Abstract
Developmental biology aims to understand how the dynamics of embryonic shapes and organ functions are encoded in linear DNA molecules. Thanks to recent progress in genomics and imaging technologies, systemic approaches are now used in parallel with small-scale studies to establish links between genomic information and phenotypes, often described at the subcellular level. Current model organism databases, however, do not integrate heterogeneous data sets at different scales into a global view of the developmental program. Here, we present a novel, generic digital system, NISEED, and its implementation, ANISEED, to ascidians, which are invertebrate chordates suitable for developmental systems biology approaches. ANISEED hosts an unprecedented combination of anatomical and molecular data on ascidian development. This includes the first detailed anatomical ontologies for these embryos, and quantitative geometrical descriptions of developing cells obtained from reconstructed three-dimensional (3D) embryos up to the gastrula stages. Fully annotated gene model sets are linked to 30,000 high-resolution spatial gene expression patterns in wild-type and experimentally manipulated conditions and to 528 experimentally validated cis-regulatory regions imported from specialized databases or extracted from 160 literature articles. This highly structured data set can be explored via a Developmental Browser, a Genome Browser, and a 3D Virtual Embryo module. We show how integration of heterogeneous data in ANISEED can provide a system-level understanding of the developmental program through the automatic inference of gene regulatory interactions, the identification of inducing signals, and the discovery and explanation of novel asymmetric divisions.
With the exponential growth of biological data, databases have taken center stage in the biological sciences. The first biological databases were “monotype,” publishing a single type of information (e.g., PubMed for literature, GenBank for sequences). More recently, systems started hosting heterogeneous data sets related to the developmental biology of most major model organisms, e.g., FlyBase (The FlyBase Consortium 1994), WormBase (Harris et al. 2004), Zfin (Sprague et al. 2001), MGD (Blake et al. 1997), Dyctibase (Chisholm et al. 2006), and TAIR (Huala et al. 2001). These databases integrate the anatomy of the organism and its evolution in time with functional gene annotations, genetic mutations, phenotypes, expression patterns, and a genome browser. Usually “gene-centric,” they describe expression profiles and the effect of loss or gain of function of individual genes, thereby facilitating the planning of wet lab experiments.
The scope of these databases remains limited, however. They were designed to provide rapid answers to relatively simple queries from bench scientists on the function or expression of individual genes. They do not attempt to integrate the data they host into a global—and computable—view of the embryonic developmental program. Transparent cross-querying of heterogeneous types of data is poorly developed, and the possibility for knowledge discovery from the automatic analysis of stored data is virtually nonexistent. Integration of the results of “gene-centric” experiments into higher order gene regulatory networks (GRNs) would constitute a first step toward a global representation of the regulatory code underlying metazoan development (Davidson 2009). The representation of embryonic anatomy is also unsatisfactory in current systems. In particular, they do not integrate the relative positions and shape of anatomical structures, which can now be efficiently imaged (Tassy et al. 2006; Keller et al. 2008). This restricts the extent to which morphogenesis and cell communication can be digitally represented.
The embryos of land nematodes and ascidians develop with a very small number of cells and according to an invariant lineage (Conklin 1905; Sulston et al. 1983; Nishida 1987), which makes them particularly suitable to the development of integrated digital systems. We focused on ascidians. These marine invertebrate chordates are closely related to vertebrates (Delsuc et al. 2006). They are particularly suited to the study of GRNs (Imai et al. 2006, 2009; Kubo et al. 2010) and their function in the control of cellular behavior (Christiaen et al. 2008). Ascidian early embryology has been extensively studied in its cellular and molecular details (for review, see Lemaire 2009), and the functions of several hundred genes have been analyzed using loss- or gain-of-function approaches (e.g., Yamada et al. 2003; Wada et al. 2008). Central to the project described here, embryonic expression patterns by in situ hybridization are available for several thousand genes (Satou et al. 2002), including most transcription factor genes and signaling ligands (Imai et al. 2004; Miwata et al. 2006). Several specialized databases cover specific aspects of ascidian biology: genomes and coding genes (Ensembl, Birney et al. 2006; Hubbard et al. 2007; JGI, Dehal et al. 2002), embryonic anatomy (FABA, Hotta et al. 2007), gene expression (GHOST, Satou et al. 2002; MAGEST, Kawashima et al. 2002), or cis-regulatory sequences (DBTGR, Sierro et al. 2006).
The system described here is a first attempt at combining and extending available information into a generalist model organism database, with capabilities that extend the classical “gene-centric” approach used by most model organism databases. As a proof of principle of the power of this system, we identified novel asymmetric cell divisions during early embryogenesis, identified the correct neural inducer among many possible candidates, and automatically reconstructed GRNs to a significant extent.
Results
General organization of the ANISEED system
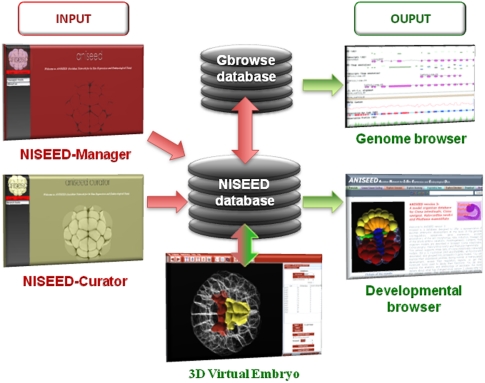
Figure 1 presents the general organization of the ANISEED system, the ascidian implementation of a generic system called NISEED (Network for in situ Expression and Embryological Data). The content of the ANISEED database can be explored via the Developmental Browser (http://aniseed-ibdm.univ-mrs.fr), which uses hub pages to organize key information about embryo anatomy, gene function, gene expression, regulatory interactions, and literature (Supplemental Figs. S1, S2). Part of the ANISEED data can be displayed in their genomic context via a Gbrowse-based genome browser (Stein 2002). In addition, a “3D Virtual Embryo” module is used both to map molecular or anatomical information onto interactive 3D embryo models, and to generate a quantitative description of the geometry of individual embryonic territories/cells and their topological arrangement, which are then imported into ANISEED (Tassy et al. 2006). The maintenance of the system is facilitated by two sets of administration and curation tools (see Methods). A Supplemental source code is provided.
Figure 1.
Overview of the architecture of the NISEED system. (Arrows) Direction of information flow. Supplemental Figures S1 and S2 present the search interfaces in more details.
Representation of embryo anatomy and its evolution with time
Advanced model organism databases represent embryonic anatomy via a hierarchical textual ontology defined for each developmental stage. As such description was lacking for ascidians, we created an Open Biomedical Ontologies (OBO)-compliant ascidian anatomical ontology for each stage of the Ciona intestinalis developmental table (Hotta et al. 2007). Development was split into three periods, for which we used different ontology logics: cleavage stages, gastrula to larval stages, and post-metamorphosis stages (Supplemental Fig. S3A). The resulting 25 ontologies for Ciona intestinalis describe 2240 anatomical territories.
In addition to Ciona, four other solitary ascidian species are used in the community: Ciona savignyi, Phallusia mammillata, Halocynthia roretzi, and Boltenia villosa. The strong developmental conservation between ascidian species was used to create parallel tentative embryonic and adult anatomical ontologies for these species. Because of the phylogenetic proximity of C. savignyi and P. mammillata to C. intestinalis, their ontologies were inferred to be identical to those of Ciona intestinalis. The ontologies of the more distantly related H. roretzi and B. villosa slightly deviate from those of Ciona. Supplemental Figures S3B,C and S4 describe the deviations that were incorporated as a result of new findings (division patterns of A7.6 and B7.6; fates of B7.5) and interspecies variability (b8.19) using the dedicated ontology editing tools of the NISEED-manager.
Integration of ontologies at successive stages was achieved via lineage links between mother and daughter territories. Around 2000 such lineage links were established manually for each supported species, via a dedicated interface of the ANISEED-manager. Besides classical studies, this work integrated three recent studies of the Ciona intestinalis tail epidermis lineage (Pasini et al. 2006) and posterior neural plate lineages (Nicol and Meinertzhagen 1988; Cole and Meinertzhagen 2004). Lineage relationships were used to associate one or several larval fates with each embryonic territory.
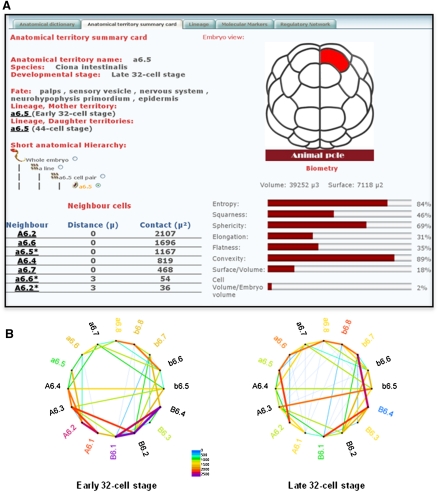
Finally, the ANISEED description of ascidian anatomy integrates the three-dimensional (3D) topology of the cells and tissues of the embryo, computed from reconstructed 3D embryo models (Supplemental Fig. S4A; Tassy et al. 2006). This information is currently available in Ciona for all cleavage stages, up to the early gastrula stage. Phallusia mammillata stages between the 64-cell and the early gastrula stages are covered. For each territory, the system indicates the physical distances separating cells or structures, a measure of the surface of contacts between adjacent structures, as well as a quantitative description of the territory's geometry (volume, external surface, sphericity, squareness, convexity, elongation, flatness, and entropy) (Fig. 2A). The search for cells most likely to communicate and the study of the evolution of cell contacts is facilitated by cell neighbor graphs available for all pregastrula stages in Ciona intestinalis (Fig. 2B).
Figure 2.
Representation of embryonic anatomy. (A) Screenshot of an anatomical territory card representing the lineage, fate, position/contacts with neighbors, and geometry of the a6.5 cell at the early 32-cell stage. Note the tabs at the top of the screen capture that lead to the whole anatomical ontology for the stage of interest (“Anatomical Ontology”), to the precursors and progeny of the territory of interest (“Lineage”), to expression profiles restricted to this territory (“Molecular Markers”), and to the regulatory interactions that take place in the lineage leading to this territory (“Regulatory Network”). (B) Neighborhood graph showing which cells of the late 32-cell embryo contact each other. The nodes of the graph represent individual cells, the edges represent contacts. The thickness of the edge reflects the area of contact between adjacent cells. The indicated values are in square microns. Supplemental Figures S3 and S4 give more details about the description of ascidian anatomy in ANISEED.
In summary, for each stage, an anatomical structure is represented in ANISEED by six sets of parameters: its position in the hierarchical anatomical ontology defined at this stage (“is part of”), its lineage (“is progeny of”—a structure from the previous stage dictionary), its fate at larval stages, its contacts to neighboring structures, and its shape and size. Each of these parameters can be individually explored using dedicated interfaces organized in the “Explore anatomy” section of the Developmental Browser (see Supplemental Fig. S2).
Representation of genomic features
ANISEED defines a set of 20,631 Ciona intestinalis gene models obtained by clustering transcripts models predicted from JGI (Dehal et al. 2002), Ensembl (Hubbard et al. 2007), and the Kyoto genome consortium (Satou et al. 2008). These gene models are functionally annotated by running a dedicated automatic annotation pipeline, based on protein domain detection and evolutionary inference of function and biological name (Supplemental Fig. S5; Supplemental Methods). Gene models are linked to all available public ESTs and cDNAs and to experimentally validated cis-regulatory regions. All sequence features can be visualized in their genomic context in the ANISEED Genome Browser, which also displays the conservation profiles between the two sequenced Ciona genomes (Ciona intestinalis and Ciona savignyi) and predictions of local nucleosome occupancy (Khoueiry et al. 2010).
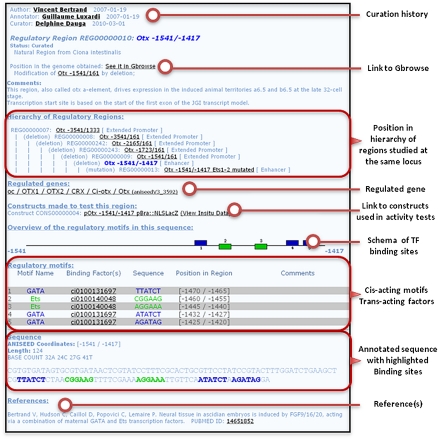
A sophisticated representation of the structure and activity of cis-regulatory elements and of their upstream regulators was designed, as these elements play a major role in the control of the developmental program and are central to the reconstruction of GRNs. To faithfully represent the cis-regulatory logic, all experimentally tested regions at one locus are organized hierarchically (Fig. 3). The type of activity of each region in in vivo reporter assays is described using a controlled vocabulary. Eight classes of activity were thus defined, five of which match Sequence Ontology terms (Supplemental Fig. S6; see Methods; Eilbeck et al. 2005). When experimentally described, individual functional transcription factor binding sites are shown and linked to the corresponding experimentally verified trans-acting factors (Fig. 3). The precision and traceability of this representation is an advance over model organism databases such as WormBase, MGI, FlyBase, or Zfin, or dedicated cis-regulatory element databases such as DBTGR (Sierro et al. 2006), ORegAnno (Montgomery et al. 2006), RedFly (Gallo et al. 2005; Halfon et al. 2008), or the VISTA enhancer browser (Visel et al. 2007).
Figure 3.
Representation of cis-regulatory information. Screenshot of the regulatory region card for the early minimal neural enhancer of Ciona intestinalis Otx. The precise pattern of activity of this region is accessed by clicking on the “view in situ data” link located in the “Constructs made to test this region” section. Supplemental Figure S6 presents the classification system for cis-regulatory regions used in ANISEED.
ANISEED currently describes 528 published or unpublished regulatory regions controlling the transcription of 158 genes. For 85 regions, functionally tested binding sites for specific transcription factors are indicated. These numbers are comparable to those of the Redfly project, which annotates the regulatory regions of the much more studied Drosophila.
Description of expression data in wild-type and experimentally manipulated contexts
Spatio-temporal gene expression is described in ANISEED with EST counts, in situ hybridizations, protein immunolocalization, and cis-regulatory element activity. ESTs counts from 28 sequenced non-normalized cDNA libraries provide a low-resolution temporal expression information for 80% of the genes (Satou et al. 2003). A digital differential display (DDD) tool (Supplemental Fig. S2) uses this information to find genes differentially represented between sets of sequenced cDNA libraries (Audic and Claverie 1997).
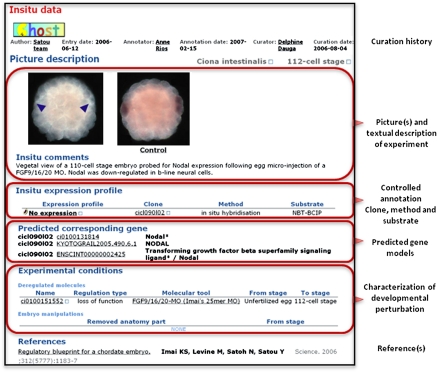
A more precise description of gene expression profiles during development is obtained via in situ hybridization, immunolocalizations, and electroporation assays of cis-regulatory elements. These spatial patterns are illustrated by standardized pictures (orientation, format) (Fig. 4) and formalized by selecting terms from the relevant anatomical ontology that describe territories of expression. ANISEED currently hosts 5853 in situ hybridization patterns from Halocynthia roretzi in wild-type conditions and 24,124 patterns from Ciona intestinalis, providing information for around 4000 genes at one or more developmental stages in wild-type conditions. In addition, 777 patterns describe the spatio-temporal activity of Ciona cis-regulatory regions.
Figure 4.
Representation of spatial expression patterns. Screenshot of the expression card describing the expression of the Ciona intestinalis Nodal gene at the early gastrula stage, in response to the inhibition of FGF9/16/20 function. Note the control picture that was taken in the same experiment; clicking on the “control” word leads to the description of the expression in wild-type conditions.
The precise description of the transcriptional consequences of experimental perturbations received particular attention. Two types of perturbations are currently supported: molecular perturbations of gene function and embryological perturbations. Molecular perturbations (Fig. 4) are formally represented by: (1) the deregulated gene, (2) the type (gain- or loss-of function) and timing of the perturbation, and (3) the “Molecular Tool” used to affect the function of the gene. “Molecular Tools” include antisense Morpholinos, overexpression constructs (electroporation or mRNA injection), or pharmacological reagents. Each tool is linked to all articles and experiments in which they were used and described. ANISEED currently hosts 619 morpholino sequences (targeting 569 genes) described in the literature. Morphological phenotypes were obtained following loss of function of 221 of these genes. These phenotypes were described textually and with a picture (Yamada et al. 2003; Hamada et al. 2007). In addition, ANISEED describes the transcriptional consequences of gene loss- or gain-of-function of 94 genes with developmental phenotypes, mostly transcription factors, signaling ligands, or genes with no previously known function. Collectively, these experiments describe the changes in the expression patterns of 232 target genes (total of 1152 patterns). Embryological perturbations, such as cell ablations or explants, are defined by the removed anatomy parts and the developmental stage of the perturbation. Eighty-seven such ablation experiments are reported, in which the expression patterns of 18 genes were analyzed. In contrast to ANISEED, advanced model organism databases such as FlyBase, WormBase, Zfin, or MGI describe the morphological or anatomical phenotype resulting from experimentally altered gene function, but provide little information about the transcriptional consequences of these mutations.
As developmental genes often have dynamic expression patterns (Sobral et al. 2009), wild-type and experimentally modified expression patterns should be compared within a given experiment. We thus associated each expression pattern in perturbed conditions with its matching wild-type control in the same experiment (Fig. 4). This curation decision is unique to ANISEED: Although Zfin describes some in situ expression profiles of target genes in response to experimental perturbations, this information is not associated with the matching wild-type control.
Impact, issues, and enhancement of manual curation
Published small-scale studies are the primary source for expression profiles in deregulated contexts and their associated controls. One-hundred-sixty manually curated articles are currently publicly available in ANISEED, a data set that covers most of the molecular literature on Ciona intestinalis. These manually extracted data represent 18.5% of all ANISEED spatial expression profiles, illustrating that the manual compilation of small- and mid-scale studies can reach a scale similar to large-scale screens.
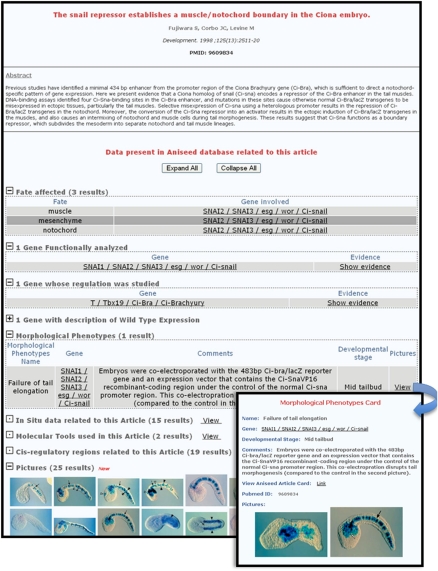
The capture of published information was streamlined by the creation of the “Article Card” concept. Each Article Card summarizes in a standardized and structured format the content of the text and figures of an article (Fig. 5). It lists, and links to the corresponding experimental evidence, all cell fates affected; all genes whose expression, regulation, or function was studied; all regulatory sequences; and all morphological phenotypes described in the article. It also provides a bird's-eye view of all pictures from the article inserted in the database. This curation strategy also allows the direct retrieval of all articles describing the expression/regulation/function of a gene of interest, or related to the specification of a given territory.
Figure 5.
The article card. Screenshot of an example of an article card showing the various types of information recapitulating the scientific message of the article. (Inset) Description of the morphological phenotype caused by the inhibition of Ci-snail function.
Manual curation was notably affected by missing information in some articles. The identity of the clones for in situ hybridization probes and the precise sequences tested in reporter assays constituted the most frequent omissions. Supplemental Information S1 presents an article minimum information standard (AMIS), which will help authors include all necessary information in future articles.
Taken together, the ANISEED ascidian data set is unique in the wide variety and volume of its data, and in the rich semantics used to closely fit the representation of individual experiments to their experimental designs. In the following sections, we exemplify how ANISEED can be used to integrate individual experiments into a higher-level understanding of the ascidian developmental program.
Integrating cellular shape and lineage: Identification of novel unequal cleavages
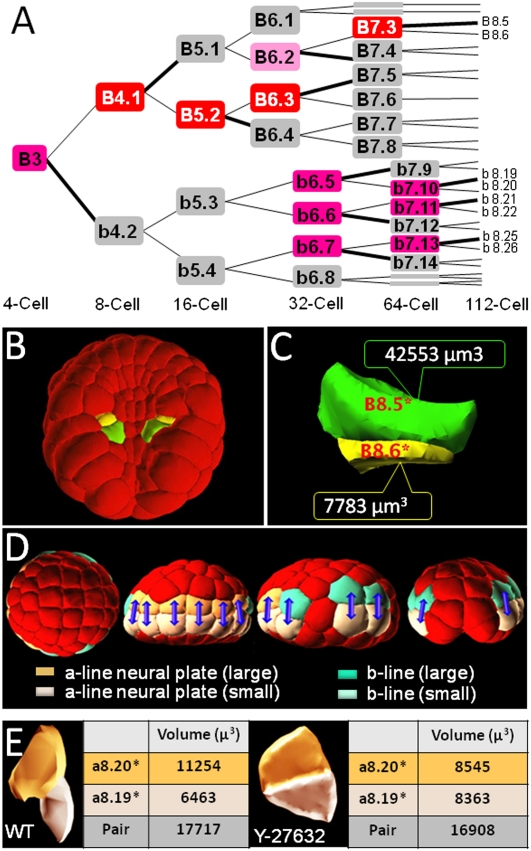
Unequal cell cleavages, leading to daughter cells of different sizes, are frequently associated with cell fate decisions (Sardet et al. 2007). We previously integrated lineage information and cell volumes to identify unequal cleavages up to the 44-cell stage in Ciona embryos (Tassy et al. 2006). This approach was extended in Ciona and Phallusia using our novel set of 3D embryo models. Up to the early gastrula stage, 23 out of 63 cell-pair divisions in Ciona are geometrically asymmetric (Fig. 6A; Supplemental Fig. S7). Most of these unequal cleavages are conserved in Phallusia (data not shown). In the vegetal hemisphere, asymmetric divisions give birth to sister cells fated to different tissue types (Fig. 6B,C) and are topologically scattered, suggesting that they were under local control. In contrast, unequal divisions in the animal hemisphere between the 64- and 112-cell stages led to sisters that sometimes shared the same fate (e.g., b8.19 and b8.20 both contribute to the tail epidermis). The stereotyped position of the mother cells at the equator of the embryo, the alignment of their division along meridians, and the position of the smaller sister below its larger sibling suggested a global control (Fig. 6D). Endoderm invagination is initiated in Ciona between the 64- and 112-cell stages. To test whether mechanical pulling forces from endodermal progenitors may affect the geometry of the division of their ectodermal neighbors, we analyzed two 112-cell Ciona embryos in which endoderm invagination had been prevented by treatment with the Rho-kinase inhibitor Y-27632 (Sherrard et al. 2010). While the orientation of the division of mother cells was unchanged, the volumes of sister cells were now equal. Thus, endoderm invagination may influence the asymmetry, but not the orientation, of animal cell cleavages (Fig. 6E).
Figure 6.
Systematic identification of unequal cell cleavages: (A) Lineage tree for the B3 (posterior) blastomere between the four-cell and the 112-cell stages in Ciona intestinalis. The only blastomeres named are in lineages where unequal divisions occur. (Gray) Cells that divide symmetrically, (light pink) cells with weak asymmetry (index > 15%; Tassy et al. 2006), (pink) cells with marked asymmetry (index > 25%), (red) cells with strong asymmetry (>50%). Supplemental Figure S7 shows all asymmetric divisions up to the 112-cell stage. (B,C) Example of asymmetry between the 64- and 112-cell stages in the vegetal hemisphere of Ciona intestinalis embryos. The yellow (B8.6) and green (B8.5) cells are daughters of the same mother cell (B7.3). Note the important difference in cell volumes between the two sisters. This unequal division is conserved in Halocynthia roretzi (Darras and Nishida 2001). (D) Position of unequally cleaving animal cells between the 76- and 112-cell stages. From the left: animal view, anterior is to the left; frontal view, animal is to the top; lateral view, anterior is to the left, animal to the top; posterior view, animal is to the top. (Blue arrows) Sister cells are linked. (Colors) Lineage and size of cells (see legend). (E) Effect of the inhibition of endoderm invagination with the Rho-kinase inhibitor Y-27632. Side views of the a8.19/a8.20 cell pair in wild-type and Y-treated conditions are shown, as well as a measure of the mean volumes of these cells in the two embryos analyzed.
Integrating functional gene annotation, expression data, and anatomy: In search of the ascidian neural inducer
In ascidians, the anterior neural tissue is induced in a6.5 blastomeres at the 32-cell stage by a signal originating from the A-line vegetal cells (for reviews, see Meinertzhagen et al. 2004; Lemaire 2009). To identify the best candidate inducer among the 54 secreted ligands from major signaling pathways expressed around this stage, we made use of the sequential query mode of ANISEED, in which the results of an initial query are used as the search space for the next one (Supplemental Fig. S8). Assuming that translation, folding, and transport of the inducer takes ∼20 min and that activation of a target gene may take an additional 15 min, we looked for genes coding for secreted proteins that were expressed at the 16-cell stage in the vegetal neighbors of the progenitors of the induced cells. We formulated two sets of parallel queries: one molecular and the other anatomical. The set of molecular queries identified genes coding for secreted proteins (Gene Ontology query) and expressed during the cleavage stages (Digital Differential Display query). The set of anatomical queries identified the ancestor of the a6.5 cell pair, and then the vegetal neighbors of these animal cells. Integration of these two sets through the “Query integration by ID” web interface (Supplemental Fig. S2) yielded two genes, FGF9/16/20 and Lefty. The former encodes the demonstrated neural inducer (Bertrand et al. 2003). Lefty, an antagonist of nodal signaling (Meno et al. 1996), does not appear to play a major role during early Ciona embryogenesis (Imai et al. 2006).
Integration of expression patterns in wild-type and deregulated conditions: Automatic inference of gene regulatory interactions
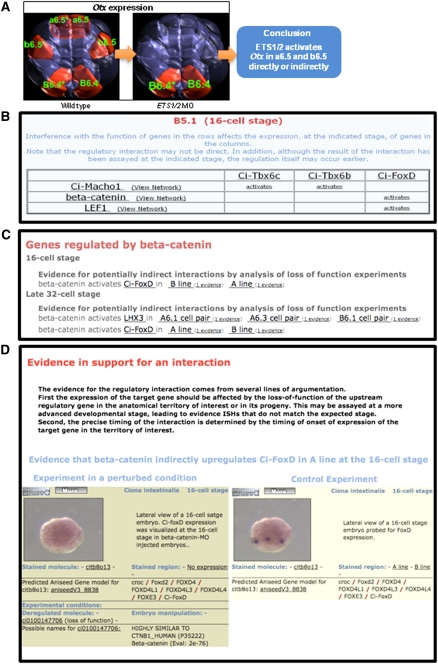
We finally sought to use the ANISEED expression data set to automatically extract transcriptional regulatory interactions underlying ascidian development. The analysis of Ci-Otx regulation (Fig. 7A) exemplifies the simple inference logic we used (described in Supplemental Methods). At the 44-cell stage, Ci-Otx is expressed in the a6.5, b6.5, and B6.4 cell pairs. When the function of the maternal ETS1/2 mRNAs is blocked by Morpholino (MO) injection, expression of Ci-Otx becomes restricted to B6.4. Comparing the two sets of expressing territories, we infer that by the 44-cell stage ETS1/2 positively regulates Otx in a6.5 and b6.5. Integration with cis-regulatory information reveals that this regulatory event is direct: The early Otx neural enhancer includes two ETS1/2 binding sites that are required for its activity (Bertrand et al. 2003).
Figure 7.
Automatic inference of transcriptional regulatory interactions. (A) Example of the inference logic that led to the establishment of a regulatory interaction between ETS1/2 and Otx in a6.5 and b6.5 neural precursors at the late 32-cell stage. Use of an antisense Morpholino oligonucleotide is interpreted as a loss-of-function experiment. Territories in which Otx expression is lost are thus territories where ETS1/2 regulates Otx expression. (B) Screenshot of the “Anatomy Regulation” card for the B5.1 precursor at the 16-cell stage, showing the regulatory interactions that take place in the lineage leading to this blastomere. (C) Screenshot of the “Downstream Target” card of beta-catenin, showing transcriptional targets of this gene at the 16-cell and late 32-cell stages. Note that the anatomical territories in which the regulatory interactions are confirmed as indicated. (D) Screenshot of the page presenting the evidence that beta-catenin upregulates Ci-FoxD in the A-line at the 16-cell stage. Supplemental Figure S9 presents the number of inferred regulatory links at each developmental stage.
By applying these rules to the whole ANISEED data sets, we obtained 498 distinct gene regulatory interactions, supported by loss-of-function assays involving 183 genes at different developmental stages (Supplemental Fig. S9). This network includes and significantly extends the networks from Imai et al. (2006, 2009) (195 interactions for 81 genes) and describes 20 direct interactions confirmed by cis-regulatory region mutational analysis.
Two classes of web interfaces were developed to navigate this information. For each anatomical territory, the “Anatomical Gene Network card” indicates the known regulatory interactions that have taken place in this territory by the stage studied (Fig. 7B). For each individual gene, the “Upstream regulators” page gives access to known upstream regulators in each territory. Conversely, the “Downstream targets” page lists targets of the gene of interest by territories and stages (Fig. 7C). In all cases, the interactions are linked to their supporting experimental evidence (Fig. 7D).
Discussion
ANISEED is the first integrated system for ascidians and is widely used in the growing community of ascidian laboratories. Over the past 2 yr the system received 51,000 unique visitors, mainly from France, Japan, the USA, and Italy—all countries with strong ascidian communities. In addition, the generic nature of the system and its design principles may hold lessons for the future of model organism databases.
The tight collaboration between experimentalists, biocurators, and computer scientists was a central aspect of the ANISEED project. Experimental biologists used their knowledge of ascidian development and experimental approaches to define the general scope of the project, and the most important types of data for the ascidian community. This led to a strong focus on transcriptional regulation, gene regulatory networks, and their integration into a precise three-dimensional representation of embryo development. A second major role of experimentalists was to define minimal information standards for each type of experiment represented, thereby achieving a well-defined and consistent set of data representations. Biocurators then defined the ontologies and controlled vocabularies necessary to formalize the representation of experiments, thus transforming the “biological interpretability” of the data into their “computability.” This close fit between experimental design, raw experimental data, and their digital representation was crucial for the successful integration of heterogeneous individual experiments to identify novel asymmetric divisions, reconstruct Gene Regulatory Networks, or predict the identity of tissue inducers. No other major model organism database currently offers interfaces that directly integrate individual experiments into a broad view of a developmental program.
NISEED was designed as a generic framework. It is thus adaptable to most model organisms amenable to embryological manipulations similar to those carried out in ascidians, including zebrafish (Felsenfeld 1996), Xenopus (Koide et al. 2005), urchins (Oliveri et al. 2008), Amphioxus (Garcia-Fernàndez et al. 2009), hemichordates (Lowe et al. 2006), annelids (Schneider and Bowerman 2007), and cnidarians (Momose and Houliston 2007). Although the stereotyped development of ascidians, based on fixed cell lineages, is an obvious advantage for the approach presented here, the system can easily be adapted to integrate the geometry of embryos that develop in a less reproducible fashion. This adaptation could involve a change of scale of analysis, measuring the generally reproducible geometry and topological arrangement of fields of cells or organ rudiments. This tissue-level representation could be complemented at the mesoscopic scale by statistical measures of individual cell geometries and arrangements within each field or rudiment (e.g., Blanchard et al. 2009; Butler et al. 2009). Implementation of parallel NISEED systems will greatly facilitate the evolutionary comparison of developmental program within a taxon or phylum (e.g., Sobral et al. 2009).
The types of data represented in NISEED currently limit its predictive power. It will be simple to represent mutant lines, thus opening the system to model systems with a strong history of genetics, including fly, Caenorhabditis elegan, and mouse. Genome-wide chromatin assays and RNA-seq data are rapidly accumulating in many model organisms. Their careful integration into NISEED will help reconstruct regulatory networks, provided the convergent support given by distinct data sets (e.g., chromatin immunoprecipitation analyses, cis-regulatory information, expression patterns) are adequately ranked and attributed specific confidence values. This will require an evolution of the database schema. The GMOD (Generic Model Organism Database) consortium recently proposed a flexible modular database schema, CHADO (Mungall and Emmert 2007), relying, like NISEED, on the widespread use of ontologies and controlled vocabularies. This system currently offers a sophisticated representation of sequence features, but limited representations of embryo anatomy and gene expression patterns. Its extension with NISEED modules describing the three-dimensional anatomy of the embryo and the transcriptional program of each territory will help to further reconstruct the developmental GRNs acting in each territory by integrating the results of short-read-based expression and chromatin assays.
Methods
Hardware and software
The system runs on all platforms supporting PostgreSQL8, JAVA, and JAVA 3D, including Windows, UNIX, Linux, and Mac OSX 10.5. Source code is available as Supplemental source code. Web pages are correctly displayed on Browsers supporting HTML level 4.0 or higher and JavaScript, including Firefox and Internet Explorer. Some display issues were reported with current versions of Chrome or Safari.
System administration, curation, and data download
Two sets of management tools are proposed. The NISEED-manager allows de novo creation of NISEED databases, the management of users, the editing of ontologies, the functional annotation of genes, and the import of large-scale data from flat files. It also centralizes scripts to update and upload data. Through the NISEED-curator, data from small-scale experiments can be inserted and manually curated. A precise description of the curation strategy can be found in the Supplemental Methods.
Three types of data can be obtained from the download section: genomic, anatomical, and expression data. Genomic data include gene and transcript models, functional gene annotations, and cis-regulatory regions. Anatomical data include anatomical ontologies in OBO format (Smith et al. 2007), the collection of available reconstructed embryos, each with its biometry data and the 3DVE installation package. Expression data include all in situ hybridization data in a MISFISHIE-compliant XML format (Deutsch et al. 2008), including images, and modified to associate expression patterns in experimentally manipulated and control situations (example available at http://aniseed-ibdm.univ-mrs.fr/exchange_format.php). The NISEED-manager also allows to import Misfishie-Compliant XML files describing large-scale in situ hybridizations data sets.
3D embryo reconstruction and treatments
Reconstructed two-cell to 44-cell Ciona embryos were obtained from Tassy et al. (2006). Reconstructed Boltenia villosa and some Phallusia embryos were obtained from (Sherrard et al. 2010). Additional Ciona and Phallusia embryos were obtained by in vitro fertilization as in Robin et al. (2010). Rho-kinase inhibition was achieved by treating embryos from the 64-cell stage with 100μM Y-27632, a specific pharmacological inhibitor of this kinase. Live or fixed embryos of Phallusia mammillata and Ciona intestinalis between the 64-cell and early gastrula stages were reconstructed using Amira and the files processed for import in 3DVE as indicated in Robin et al. (2010).
Definition, expression, and annotation of ANISEED gene models and cis-regulatory regions
ANISEED supports four previously generated independent Ciona intestinalis transcript model sets (JGIv1.0, Ensembl v2.0, KyotoGrail2005, and KH), which were found to be of highest quality and complemented each other. Short single exon models (<300 bp) that were supported neither by EST information nor by blast hits to other species, and presumably representing erroneous ab initio predictions, were not considered. Transcript models sharing at least one full exon were grouped into 20,915 ANISEED-v3.0 gene models by applying a published clustering algorithm (Gilchrist et al. 2004). Halocynthia roretzi transcript models were generated assembling 60,000 expressed sequences (ESTs, cDNAs) into contigs (Huang and Madan 1999; Quackenbush et al. 2000; Gilchrist et al. 2004). This procedure generated 14,970 tentative consensus sequences (TCs) that were used as transcript models. The NISEED automatic annotation pipeline integrates Interproscan (Quevillon et al. 2005), InParanoid v2.0 (Remm et al. 2001), and BLASTP. The pipeline is detailed in the Supplemental Methods section and illustrated in Supplemental Figure S5.
Cis-regulatory regions are by convention segments of the published genome sequence (Dehal et al. 2002). Because of high polymorphism level in ascidians, the tested sequences often depart from the published genome sequence, so that each regulatory region is associated with the specific constructs that were used to test it, with their sequence. A description of the strategy used to name, represent, and classify cis-regulatory regions and their activity can be found in the Supplemental Methods and Supplemental Figure S6.
Spatial expression data (ISH, immunohistochemistry, and reporter assays) were represented by associating the in situ probe clone (ISH) or transcript model (Immunohistochemistry) or reporter construct (cis-reg. regions) to ontology terms using a curation strategy detailed in the Supplemental Methods section.
Acknowledgments
We thank the members of our laboratories for discussion and input, and E. Jacox for his careful reading of the manuscript. E. Carmona, M. Grange, C. Degos, J. Lucchino annotated expression data, and E. Drula and R. Pardoux reconstructed several embryos. The annotation quality was much improved by suggestions from S. Irvine, W. Shi, N. Kawai, S. Shimeld, B. Davidson, T. Meedel, V. Picco, H. Yasuo, and A. Pasini. We thank the members of the community who contributed unpublished expression patterns and data, and J.F. Guillemot for server administration. This work was supported by the CNRS, the French Ministry of Research (ACI, ANR-blanc [“Chor-Reg-Net” and “Chor-Evo-Net” and ANR-SysCom “GeneShape”] programs and GIS “Génomique Marine”), the Marseille-Nice Genopôle, the ARC, and a European Network, “Embryos against Cancer (EAC)” (QLK3-CT-2001-01890). O.T. was supported by EAC and by a fellowship from the Association pour la Recherche sur le Cancer. D.D. was supported by “Chor-Reg-Net”. F.D. was supported by the Marseille-Nice Génopôle. D.S. was supported by a Fellowship from the Science and Technology Foundation of Portugal. B.L. was supported by “GeneShape.”
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108175.110.
References
- Audic S, Claverie JM 1997. The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P 2003. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115: 615–627 [DOI] [PubMed] [Google Scholar]
- Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, et al. 2006. Ensembl 2006. Nucleic Acids Res 34: D556–D561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Richardson JE, Davisson MT, Eppig JT 1997. The Mouse Genome Database (MGD). A comprehensive public resource of genetic, phenotypic and genomic data. The Mouse Genome Informatics Group. Nucleic Acids Res 25: 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard GB, Kabla AJ, Schultz NL, Butler LC, Sanson B, Gorfinkiel N, Mahadevan L, Adams RJ 2009. Tissue tectonics: Morphogenetic strain rates, cell shape change and intercalation. Nat Methods 6: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LC, Blanchard GB, Kabla AJ, Lawrence NJ, Welchman DP, Mahadevan L, Adams RJ, Sanson B 2009. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nat Cell Biol 11: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm RL, Gaudet P, Just EM, Pilcher KE, Fey P, Merchant SN, Kibbe WA 2006. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res 34: D423–D427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M 2008. The transcription/migration interface in heart precursors of Ciona intestinalis. Science 320: 1349–1352 [DOI] [PubMed] [Google Scholar]
- Cole AG, Meinertzhagen IA 2004. The central nervous system of the ascidian larva: Mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev Biol 271: 239–262 [DOI] [PubMed] [Google Scholar]
- Conklin E 1905. The organization and cell lineage of the ascidian egg. J Acad Nat Sci Phila 13: 1 [Google Scholar]
- Darras S, Nishida H 2001. The BMP signaling pathway is required together with the FGF pathway for notochord induction in the ascidian embryo. Development 128: 2629–2638 [DOI] [PubMed] [Google Scholar]
- Davidson EH 2009. Network design principles from the sea urchin embryo. Curr Opin Genet Dev 19: 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157–2167 [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439: 965–968 [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen JH, Daian F, et al. 2008. Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE). Nat Biotechnol 26: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Lewis S, Mungall C, Yandell M, Stein L, Durbin R, Ashburner M 2005. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol 6: R44 doi: 10.1186/gb-2005-6-5-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld AL 1996. Defining the boundaries of zebrafish developmental genetics. Nat Genet 14: 258–263 [DOI] [PubMed] [Google Scholar]
- The FlyBase Consortium 1994. FlyBase—the Drosophila database. Nucleic Acids Res 22: 3456–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo SM, Li L, Hu Z, Halfon MS 2005. REDfly: A regulatory element database for Drosophila. Bioinformatics 22: 381–383 [DOI] [PubMed] [Google Scholar]
- Garcia-Fernàndez J, Jiménez-Delgado S, Pascual-Anaya J, Maeso I, Irimia M, Minguillón C, Benito-Gutiérrez E, Gardenyes J, Bertrand S, D'Aniello S 2009. From the American to the European amphioxus: Towards experimental Evo-Devo at the origin of chordates. Int J Dev Biol 53: 1359–1366 [DOI] [PubMed] [Google Scholar]
- Gilchrist MJ, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E 2004. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev Biol 271: 498–516 [DOI] [PubMed] [Google Scholar]
- Halfon MS, Gallo SM, Bergman CM 2008. REDfly 2.0: An integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res 36: D594–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Wada S, Kobayashi K, Satoh N 2007. Novel genes involved in Ciona intestinalis embryogenesis: Characterization of gene knockdown embryos. Dev Dyn 236: 1820–1831 [DOI] [PubMed] [Google Scholar]
- Harris TW, Chen N, Cunningham F, Tello-Ruiz M, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Chan J, et al. 2004. WormBase: A multi-species resource for nematode biology and genomics. Nucleic Acids Res 32: D411–D417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K 2007. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn 236: 1790–1805 [DOI] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. 2001. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A 1999. CAP3: A DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. 2007. Ensembl 2007. Nucleic Acids Res 35: D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y 2004. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Towards a comprehensive understanding of gene networks. Development 131: 4047–4058 [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y 2006. Regulatory blueprint for a chordate embryo. Science 312: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y 2009. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136: 285–293 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Kawashima S, Kohara Y, Kanehisa M, Makabe KW 2002. Update of MAGEST: Maboya Gene Expression patterns and Sequence Tags. Nucleic Acids Res 30: 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK 2008. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322: 1065–1069 [DOI] [PubMed] [Google Scholar]
- Khoueiry P, Rothbächer U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P 2010. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol 20: 792–802 [DOI] [PubMed] [Google Scholar]
- Koide T, Hayata T, Cho KWY 2005. Xenopus as a model system to study transcriptional regulatory networks. Proc Natl Acad Sci 102: 4943–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, Satou Y 2010. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development 137: 1613–1623 [DOI] [PubMed] [Google Scholar]
- Lemaire P 2009. Unfolding a chordate developmental program, one cell at a time: Invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol 332: 48–60 [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, et al. 2006. Dorsoventral patterning in hemichordates: Insights into early chordate evolution. PLoS Biol 4: e291 doi: 10.1371/journal.pbio.0040291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA, Lemaire P, Okamura Y 2004. The neurobiology of the ascidian tadpole larva: Recent developments in an ancient chordate. Annu Rev Neurosci 27: 453–485 [DOI] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H 1996. Left–right asymmetric expression of the TGFβ-family member lefty in mouse embryos. Nature 381: 151–155 [DOI] [PubMed] [Google Scholar]
- Miwata K, Chiba T, Horii R, Yamada L, Kubo A, Miyamura D, Satoh N, Satou Y 2006. Systematic analysis of embryonic expression profiles of zinc finger genes in Ciona intestinalis. Dev Biol 292: 546–554 [DOI] [PubMed] [Google Scholar]
- Momose T, Houliston E 2007. Two oppositely localised frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol 5: e70 doi: 10.1371/journal.pbio.0050070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Griffith OL, Sleumer MC, Bergman CM, Bilenky M, Pleasance ED, Prychyna Y, Zhang X, Jones SJM 2006. ORegAnno: An open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics 22: 637–640 [DOI] [PubMed] [Google Scholar]
- Mungall CJ, Emmert DB 2007. A Chado case study: An ontology-based modular schema for representing genome-associated biological information. Bioinformatics 23: i337–i346 [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA 1988. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev Biol 130: 737–766 [DOI] [PubMed] [Google Scholar]
- Nishida H 1987. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol 121: 526–541 [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH 2008. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci 105: 5955–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini A, Amiel A, Rothbächer U, Roure A, Lemaire P, Darras S 2006. Formation of the ascidian epidermal sensory neurons: Insights into the origin of the chordate peripheral nervous system. PLoS Biol 4: e225 doi: 10.1371/journal.pbio.0040225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J 2000. The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28: 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R 2005. InterProScan: Protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Robin F, Dauga D, Tassy O, Sobral D, Daian F, Lemaire P 2010. From confocal imaging to 3D model: A protocol for creating 3D digital replica of ascidian embryos. In Imaging in Developmental Biology: A laboratory manual (ed. Wong R, Sharpe J). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: (in press) [Google Scholar]
- Sardet C, Paix A, Prodon F, Dru P, Chenevert J 2007. From oocyte to 16-cell stage: Cytoplasmic and cortical reorganizations that pattern the ascidian embryo. Dev Dyn 236: 1716–1731 [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Fujiwara S, Nishikata T, Saiga H, Kusakabe T, Shin-i T, Kohara Y, Satoh N 2002. Ciona intestinalis cDNA projects: Expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene 287: 83–96 [DOI] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Kohara Y, Satoh N 2003. Large scale EST analyses in Ciona intestinalis: Its application as Northern blot analyses. Dev Genes Evol 213: 314–318 [DOI] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, et al. 2008. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol 9: R152 doi: 10.1186/gb-2008-9-10-r152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SQ, Bowerman B 2007. Beta-catenin asymmetries after all animal/vegetal- oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell 13: 73–86 [DOI] [PubMed] [Google Scholar]
- Sherrard K, Robin F, Lemaire P, Munro EM 2010. Sequential activation of apical and basolateral contractility drives ascidian endoderm invagination. Curr Biol (in press). doi: 10.1016/j.cub.2010.06.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Kusakabe T, Park K, Yamashita R, Kinoshita K, Nakai K 2006. DBTGR: A database of tunicate promoters and their regulatory elements. Nucleic Acids Res 34: D552–D555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. 2007. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral D, Tassy O, Lemaire P 2009. Highly divergent gene expression programs can lead to similar chordate larval body plans. Curr Biol 19: 2014–2019 [DOI] [PubMed] [Google Scholar]
- Sprague J, Doerry E, Douglas S, Westerfield M 2001. The Zebrafish Information Network (ZFIN): A resource for genetic, genomic and developmental research. Nucleic Acids Res 29: 87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. 2002. The Generic Genome Browser: A building block for a model organism system database. Genome Res 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Tassy O, Daian F, Hudson C, Bertrand V, Lemaire P 2006. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol 16: 345–358 [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Hamada M, Kobayashi K, Satoh N 2008. Novel genes involved in canonical Wnt/beta-catenin signaling pathway in early Ciona intestinalis embryos. Dev Growth Differ 50: 215–227 [DOI] [PubMed] [Google Scholar]
- Yamada L, Shoguchi E, Wada S, Kobayashi K, Mochizuki Y, Satou Y, Satoh N 2003. Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development 130: 6485–6495 [DOI] [PubMed] [Google Scholar]