Abstract
Background:
Several studies hinted about the clustering of risk variables of the metabolic syndrome (MS) and suggested that the underlying genetic polymorphisms could be responsible for the increasing incidence of coronary heart disease (CHD) in people of Indian origin. Therefore, identification of the components of the MS along with the genetic factors could be one of the aspects to make an attempt to prevent the increasing incidence of CHD.
Materials and Methods:
Principal component factor analysis (PCFA) was undertaken to identify the components or factors of the MS among the adult (≥30 years) Asian Indians living in and around Calcutta, India. The study comprised 350 adult Asian Indians. Anthropometric measurements were taken, and lipid profiles, blood pressure and fasting blood glucose were measured for each participant. Two genetic polymorphisms, namely, angiotensin converting enzyme (ACE) gene polymorphism (insertion/deletion [I/D]) or ACE (I/D) and apolipoproteinE (Hha I) were also studied.
Results:
PCFA revealed 3 factors that cumulatively explained 65.39% of the observed variance of the MS by measured variables. The 3 factors identified were lipids and lipoprotein (Factor 1), centripetal fat and blood pressure (Factor 2), and ACE (I/D) polymorphism with blood pressure (Factor 3). Moreover, the first 2 factors, that is, lipids, lipoprotein, centripetal fat, and blood pressures cumulatively explained ~46% (45.94%) of the observed variance of MS in this population.
Conclusions:
Since more than 1 factor was identified for the MS phenotype, more than 1 physiogenetic mechanism could be accounted for MS in the Asian Indian population.
Keywords: Asian Indians, factors, gene polymorphism, metabolic syndrome, obesity
INTRODUCTION
The prevalence of coronary heart disease (CHD) is known to be very high among Indians, both in India and abroad. Moreover, among Indians, CHD occurs at least a decade or 2 earlier compared with Europeans.[1,2] The reason for the increased susceptibility of Indians to CHD is yet to be completely understood. However, several studies have hinted that the clustering of risk variables (mechanism of which is still unknown) of metabolic syndrome (MS) could be responsible for the increasing incidence of CHD among Indians. This includes central obesity, hypertriglyceridemia, less levels of high-density lipoprotein cholesterol, high blood pressure, and high levels of fasting blood glucose,[1‐4] along with certain genetic factors (genetic polymorphisms) that adversely affect the levels of such variables, for example, angiotensin converting enzyme (ACE) gene polymorphism (insertion/deletion [I/D]) or ACE (I/D) and ApolipoproteinE gene (ApoE) polymorphisms.[5]
Throughout the Asia-Pacific region, there are differences in obesity prevalence as well as in body fat distribution[6] In Asian populations, morbidity and mortality from cardiovascular diseases (CVD) is occurring also in people with lower body mass index (BMI) and smaller waist circumference.[7,8] Thus, they tend to accumulate intra-abdominal visceral fat without developing generalized obesity.[1,7‐9] South Asians have a more centralized distribution of body fat and markedly higher mean waist–hip ratio for a given level of BMI compared with Europeans and Americans.[5‐9] The MS, which can be defined as the constellation of CVD risk factors, is one of the growing public health burdens in the Asia-Pacific region, although people of this region are no more overweight than Europeans and Americans.[6,9]
Several statistical techniques could be applied to identify the components of the MS. Principal component factor analysis (PCFA) is one such approach that groups quantitatively measured variables into clusters known as factors, on the basis of the correlation between variables.[10] PCFA was used to identify the domains of the risk variables of the MS. For example, if there is a single underlying cause for the clustering of the risk variables of the MS, then factor analysis should produce only 1 major factor or component. Therefore, identification of component(s) of the MS (considered to be the leading cause of CHD) is most essential for the etiology of CHD[7] However, a very few studies have so far been undertaken to identify the components of the MS in Asian Indian population[7‐9,11‐15] These studies suggested that there existed no single or central etiological factor for the clustering of MS phenotypes.[7,8] Therefore, it seems reasonable to argue that several underlying abnormalities do exist that might have relatively greater genetic basis.[7,8]
However, to the best of the authors' knowledge, no study has been undertaken on Asian Indians incorporating the genetic polymorphism(s), lipids, blood glucose, blood pressure, and body fat patterns simultaneously to identify the components of MS in this ethnic group. Keeping this view in mind, the present investigation is an attempt to find out the physiogenetic factors responsible for the observed variation of MS in the Asian Indian population living in the eastern part of India.
MATERIALS AND METHODS
Study population
The present community-based cross-sectional study comprised adult (≥30 years) Asian Indians living in and around Calcutta, India. A total of 350 (male = 184 and female = 166) individuals participated in the study. Pregnant women, women undergoing hormone therapy, as well as individuals with known illnesses, such as ischemic heart disease, type 2 diabetes mellitus, and hypertension were not included in the study. Prior to participation, public advertisement was given about the study with the help of the local officials. Individuals who responded to the advertisement were selected randomly. It is noteworthy that only unrelated adults from a household were included as participants to avoid the effects of intra-household clusters of CVD risk factors. The Institutional Ethics Committee (IEC) of the “Human Genetic Engineering Research Center” (HGERC), Calcutta, India, has approved the study. Written consent from the participants was also obtained prior to the actual commencement of the study.
Anthropometric measurements
Anthropometric measurements, namely, height, weight, waist circumference, and subcutaneous skinfold were obtained using standard techniques.[16] Height and weight (in light clothing) were measured to the nearest 0.1 cm and 0.5 kg, respectively. Waist circumference (WC) was measured to the nearest 0.1 cm using an inelastic tape. The minimum WC was measured at the level of natural waist, which was the narrowest part of the torso. Subcutaneous skinfolds at biceps, triceps, suprailiac and subscapular sides were measured and the sum of the 4 skinfolds was computed subsequently.
Blood pressure
Left arm systolic and diastolic blood pressure measurements were taken twice using sphygmomanometer and stethoscope and were averaged for analyses. A third measurement was taken only when the difference between the 2 measurements was >5 mmHg. Previous medical records for blood pressure were also taken into consideration.
Metabolic profiles
A fasting blood sample (~7 mL) was collected from each subject for determining the metabolic profiles. All the subjects maintained an overnight fast of ≥12 h prior to blood collection. Estimation of total cholesterol (TC), triglycerides (TG), high-density lipoprotein (HDL), and fasting blood glucose was carried out on separated serum by means of a semi-auto analyzer. All biochemical analyses were estimated in mg/dL (mg%) unit.
Genotyping
Two genetic polymorphisms, namely, ACE (I/D) and ApoE (Hha I) polymorphisms were studied in 138 participants. To study ACE (I/D) and ApoE (Hha I) polymorphisms, DNA samples of participants belonging to the highest (90th) and lowest (10th) percentiles of blood pressure centiles (percentiles) and/or lipids were considered. The detailed procedures of genotyping have been mentioned elsewhere.[17,18]
Statistical analyses
Descriptive statistics, such as mean and standard deviation (SD), of all the variables were calculated. Frequencies (%) of different alleles of ACE (I/D) and ApoE (HhaI) polymorphisms were also calculated. Factor analysis was undertaken to group quantitatively measured variables into clusters known as factors. It was done in 3 steps: computation of a correlation matrix for all variables included; factor extraction; and orthogonal rotation to make factors readily interpretable. The factors were extracted by PCFA in which the linear combinations of the variables were formed with the first component accounting for the largest amount of variance in the sample. Varimax rotation, an orthogonal rotation in which the factors are assumed to act independently (maximum likelihood), was used in the study. The components were all uncorrelated. Variables with a factor loading of at least 0.3 have generally been considered for interpretation, although it is suggested that only loading ≥0.4, which therefore shares at least 15% of the variance with a factor, should be used in the interpretation.[19] A factor loading of ≥0.4 was used to interpret the factors in the study. Previous studies have also used a factor loading of ≥0.4 to interpret the final rotated factor pattern.[7,8,19‐24]
All statistical analyses were performed using SPSS (PC+ version 10). A P value of < 0.05 (two-tailed) was considered as statistically significant.
RESULTS
The distribution of 184 males and 166 females by age groups and sex is presented in Table 1. It was observed that the participants were distributed more or less equally across the age groups and sex.
Table 1.
Distribution of study population by age group and sex
| Age group (years) | Male | Female |
|---|---|---|
| 30–39 | 20 | 39 |
| 40–49 | 47 | 48 |
| 50–59 | 63 | 43 |
| ≥60 | 57 | 36 |
The mean and standard deviation (SD) of anthropometric, lipids profile, blood glucose, and blood pressure measures are presented in Table 2. The mean ± SD WC in the study population was 89.38 ± 9.87. The mean (SD) triglyceride in the study was 141.95 (25.30). When the known South Asians' specific cutoffs were taken into consideration, the prevalence of MS in the study was 31.4%.
Table 2.
Descriptive statistics of study population (n = 350)
| Variables | Mean | SD |
|---|---|---|
| Waist circumference (cm) | 89.38 | 9.87 |
| Sum of four skinfolds (mm) | 97.09 | 26.58 |
| Total cholesterol (mg%) | 202.55 | 25.76 |
| Triglyceride (mg%) | 141.95 | 25.30 |
| High-density lipoprotein (mg%) | 44.35 | 4.72 |
| Fasting blood glucose (mg%) | 90.98 | 20.73 |
| Systolic blood pressure (mmHg) | 134.98 | 24.31 |
| Diastolic blood pressure (mmHg) | 82.82 | 11.01 |
The frequency of ACE (I/D) and ApoE (Hha I) gene polymorphisms is presented in Table 3. The frequency (%) of Insertion/Insertion (I/I) polymorphism for ACE gene was found to be the highest (37%) in the study. On the other hand, epsillion 3/ epsillion 3 for ApoE gene (Hha I) was the most frequent (60.9%) in the study population.
Table 3.
Frequency of ACE and ApoE genotypes (n = 138)
| Gene | Polymorphic type | n (%) |
|---|---|---|
| ACE | Insertion/Insertion (I/I) | 51 (37.0) |
| Insertion/Deletion (I/D) | 47 (34.0) | |
| Deletion/Deletion (D/D) | 40 (29.0) | |
| ApoE | epsillion 2/ epsillion 3 | 23 (16.7) |
| epsillion 2/ epsillion 4 | 09 (6.5) | |
| epsillion 3/ epsillion 3 | 84 (60.9) | |
| epsillion 3/ epsillion 4 | 18 (13.0) | |
| epsillion 4/ epsillion 4 | 04 (2.9) |
ACE, angiotensin converting enzyme; ApoE, apolipoproteinE
The factor-loading pattern of the 3 factors (components) identified in the study is presented in Table 4. Only variables with loading ≥0.4 were considered for interpretation. The loading of individual risk variable varied from 0.413 to 0.915. The factor 1 (lipids, 25.53%); factor 2 (centripetal fat with blood pressure, 20.41%) and factor 3 (ACE gene along with blood pressure, 19.44%) cumulatively explained 65.39% of the total variation of MS in the study [Figure 1]. Most importantly, the first two factors (lipids, centripetal fat along with blood pressure) cumulatively explained ~46% (45.94%) of the total variation of MS in the study population.
Table 4.
Factor loading pattern of cardiometabolic risk factors
| Factors | ||||
|---|---|---|---|---|
| Variables | Factor 1 | Factor 2 | Factor 3 | |
| Waist circumference | 0.196 | 0.868* | 0.207 | |
| Sum of 4 skinfolds | 0.108 | 0.915a | 0.060 | |
| Total cholesterol | 0.824* | –0.069 | 0.002 | |
| Triglyceride | 0.898* | 0.251 | 0.090 | |
| High-density lipoprotein | –0.867* | –0.233 | 0.051 | |
| Fasting blood glucose | 0.314 | 0.018 | 0.197 | |
| Systolic blood pressure | –0.114 | 0.281 | 0.866* | |
| Diastolic blood pressure | –0.028 | 0.413* | 0.808* | |
| ACE gene polymorphism | 0.280 | –0.283 | 0.588a | |
| ApoE gene polymorphism | 0.275 | 0.000 | 0.316 | |
| Variance explained | 25.53(%) | 20.41 | 19.44 | |
| Cumulative variance | 25.53(%) | 45.94 | 65.39 | |
ACE, angiotensin converting enzyme; ApoE, apolipoproteinE. a Loading with absolute value ≥ 0.4.
Figure 1.
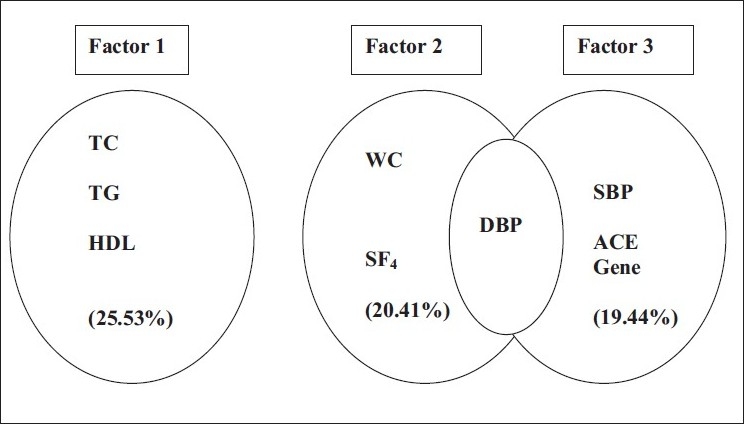
Clustering patterns of cardiometabolic risk variables in the study
DISCUSSION
The association of central obesity, glucose intolerance, hypertension, dyslipidemia, and hyperinsulinemia known as MS, has been observed in a number of ethnic groups worldwide. Studies across populations demonstrate that MS plays a pivotal role in the occurrence of CVD, including CHD. Therefore, identification of the components of the MS, including the genetic factors would be helpful in understanding the etiology of CHD. A very few studies have so far been undertaken to identify the underlying factors of MS in the Asian Indian population.[7‐9,11‐15] However, virtually no study has been undertaken on Asian Indians incorporating the genetic polymorphism(s), lipids, blood glucose, blood pressure, and body fat patterns simultaneously to identify the components of MS in this ethnic group. The present investigation was aimed at identifying the physiogenetic factors responsible for the observed variation of MS in Asian Indian population living in the eastern part of India.
PCFA had identified 3 factors with 65.39% that explained variance of the MS among the adult Asian Indians of Calcutta. Neither of the variables loaded on all the 3 components. These 3 factors could be identified as lipid (factor 1), centripetal fat with blood pressure (factor 3), and ACE gene along with blood pressure (factor 3). The first 2 factors, that is, lipids, centripetal fat and blood pressure cumulatively explained ~47% of the total variance of the MS in the study population. Except diastolic blood pressure, no overlapping of variables on more than 1 factor indicated that more than 1 variable is responsible for the ultimate phenotype of the MS. The present factor analysis confirmed the general findings from other factor analyses of the MS on different ethnic groups that had 3–4 factors identified [Table 5].
Table 5.
Factors of cardiometabolic risk variables across the ethnic groups
| Author(s) | Population | Findings |
|---|---|---|
| Bhagat et al., (2010)[25] | Adult Asian Indian (women) | Four factors in pre-and postmenopausal women were found and the factors were uncorrelated. It was suggested that a single risk axis for clustering of cardiometabolic phenotypes was highly unlikely. |
| Oliveira et al., (2010)[26] | Portugal (men and women) | Three factors were identified suggesting that more than one physiological mechanism is associated with high-sensitivity C-reactive protein in both men and women |
| Deshmukh et al., (2009)[27] | Bogalusa Heart Study (U.S. blacks, white men and women) | Effect of western dietary pattern (WDP) rich in refined grains, high-fat, dairy products, meat and sweets, and PDP consisted of whole grain, legumes, vegetables, fruits etc., were analyzed. Unlike WDP, diet rich in PDP had had inverse association with WC, triceps skinfold, plasma insulin, and MS. |
| Wu et al., (2008)[28] | Chinese (normal; IGT; type 2 diabetes mellitus) | Three factors were identified: I – blood pressure, II – insulin resistance III – adiposity/glucose. Therefore, it was considered that MS was not unified by a single underlying etiology, that is, insulin resistance. |
| Harriss et al., (2007)[29] | Australians (native and nonnative) | Dietary pattern and cardiovascular mortality; four dietary factors were identified: I – Mediterranean factor; II and III were vegetables and fruits, respectively; and IV – meat factor not associated with CVD mortality. It was suggested that traditional Mediterranean foods were associated with reduced cardiovascular mortality. |
| Ghosh (2005)[7] | Adult Asian Indian (men) | Four uncorrelated factors were identified: I – central obesity; II – centralized subcutaneous fat; III – lipid profi le, blood glucose; IV – blood pressure. Since no observed variable loaded on all the 4 factors, it was suggested that more than one physiological mechanism could be accounted for risk variables of the MS. |
| Hanley et al., (2004)[30] | Adult U.S. (African-American, Hispanic, and non-Hispanic whites) | Three factors were identified underlying among a group of infl ammation and MS variables: I – metabolic factor; II – inflammation factor; III – blood pressure factor. Insulin sensitivity was loaded on both the metabolic and inflammation variable clusters. Each factor significantly predicted diabetes and therefore it was supported by the emerging hypothesis that chronic subclinical inflammation is associated with insulin resistance and comprises a component of the MS. |
| Howard et al., (2003)[31] | Adult Women (white, black, Hispanic, Asian/Pacific islander women) | Four factors were identified: I – obesity factor; II – dyslipidemia factor; III – TC and LDL; and IV – blood pressure. It indicated that the components of insulin resistance syndrome was associated with CVD in postmenopausal women, although the magnitude of these relationships differed by ethnicity. |
| Lehto et al., (2000)[32] | Finland (adult men and women with type II diabetes) | The hyperinsulinemia cluster (a factor having high-positive loadings for BMI, TG, and insulin; and a high-negative loading for HDL) was predictive of death from CHD in patients with type 2 diabetes. Hence, it was mentioned that CVD risk factors clustering with endogenous hyperinsulinemia increase the risk of death from CHD in patients with type II diabetes not treated with insulin. |
BMI, body mass index; CVD, cardiovascular diseases; CHD, coronary heart disease; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TC, total cholesterol; TG, triglycerides; WDP, western dietary pattern; PDP, prudent dietary pattern; WC, waist circumference; IGT, impaired glucose tolerance; MS, metabolic syndrome.
The major limitation of this study is that it was performed on a relatively small sample size, and therefore is not representative of the Asian Indian population. Owing to considerable ethnic and cultural heterogeneity in the Asian Indian population, it is necessary to study other ethnic groups to see if the trends observed here also exist among them. However, it is noteworthy that results from different factor analysis are limited by differences in the ethnic group, sex, and age composition of the study samples, in the number of risk variables included, sample size, and cutoff points of loadings set by the investigators.[7] At the same time, to the best of our knowledge, no PCFA of MS has been undertaken so far, incorporating data on the angiotensin gene and the apolipoproteinE gene, along with the other confounding factors related to the MS in this part of the world. As Indian Diaspora offers a unique opportunity to study the “gene–environment” interaction involved in the etiology of CHD, further comparative studies between Indians living in India and Indians settled elsewhere could yield valuable information on the reasons behind the ethnic susceptibility to CHD among Indians.[7]
This model suggests that the clustering of the variables in MS is a result of multiple factors, including genetic polymorphisms with centripetal fat, lipids, and blood pressure playing key roles. Moreover, all the loaded risk variables, apart from the genetic polymorphism, are modifiable in nature. Therefore, it seems reasonable to argue that early prevention and proper intervention strategies to promote a healthy lifestyle could reduce the burden of MS in this part of the world.
Acknowledgments
AG received financial support (Ref. No. 5/9/48/2006-RHN vide RFC No. RHN/Adhoc/1/2009-10) from the Indian Council of Medical Research (ICMR), Government of India, New Delhi. MD received partial funding [Ref. No. F.PSW-176/09-10(ERO)] from the University Grants Commission (UGC), Government of India, New Delhi. The authors are grateful to the staff and technicians of the HGERC, Calcutta, India, for their sincere help in analyzing the metabolic profiles and genotyping. The authors are also indebted to all the subjects participated in the study.
Footnotes
Source of Support: AG received fi nancial support (Ref. No. 5/9/48/2006-RHN vide RFC No. RHN/Adhoc/1/2009-10) from the Indian Council of Medical Research (ICMR), Government of India, New Delhi. MD received partial funding [Ref. No. F.PSW- 176/09-10(ERO)] from the University Grants Commission (UGC), Government of India, New Delhi.,
Conflict of Interest: None declared.
REFERENCES
- 1.McKeigue PM, Shah B, Marmot MG. Relation of central obesity and insulin resistance with high diabetes prevalence and cardiovascular risk in South Asians. Lancet. 1991;337:382–6. doi: 10.1016/0140-6736(91)91164-p. [DOI] [PubMed] [Google Scholar]
- 2.Enas EA, Yusuf S, Mehta JL. Prevalence of coronary artery disease in Asian Indians. Am J Cardiol. 1992;70:945–9. doi: 10.1016/0002-9149(92)90744-j. [DOI] [PubMed] [Google Scholar]
- 3.Rajmohan L, Deepa R, Mohan V. Risk factors for coronary artery disease in Indians: emerging trends. Indian Heart J. 2000;52:221–5. [PubMed] [Google Scholar]
- 4.Misra A, Vikram NK. Insulin resistance syndrome (metabolic syndrome) and obesity in Asian Indian: evidence and implication. Nutrition. 2004;20:482–91. doi: 10.1016/j.nut.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Das M, Pal S, Ghosh A. Synergistic effects of ACE (I/D) and Apo E (HhaI) gene polymorphisms among the adult Asian Indians with and without metabolic syndrome. Diabetes Res Clin Pract. 2009;86:e58–61. doi: 10.1016/j.diabres.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 6.The Asia-Pacifi c Perspective: Redefi ning Obesity and its Treatment. Health Communication. Australia: 2000. WHO/IASO/IOTF. [Google Scholar]
- 7.Ghosh A. Factor analysis of metabolic syndrome among the middle-aged Bengalee Hindu men of Calcutta, India. Diabetes Metab Res Rev. 2005;21:58–64. doi: 10.1002/dmrr.481. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh A. Factor analysis of risk variables associated with metabolic syndrome in Asian Indian adolescents. Am J Hum Biol. 2007;19:34–40. doi: 10.1002/ajhb.20570. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A. Comparison of risk variables associated with the metabolic syndrome in pre-and post menopausal Bengalee women. Cardiovasc J Afr. 2008;19:183–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens J. Applied Multivariate Statistics for the Social Sciences. Mahwah, NJ: Lawrence Erlbaum; 1996. [Google Scholar]
- 11.Ramachandran A, Snehalatha C, Satyavani K, Sivasankari S, Vijay V. Cosegregation of obesity with familial aggregation of type 2 diabetes mellitus. Diabetes Obes Metab. 2000;2:149–54. doi: 10.1046/j.1463-1326.2000.00067.x. [DOI] [PubMed] [Google Scholar]
- 12.Snehalatha C, Sivasankari S, Satyavani K, Vijay V, Ramachandran A. Insulin resistance alone does not explain the clustering of cardiovascular risk factors in southern India. Diabet Med. 2000;17:152–7. doi: 10.1046/j.1464-5491.2000.00239.x. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran A, Sathyamurthy I, Snehalatha C, Satyavani K, Sivasankari S, Misra J, et al. Risk variables for the coronary artery disease in Asian Indians. Am J Cardiol. 2001;87:267–71. doi: 10.1016/s0002-9149(00)01356-4. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R. Burden of coronary heart disease in India. Indian Heart J. 2005;25:126–31. [PubMed] [Google Scholar]
- 15.Vikram NK, Misra A, Pandey RM, Luthra K, Wasir JS, Dhingra V. Heterogeneous phenotypes of insulin resistance and its implications for defining metabolic syndrome in Asian Indian adolescents. Atherosclerosis. 2006;186:193–9. doi: 10.1016/j.atherosclerosis.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Lohman TG, Roche AF, Martorell R, editors. Anthropometric standardization references manual. Chicago: Human Kinetics; 1998. [Google Scholar]
- 17.Das M, Pal S, Ghosh A. Angiotensin converting enzyme gene polymorphism (insertion/deletion) and hypertension in adult Asian Indians: a population-based study from Calcutta, India. Hum Biol. 2008;80:303–12. doi: 10.3378/1534-6617-80.3.303. [DOI] [PubMed] [Google Scholar]
- 18.Das M, Pal S, Ghosh A. Apolipoprotein E gene polymorphism and dyslipidaemia in adult Asian Indians: a population based study from Calcutta, India. Indian J Hum Genet. 2008;14:80–4. doi: 10.4103/0971-6866.45000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge AM, Boyko EJ, de Courten M, Zimmet PZ, Chitson P, Tuomilehto J, et al. Leptin and other components of the metabolic syndrome in Mauritius— a factor analysis. Int J Obes Relat Metab Disord. 2001;25:126–31. doi: 10.1038/sj.ijo.0801522. [DOI] [PubMed] [Google Scholar]
- 20.Kue Young T, Chateau D, Zhang M. Factor analysis of ethnic variation in the multiple metabolic (insulin resistance) syndromes in three Canadian populations. Am J Hum Biol. 2002;14:649–58. doi: 10.1002/ajhb.10083. [DOI] [PubMed] [Google Scholar]
- 21.Meigs JB, D'Agostino RB, Sr, Wilson PW, Cupples LA, Nathan DM, Singer DE. Risk variables clustering in the insulin resistance syndrome.The Framingham Offspring Study. Diabetes. 1997;46:1594–600. doi: 10.2337/diacare.46.10.1594. [DOI] [PubMed] [Google Scholar]
- 22.Gray RS, Fabsitz RR, Cowan LD, Lee ET, Howard BV, Savage PJ. Risk factor clustering in the insulin resistance syndrome.The Strong Heart Study. Am J Epidemiol. 1998;148:869–78. doi: 10.1093/oxfordjournals.aje.a009712. [DOI] [PubMed] [Google Scholar]
- 23.Edwards KL, Burchfiel CM, Sharp DS, Curb JD, Rodriguez BL, Fujimoto WY, et al. Factors of the insulin-resistance syndrome in non-diabetic and diabetic elderly Japanese-American men. Am J Epidemiol. 1998;147:441–7. doi: 10.1093/oxfordjournals.aje.a009469. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Srinivasan SR, Elkasabany A, Berenson GS. Cardiovascular risk factors clustering factors of insulin resistance syndrome (syndrome X) in a biracial (black and white) population of children, adolescent and young adults. Am J Epidemiol. 1999;150:667–74. doi: 10.1093/oxfordjournals.aje.a010069. [DOI] [PubMed] [Google Scholar]
- 25.Bhagat M, Mukherjee S, De P, Goswami R, Pal S, Das M, et al. Clustering of cardiometabolic risk factors in Asian Indian women: Santiniketan women study. Menopause. 2010;17:359–64. doi: 10.1097/gme.0b013e3181bfac28. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira A, Lopes C, Severo M, Rodríguez-Artalejo F, Barros H. Nutr Metab Cardiovasc Dis 2010 In press; Body fat distribution and C-reactive protein: a principal component analysis. [DOI] [PubMed] [Google Scholar]
- 27.Deshmukh-Taskar PR, O'Neil CE, Nicklas TA, Yang SJ, Liu Y, Gustat J, et al. Dietary patterns associated with metabolic syndrome, sociodemographic and lifestyle factors in young adults: the Bogalusa Heart Study. Public Health Nutr. 2009;12:2493–503. doi: 10.1017/S1368980009991261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu CZ, Lin JD, Li JC, Hsiao FC, Hsieh CH, Kuo SW, et al. Factor analysis of metabolic syndrome using direct measurement of insulin resistance in Chinese with different degrees of glucose tolerance. Indian J Med Res. 2008;127:336–43. [PubMed] [Google Scholar]
- 29.Harriss LR, English DR, Powles J, Giles GG, Tonkin AM, Hodge AM, et al. Dietary patterns and cardiovascular mortality in the Mediterranean Collaborative Cohort Study. Am J Clin Nutr. 2007;86:221–9. doi: 10.1093/ajcn/86.1.221. [DOI] [PubMed] [Google Scholar]
- 30.Hanley AJ, Festa A, D'Agostino RB, Jr, Wagenknecht LE, Savage PJ, Tracy RP, et al. Metabolic and inflammation variable clusters and prediction of type 2 diabetes: factor analysis using directly measured insulin sensitivity. Diabetes. 2004;53:1773–81. doi: 10.2337/diabetes.53.7.1773. [DOI] [PubMed] [Google Scholar]
- 31.Howard BV, Criqui MH, Curb JD, Rodabough R, Safford MM, Santoro N, et al. Risk factor clustering in the insulin resistance syndrome and its relationship to cardiovascular disease in postmenopausal white, black, Hispanic, and Asian/Pacific Islander women. Metabolism. 2003;52:362–71. doi: 10.1053/meta.2003.50057. [DOI] [PubMed] [Google Scholar]
- 32.Lehto S, Rönnemaa T, Pyörälä K, Laakso M. Cardiovascular risk factors clustering with endogenous hyperinsulinaemia predict death from coronary heart disease in patients with type II diabetes. Diabetologia. 2000;43:148–55. doi: 10.1007/s001250050023. [DOI] [PubMed] [Google Scholar]


