads into contigs but rather uses the contigs constructed by a de novo assembler under different k-mer lengths, we did not evaluate the misassembly rate, which depends on the assembler used. We rather determined the improvements by looking at the assembly statistics and the number of reference transcripts recovered as compared to the single-k Velvet assembly (obtained with the optimal k-value). The number of reference transcripts recovered was calculated by comparing the resulting contigs of the de novo assembly to the Ae. aegypti reference transcriptome (Nene et al. 2007) using BLASTN. We considered as being correctly identified the hits covering at least 95% of the query sequence and having at least 99% identity with the reference transcript. We estimated the number of bases of the reference transcriptome covered by our assembly by summing the lengths of these good hits.
To investigate the behavior of the Multiple-k method, we conducted the same analyses on a simulated RNA-seq data set. First, we simulated a transcriptome from the Ensembl set of D. rerio cDNA (32,337 transcripts including splice variants). Different relative abundances were randomly assigned to each of these transcripts to mimic the variation in gene expression level observed in a real data set (the abundance profile was set according to the distribution of D. rerio ESTs density found in the Unigene database). This simulated transcriptome contained a total of 317,272 transcripts for a total length of 514,277,767 bp. An RNA-seq experiment was then simulated from this transcriptome. We generated 10 million 35-bp reads in a shotgun process using the simreads program of the Rmap package (Smith et al. 2009) and applied an error rate of 1% to mimic sequencing errors.
Similarly, to test the accuracy of the STM method in highly controlled conditions, we simulated a simplified next-generation sequencing experiment of the zebrafish coding transcriptome (32,337 coding transcripts, representing 51,837,753 bp) by generating 4 million random single-end reads of 76 bp in size (representing an ∼6× coverage of the transcriptome with homogeneous gene expression level). The simulation was performed with simreads of the Rmap package (Smith et al. 2009). This data set was first subjected to a Velvet de novo assembly with an arbitrary k-mer length of 41. It was then used to determine the scaffolding misassembly rate of the STM method. We first calculated the error rate due to the de novo assembler, and then due to the STM method, by comparing the contigs assembled to the reference transcriptome of D. rerio (the same from which the reads were simulated), using the same procedure as described for the Multiple-k method.
The scripts implementing the Multiple-k and STM methods are available in the Supplemental Material and can also be downloaded from http://www.surget-groba.ch/downloads/stm.tar.gz.
Quantification of transcripts from the RNA-seq experiments
To quantify the abundance of transcripts assembled in both Ae. aegyptii and L. gr. cataphracta, we mapped the reads from the RNA-seq experiments onto the assembled contigs using Maq v0.7.1 (Li et al. 2008). Reads mapping with a quality below 20 were discarded, and the number of reads mapping on a given transcript were corrected by the transcript size and the total number of reads to obtain the number of “reads per kilobase per million” (rpkm).
Illumina sequencing and de novo assembly of Loricaria transcriptome
To test our methods in real conditions, we conducted a complete experiment of next-generation transcriptome sequencing and de novo assembly using our methods for a non-model organism, the catfish L. gr. cataphracta.
Total RNA was extracted from fresh L. gr. cataphracta full embryos of 2–8 d post-fecundation (stages ranging from end of gastrula to hatching) using TRIzol reagent (GIBCO). After quantification and quality verification of the total RNA, mRNA was isolated using the mRNA Isolation Kit (Roche Diagnostics) according to the manufacturer's instructions. We used the “mRNA-SEQ” Transcriptome Shotgun procedure and Kit (Roche) for preparing the cDNA for Illumina sequencing. The sequencing experiment was performed by the company Fasteris SA. First, 1 μg of embryo mRNA was zinc-fragmented to reach sizes ranging from 200 to 500 bases. First-strand cDNA was synthesized using random hexamer primers. Second-strand synthesis was performed by treatment with RNase H and DNA polymerase I for strand elongation, according to the manufacturer's instructions. Double-strand cDNA ends were repaired using T4 DNA polymerase, Large (Klenow) fragment of DNA polymerase I, and T4 polynucleotide kinase in the presence of ATP and the four dNTPs. After purification, adenine nucleotides were added at the 3′ side of the blunt-ended DNA fragments with Klenow fragment (exo−) and then purified. Forked Illumina adapters were ligated to the cDNA overnight at 15°C, using T4 DNA ligase in the presence of ATP, and then purified. The cDNA–adapter complexes were loaded onto a well-resolved 3% agarose TBE gel, and complexes of 250–350 bp in size were extracted by excising the corresponding region of the gel and purifying the complexes with the High Pure PCR Product Purification Kit (Roche). Finally, the cDNA–adapter complexes were PCR-amplified for 15 cycles. The prepared cDNA library was sequenced with 71-bp single-end reads on one lane of the Illumina Genome Analyzer II platform and processed using the Illumina Pipeline Software v1.4.0, according to the manufacturer's instructions (Illumina). The reads data set was deposited at the NCBI Sequence Read Archive (SRA) under accession number SRA010189.
The 9.56 million reads of 71 bp that were generated were subjected to a series of de novo assemblies using k-mer lengths ranging from 37 to 61. The summary statistics were used to determine the optimal k-mer length (Supplemental Table S4). This data set was then subjected sequentially to the additive Multiple-k and then either to the STM− or the STM+ method.
In this experiment, we estimated the number of different genes recovered by comparing the resulting contigs to the proteome of D. rerio, using BLASTX, and kept only hits with an E-value ≤10−10. We then counted the number of distinct genes (unigenes) identified.
Public transcriptomes and proteomes
Full public transcriptomes or proteomes used for this study were retrieved from the following databases: Aedes aegypti transcriptome (http://www.vectorbase.org); Gasterosteus aculeatus proteome (http://www.ensembl.org/Gasterosteus_aculeatus); and Danio rerio transcriptome and proteome (http://www.ensembl.org/Danio_rerio).
Acknowledgments
We thank Antonis Rokas for sharing with us his Ae. aegypti next-generation short-read data set. We thank Ilham Bahechar for her help with laboratory work, and Marta Burgos and Alison R. Davis for revising the manuscript. We thank Patrick Descombes, Laurent Farinelli, and three anonymous reviewers for their useful discussions and comments. This work was supported by funds from the Canton de Genève, the Swiss National Research Fund (Number 3100A0-122303/1), and the G & A Claraz Foundation.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA010189. Source code for the Multiple-k and STM methods can be downloaded from http://www.surget-groba.ch/downloads/stm.tar.gz.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103846.109.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS 2007. SNP discovery via 454 transcriptome sequencing. Plant J 51: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J 2008. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett 4: 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J, MacCallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, Nusbaum C, Jaffe DB 2008. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res 18: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P 2008. Hunting hidden transcripts. Nat Methods 5: 587–589 [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA 2008. Short read fragment assembly of bacterial genomes. Genome Res 18: 324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LJ, Biggs PJ, Voelckel C, Joly S 2008. An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform 21: 3–14 [PubMed] [Google Scholar]
- Disset A, Cheval L, Soutourina O, Duong Van Huyen JP, Li G, Genin C, Tostain J, Loupy A, Doucet A, Rajerison R 2009. Tissue compartment analysis for biomarker discovery by gene expression profiling. PLoS ONE 4: e7779 doi: 10.1371/journal.pone.0007779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749 [DOI] [PubMed] [Google Scholar]
- Elling AA, Deng XW 2009. Next-generation sequencing reveals complex relationships between the epigenome and transcriptome in maize. Plant Signal Behav 4: 760–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT 2009. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol 7: 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A 2009. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol 26: 2731–2744 [DOI] [PubMed] [Google Scholar]
- Hahn DA, Ragland GJ, Shoemaker DD, Denlinger DL 2009. Gene discovery using massively parallel pyrosequencing to develop ESTs for the flesh fly Sarcophaga crassipalpis. BMC Genomics 10: 234 doi: 10.1186/1471-2164-10-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MC, McCormick CR, Jackson JR, DeWoody JA 2009. Next-generation pyrosequencing of gonad transcriptomes in the polyploid lake sturgeon (Acipenser fulvescens): The relative merits of normalization and rarefaction in gene discovery. BMC Genomics 10: 203 doi: 10.1186/1471-2164-10-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus Z, Zakrzewska A, Agoston VC, Ordas A, Racz P, Mink M, Spaink HP, Meijer AH 2009. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol 46: 2918–2930 [DOI] [PubMed] [Google Scholar]
- Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J 2008. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res 18: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ 1992. A contig assembly program based on sensitive detection of fragment overlaps. Genomics 14: 18–25 [DOI] [PubMed] [Google Scholar]
- Hughes J, Longhorn SJ, Papadopoulou A, Theodorides K, de Riva A, Mejia-Chang M, Foster PG, Vogler AP 2006. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles). Mol Biol Evol 23: 268–278 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway LA, Gnirke A 2009. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol 10: R115 doi: 10.1186/gb-2009-10-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A 2006. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter IA, Gnirke A, Malek J, McKernan K, Ranade S, Terrance PS, et al. 2009. ALLPATHS 2: Small genomes assembled accurately and with high continuity from short paired reads. Genome Biol 10: R103 doi: 10.1186/gb-2009-10-10-r103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV 2009. Allele-specific expression assays using Solexa. BMC Genomics 10: 422 doi: 10.1186/1471-2164-10-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Burgos JI, Foulon A, Bahechar I 2010. Transcriptome screen for fast evolving genes by Inter-Specific Selective Hybridization (ISSH). BMC Genomics 11: 126 doi: 10.1186/1471-2164-11-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA 2009. Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi ZY, Megy K, Grabherr M, et al. 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner PA, Tang HX 2001. Fragment assembly with double-barreled data. Bioinformatics 17: S225–S233 [DOI] [PubMed] [Google Scholar]
- Pevzner PA, Tang HX, Waterman MS 2001. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci 98: 9748–9753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. 2007. Multiplex amplification of large sets of human exons. Nat Methods 4: 931–936 [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL 2009. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res 19: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Puiu D, Lee VT 2008. Gene-boosted assembly of a novel bacterial genome from very short reads. PLoS Comp Biol 4: e1000186 doi: 10.1371/journal.pcbi.1000186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC 2008. Next-generation sequencing transforms today's biology. Nat Methods 5: 16–18 [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I 2009. ABySS: A parallel assembler for short read sequence data. Genome Res 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire JY 2001. Teeth outside the mouth in teleost fishes: How to benefit from a developmental accident. Evol Dev 3: 104–108 [DOI] [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan ZY, Zhang MQ 2009. Updates to the RMAP short-read mapping software. Bioinformatics 25: 2841–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Meyer A 2006. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J Mol Evol 62: 772–784 [DOI] [PubMed] [Google Scholar]
- Tang FC, Barbacioru C, Wang YZ, Nordman E, Lee C, Xu NL, Wang XH, Bodeau J, Tuch BB, Siddiqui A, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382 [DOI] [PubMed] [Google Scholar]
- Torres TT, Metta M, Ottenwalder B, Schlotterer C 2008. Gene expression profiling by massively parallel sequencing. Genome Res 18: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AL, Varala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME, et al. 2007. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 318: 441–444 [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH 2008. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17: 1636–1647 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J 2008. Dynamic repertoire of a transcriptome surveyed at single-nucleotide resolution. Nature 454: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Yamanoue Y, Miya M, Inoue JG, Matsuura K, Nishida M 2006. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei : Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet Syst 81: 29–39 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]