Abstract
Human embryonic stem cells (hESCs) hold great promise in regenerative medicine. However, before the full potential of these cells is achieved, major basic biological questions need to be addressed. In particular, there are still gaps in our knowledge of the molecular mechanisms underlying the derivation of hESCs from blastocysts, the regulation of the undifferentiated, pluripotent state, and the control of differentiation into specific lineages. Furthermore, we still do not fully understand the tumorigenic potential of hESCs, limiting their use in regenerative medicine. The RB pathway is a key signaling module that controls cellular proliferation, cell survival, chromatin structure, and cellular differentiation in mammalian cells. Members of the RB pathway are important regulators of hESC biology and manipulation of the activity of this pathway may provide novel means to control the fate of hESCs. Here we review what is known about the expression and function of members of the RB pathway in hESCs and discuss areas of interest in this field.
Keywords: Retinoblastoma, RB pathway, cell cycle, human embryonic stem cells (hESCs)
Human embryonic stem cells (hESCs) are a unique cell type that is widely touted for its potential use in regenerative medicine [Carpenter et al., 2003; Luong et al., 2008; Muller and Lengerke, 2009; Pera et al., 2000; Yu and Thomson, 2008]. However, before hESCs can be safely used in medical applications, their basic biology must be better understood and their tumorigenicity must be controlled [Knoepfler, 2009]. hESCs are characterized by the ability to proliferate while retaining their pluripotent potential. This self-renewal potential requires different gene networks and a different cell cycle structure than adult cells, which cycle more slowly or remain quiescent. However, most of our knowledge of the cell cycle in ESCs still comes from work done on cells from mice [Ciemerych and Sicinski, 2005; Savatier et al., 2002; Ying et al., 2008] and accumulating evidence indicates that murine embryonic stem cells (mESCs), while similar, are not necessarily equal to primate ESCs [Burdon et al., 2002; Fluckiger et al., 2006; Ginis et al., 2004; Sato et al., 2004; Thomson et al., 1998]. Therefore, it is important to study central regulators of the cell cycle directly in hESCs.
The regulation of the cell cycle in ESCs may be critical for maintaining a delicate balance between unrestricted proliferation, which could allow DNA damage and mutations to accumulate, and a slower cell cycle that may render the cells more susceptible to differentiation-inducing signals [Ying et al., 2008]. It is possible that the rapid cell cycle of ESCs is necessary to maintain continual self-renewal and to resist differentiation. In particular, recent reports suggest the length of the G1 phase of the cell cycle is critical for determining the fate of mESCs as it may be a key period when a cell decides whether to proliferate, differentiate, senesce, enter quiescence, or initiate apoptosis [Blomen and Boonstra, 2007; Orford and Scadden, 2008; White and Dalton, 2005]. The longer the G1 phase, the more likely a cell may become susceptible to a differentiation inducing signal, such as MAPK signaling [Burdon et al., 1999; Orford and Scadden, 2008]. Thus, tight regulation of cell cycle progression, especially at the G1/S transition, is probably critical to maintain the self-renewal potential of hESCs, and a key issue in this field is to understand how hESCs continuously proliferate without differentiating.
The product of the human retinoblastoma tumor suppressor gene (RB) belongs to a cellular pathway (Figure 1) that has been implicated in cell cycle control, differentiation, chromosome stability, chromatin structure, and a multitude of other functions [Burkhart and Sage, 2008; Knudsen and Knudsen, 2008; Macleod, 2008; Sun et al., 2007]. Briefly, the RB protein plays a key role in restricting the G1 to S transition in the cell cycle and its loss of function in mammalian cells is most often associated with unrestricted and aberrant proliferation. RB controls cell cycle progression through several mechanisms, including through its ability to bind to the E2F family of transcription factors. During normal cell cycle progression, phosphorylation of RB by Cyclin/CDK complexes in G1 and S changes RB structure and inhibits the interaction between RB and E2Fs. This allows the freed E2Fs to activate expression of their targets, which include key components of the machinery needed for DNA replication and S phase progression. Some evidence suggests that RB may also be important to maintain chromosomal stability as loss of RB function in mESCs results in an increased loss of a selectable chromosomal marker compared to wild-type cells in culture [Zheng et al., 2002]. Finally, the RB protein has been shown to bind to various transcription factors to influence differentiation. For example, RB binds to MyoD to induce expression of late markers of muscle differentiation and has also been shown to bind to and repress inhibitor of differentiation 2 (ID2) to promote differentiation [De Falco et al., 2006; Lasorella et al., 2000]. Thus, the can regulate both cellular growth and differentiation. Below, we will highlight the research that has given us crucial insight into the regulation of the cell cycle in hESCs and identify the gaps that remain in our knowledge. Specifically, after describing what is known about the pattern of expression of key members of the RB pathway in hESCs, we will investigate the potential function(s) of the RB pathway in hESCs, and we will conclude by discussing key questions that remain unanswered in this field.
Figure 1. Schematic representation of the RB pathway in hESCs.
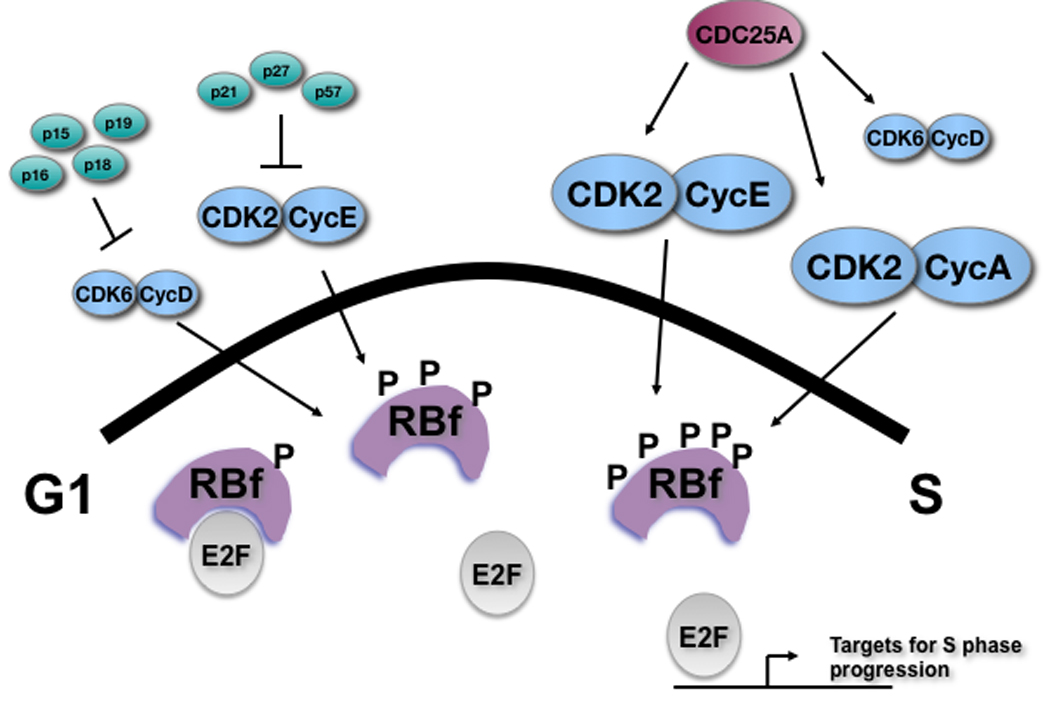
Members of the RB pathway that are expressed in hESCs are shown. CKIs (INK4 family and Cip/Kip family) are expressed at low or undetectable levels. In contrast to mESCs where RB family members (RBf) are thought to be always hyperphosphorylated (P represents a phosphorylated residue), RB is found in a hypophosphorylated state in G1 in hESCs and becomes hyperphosphorylated for the rest of the cell cycle. Increased size of the Cyclin/CDK complexes reflects an increase of Cyclin E and Cyclin A protein levels and activity near the G1/S transition.
Expression of RB pathway members in hESCs
Compared to most other cell types, both mouse and human ESCs have an abbreviated G1 phase and display a higher percentage of cells in S phase: mESCs have a very short cell cycle, completing an entire generation in about 8–10 hours [White and Dalton, 2005]. Primate and human ESCs exhibit a slightly longer cell cycle (~12–16 hours) that is still significantly shorter than that of somatic cells [Becker et al., 2006; Fluckiger et al., 2006]. This observation suggests that the control of cell cycle progression in mESCs and hESCs is different than in other cell types, which has led several groups to analyze expression of central cell cycle regulators in hESCs.
At the top of the RB pathway, Cyclin-dependent kinase inhibitors (CKIs) are found to be expressed at very low levels or absent in hESCs. Members of the Cip/Kip family of CDK inhibitors, p21, p27, and p57 are barely detectable at the RNA level in hESCs [Becker et al., 2006]. At the protein level, p57 is detectable but p21 and p27 are low or absent [Sengupta et al., 2009]. RNA from members of the INK4 family of inhibitors, p16, p18, and p19, whose products inhibit CDK4 and CDK6, is not expressed or expressed at very low levels in hESCs [Miura et al., 2004]. At the protein level, p15INK4b and p16 INK4a are not detectable and p18 INK4c and p19 INK4d have very low expression in hESCs [Zhang et al., 2009]. These observations correlate well with what has been shown in mESCs [Faast et al., 2004; Savatier et al., 1996; Stead et al., 2002; White and Dalton, 2005] and would be expected given the abbreviated cell cycle of hESCs.
CDKs are the targets of the CKIs. At the mRNA level in hESCs, CDK4 was shown to have a higher expression level compared to CDK2 and CDK6 [Becker et al., 2006]. There also appears to be some cell cycle dependent regulation at the RNA level; CDK2, CDK4, and CDK6 have increased mRNA levels in G1 and CDK1 in G2 [Becker et al., 2006; Neganova et al., 2009]. In mESCS, CDK2 protein levels were found to be high and lack cell cycle periodicity [Stead et al., 2002]. At the protein level, CDK6 is expressed at a low level in hESCs [Card et al., 2008; Neganova et al., 2009]. CDK4 and CDK1 display unchanging expression of their proteins throughout all phases of the cell cycle [Neganova et al., 2009]. Thus, all CDKs may participate in the rapid cell cycle of hESCs.
One of the most striking differences between mESCs and hESCs is the cell cycle dependent expression of Cyclins, the partners of CDKs (Figure 2). In mESCs, all Cyclins are constitutively expressed except Cyclin B1, whose expression peaks during M phase [White and Dalton, 2005]. Transcriptionally, Cyclin D1, D2, and D3 have been shown to be upregulated in G1 and Cyclin A, E, B1 and D2 to be upregulated at G2 in hESCs [Becker et al., 2006; Neganova et al., 2009]. In a comparison of mRNA levels using quantitative RT-PCR, Cyclins A2, B1, and B2 have the highest expression levels compared to the other Cyclins [Becker et al., 2006]. However, Cyclins can be regulated post-transcriptionally and in fact, hESCs show slightly different Cyclin expression patterns when examined at the protein level: the Cyclin B1 pool of proteins is upregulated around G2/M, similar to mESCs [Ghule et al., 2007; Neganova et al., 2009; White et al., 2005]. However, in contrast to mESCs, where Cyclin E levels remain constant throughout the cell cycle, Cyclin E protein levels increase around the G1/S transition and Cyclin A protein levels are upregulated in late G1/S through G2/M in hESCs [Ghule et al., 2007; Neganova et al., 2009]. In another study, FACS analysis and immunofluorescence staining showed that Cyclin E is constitutively expressed but Cyclin A is upregulated in S and G2/M in hESCs [Filipczyk et al., 2007]. This cyclical nature of Cyclin E and Cyclin A expression is more similar to what is seen in adult cells than in mESCs, and correlates to the observation that hESCs cycle more slowly than mESCs. At the protein level, Cyclin D appears to be present and expressed constitutively throughout the cell cycle of hESCs using Western blot analysis [Ghule et al., 2007; Neganova et al., 2009]. However, one research group reported being unable to detect Cyclin D1, D2, and D3 by immunofluorescence in undifferentiated hESCs [Filipczyk et al., 2007]. The discrepancy between these reports may be due to a higher sensitivity of the western blot assay, some contribution from spontaneously differentiated cells, or some variability between hESC lines as each of the three groups used different hESCs in their experiments.
Figure 2. Expression of G1/S Cyclins in embryonic stem cells and adult cells.
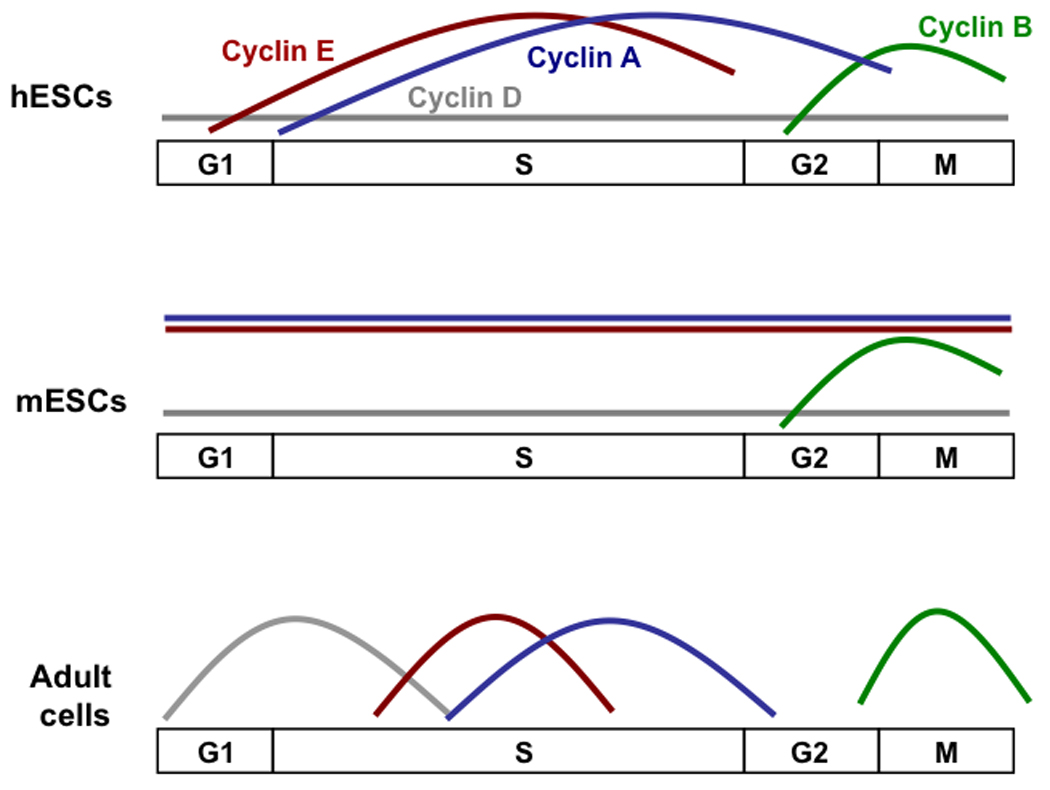
mESCs display little cell cycle dependent regulation of Cyclins except for Cyclin B. In contrast, Cyclins in hESCs do show cell cycle-dependent expression, except for the D Cyclins, which are expressed at a low level. This suggests that the regulation of the cell cycle of hESCs differs from what has been shown for mESCs in culture, and may more closely resemble mouse epiblast stem cells than mESCs.
Cyclins form complexes with CDKs to phosphorylate RB and other cellular targets. In mESCs, the predominantly active complex is Cyclin E/CDK2, with high Cyclin D3/CDK6 activity but almost undetectable activity from Cyclin D1/CDK4 [White and Dalton, 2005]. Using immunoprecipitation studies, multiple Cyclin/CDK complexes have been detected in hESCs. Cyclin D1 was shown to be associated mainly with CDK6 and to a lesser extent with CDK4 [Neganova et al., 2009]. In contrast, Cyclin D2 is mostly associated with CDK4 and Cyclin D3 with both CDK4 and CDK6 [Neganova et al., 2009]. Cyclin A/CDK2 and Cyclin E/CDK2 complexes are present in hESCs as well as complexes between CDK2 and CDC25A (a phosphatase that can activate CDKs) and CDK2 with c-Myc (a cellular oncogene) [Neganova et al., 2009]. CDK2 was shown to have the highest kinase activity in S phase, while CDK4 and CDK6 displayed the highest kinase activity in the G1 phase in hESCs [Neganova et al., 2009]. In asynchronously cycling hESCs, CDK2 displayed the highest kinase activity overall [Neganova et al., 2009]. CDC25A is upregulated at the protein level in G1 [Neganova et al., 2009]. These finding underscore the differences between mouse and human ESCs; mESCs rely on high CDK2 activity with little or no CDK4 activity, while hESCs display CDK2, 4, and 6 activity in a cell cycle regulated manner.
RB and its two family members, p107 and p130, have been shown to be expressed in hESCs [Becker et al., 2007] and mESCs [Sage et al., 2000]. In mESCs, RB and its family members p107 and p130 are hyperphosphorylated and do not associate with E2Fs [White and Dalton, 2005]; mESCs deficient for the three RB family members have no reported cell cycle phenotype [Dannenberg et al., 2000; Sage et al., 2000]. In hESCs, p130 seems to be the predominantly expressed family member in hESCs at the mRNA level [Becker et al., 2007]; however, p130 protein levels are highly regulated and high levels of p130 RNA may not correlate with protein levels [Tedesco et al., 2002]. In response to DNA damage, RB mRNA is modestly increased while p107 and p130 mRNA expression is decreased. At the protein level in hESCs, RB exists in both the hyperphosphorylated and hypophosphorylated forms, as assessed by western blot and immunofluorescence staining, while p107 and p130 levels have not been studied [Filipczyk et al., 2007]. Further analysis showed that a pool of hypophosphorylated RB exists, predominantly in the G1 phase of the cell cycle [Filipczyk et al., 2007]. Future studies should comprehensively examine the expression levels and the phosphorylation state of RB and its family members at each stage of the cell cycle using multiple methods.
A comparison of mRNA expression for E2F family members in hESCs indicates that the E2F4 and E2F5 repressors have the highest expression levels [Becker et al., 2007]. After DNA damage, an increase in E2F5 and E2F6 mRNA was observed [Becker et al., 2007]. Nothing is known in hESCs about the expression of the three E2F family members that can bind DNA and control gene expression independently of RB family members, E2F6 [Cartwright et al., 1998; Gaubatz et al., 1998], E2F7, and E2F8 [Li et al., 2008; Moon and Dyson, 2008]. Because of the high activity of Cyclin/CDK complexes in hESCs, one would expect that RB family members would be largely unable to bind to E2Fs, allowing E2F1-3 to activate their target genes and leaving E2F4-5 off the DNA. However, the DNA binding activity of the three RB family members and eight E2F family members in hESCs has not been investigated. It also remains to be shown whether E2F dependent transcription is cell cycle independent, as in mESCs, or cell cycle dependent [White and Dalton, 2005].
Other major regulators of the cell cycle whose activity is connected to the RB pathway include p53, c-Myc, and telomerase. At the RNA level, p53 is not expressed or expressed at very low levels in hESCs [Miura et al., 2004]. p53 is also low at the protein level, but its expression can increase upon exposure to UV or gamma-irradiation [Qin et al., 2007]. At the RNA level, c-Myc expression is the highest in the G1 phase of the cell cycle in hESCs, but has been shown to increase in S and G2 at the protein level [Neganova et al., 2009]. Telomerase activity has also been linked to the cell cycle, having been shown to both stimulate cell proliferation and mediate expression of growth-promoting and growth-inhibiting genes [Jagadeesh and Banerjee, 2006; Yang et al., 2008]. In hESCs, both the telomerase reverse transcriptase (TERT) and the RNA component (TR) were found to be expressed, and high amounts of telomerase activity was detected [Saretzki et al., 2008]. The TERT protein has also been shown to control the activity of adult stem cells [Sarin et al., 2005]. This suggests that telomerase may be important to regulate the cell cycle of hESCs, an idea that has not been thoroughly tested yet.
In conclusion, major cell cycle regulators in hESCs appear to display unique expression patterns compared to mESCs and adult cells. In adult cells, dramatic cell cycle dependent expression changes are important for progression through the different phases of the cell cycle. hESCs lack this marked periodicity in expression of cell cycle regulators and CDK activity is much higher than that observed in adult cells. In contrast to mESCs, CDK2 activity decreases in activity briefly during G1, which may result in the de-phosphorylation of RB, potentially slowing cell cycle progression and creating a time window when hESCs in culture may be more sensitive to differentiation signals. Because both mESC and hESCs proliferate indefinitely while retaining their undifferentiated status, these differences may not be critical for the maintenance of self-renewal. It is also possible that the culture conditions for hESCs are still not as optimal as for mESCs, which would lead to some culture stress and may affect cell cycle progression and explain why the two ESC types show these differences. Alternatively, it has been suggested that mESCs and hESCs represent two different stages of embryo development [Brons et al., 2007; Tesar et al., 2007]. Therefore, the cell cycle differences between the mESCs and hESCs may reflect the properties of two developmentally distinct pluripotent cell populations.
Strikingly, the pattern of expression of members of the RB pathway changes dramatically upon differentiation. Cyclin D1, 2, 3, and Cyclin B1 all show increased protein levels upon differentiation while Cyclin E and Cyclin A levels decrease [Card et al., 2008; Filipczyk et al., 2007; Neganova et al., 2009]. CDK4 and CDK6 protein levels also decrease but CDK2 may increase upon differentiation [Neganova et al., 2009]. CKIs p16, p18, and p21 mRNAs increase upon differentiation, and p27 protein levels increase while levels of its inhibitor, SKP2, decrease as differentiation progresses, thereby re-instating the regulation seen in somatic cells [Egozi et al., 2007; Miura et al., 2004]. Telomerase activity, TERT and TR also decrease upon differentiation [Saretzki et al., 2008]. The specific patterns of expression of cell cycle regulators in hESCs suggest that some of these cell cycle regulators have key functions in the proliferative capacity of these cells, the maintenance of self-renewal potential, and the prevention of untimely differentiation.
Functional studies of cell cycle regulators in hESCs
While there is no published report of any direct functional studies for RB and its family members p107 and p130 in hESCs, many groups have begun to tease out the role of the RB pathway by assessing the function of critical upstream regulators of the G1/S transition of the cell cycle (Figure 1). The only CKI that has been manipulated thus far in hESCs is p21. As expected from its role in somatic cells, overexpression of p21 leads to an increase in the percentage of hESCs in G1 [Wang et al., 2008]. Another study showed an increase in markers of differentiation upon activation of p21, both in p53-dependent and independent manners [Maimets et al., 2008]. p21 normally acts to inhibit Cyclin E/CDK2 complexes so this data suggests that this kinase activity is important for cell cycle progression and prevention of differentiation in hESCs. Future experiments should compare these results to studies of the INK4 family and the other Cip/Kip family members to determine if certain Cyclin/CDK complexes are more critical for cell cycle progression than others.
Two independent groups showed that hESCs arrest in the G1 phase of the cell cycle upon inhibition of CDK2 activity [Filipczyk et al., 2007; Neganova et al., 2009]. Treatment with CDK2 inhibitor roscovitine also revealed an increase in the number of cells expressing hypophosphorylated RB, as assessed by immunofluorescence [Filipczyk et al., 2007]. Concomitantly to G1 arrest, knockdown of CDK2 expression by siRNAs induced differentiation toward the extra-embryonic lineage accompanied by a corresponding increase in Cyclin D2, p21 and p27 protein levels [Neganova et al., 2009]. This G1 arrest upon decreased CDK2 activity is consistent with what was seen with p21 overexpression. These data also correlate with what is seen in mESCs where inhibition of CDK2 results in a lengthening of the cell cycle [White and Dalton, 2005], but hESCs show a more dramatic phenotype with an arrest in G1. CDK2 is also involved in the progression of the cell cycle in a process independent from RB. p220NPAT, a substrate for CDK2/Cyclin E-mediated phosphorylation interacts with its cofactor HiNF-P to modulate histone H4 expression necessary for cell cycle progression in hESCs [Becker et al., 2007; Ghule et al., 2007]. It was recently shown that p57 was the most effective Cip/Kip family member at blocking the p220NPAT/HiNF-P pathway by forming a complex with p220NPAT and suppressing its phosphorylation by CDK2/Cyclin E complexes [Mitra et al., 2009]. This suggests that members of the RB pathway may act through RB independent means to control cell cycle progression. Together, these results support a role for CDK2 in the G1/S transition in hESCs and also generate evidence to support a model where a change in the length of the G1 phase shifts the delicate balance between self-renewal and differentiation.
In contrast to CDK2, inhibition of CDK4 activity elicits no obvious changes in the morphology or the cell cycle profile of hESCs [Filipczyk et al., 2007]. Overexpression of Nanog in hESCs revealed an increase in the cell cycle regulators CDK6 and CDC25A at both the RNA and protein level, and a corresponding decrease in response to Nanog knockdown [Zhang et al., 2009]. Further analysis revealed the ability of the C terminus of Nanog to bind to and regulate the expression of both CDK6 and CDC25A, adding another piece of evidence that self-renewal/pluripotency and cell cycle control are entwined [Zhang et al., 2009]. Functional analyses revealed that overexpression of either CDK6 or CDC25A results in a faster S phase (~2 hours) than in control cells, and this overexpression rescued the delay in cell cycle entry after synchronization seen in hESCs with Nanog knockdown [Zhang et al., 2009]. CDK6 knockdown also showed a delay in S phase entry or progression after release from nocodazole synchronization and similarly, CDC25A knockdown resulted in retention of cells in G1 [Zhang et al., 2009]. These results suggest CDK6 and CDC25A are, at least in part, acting downstream of Nanog to control the G1 to S transition in hESCs, thereby linking the regulatory networks critical for the maintenance of stemness to the cell cycle machinery. A work-in-progress model of these cell cycle regulators in the G1 to S transition of hESCs is shown in Figure 1. Cyclins have not been functionally studied in hESCs, but recently Cyclin A has been shown to be essential for cell cycle progression in mESCs, suggesting these cell cycle regulators may be critical to the biology of ESCs [Kalaszczynska et al., 2009].
The p53 pathway directly interacts with the RB pathway and also has a role in cell cycle regulation. Knockdown of p53 in hESCs results in an increase in proliferation and single cell survival, with decreased spontaneous apoptosis and a decreased ability to differentiate into definitive endoderm using low concentrations of Activin A [Qin et al., 2007]. This correlates well with the phenotype of overexpression of HDM2, a direct inhibitor of p53. HDM2 overexpression increased single cell survival in hESCs, while decreasing spontaneous differentiation and also decreasing the RNA levels two p53 target genes, Bax and Nova [Qin et al., 2007]. These results highlight the role of p53 in the survival of hESCs. Another group studied the result of p53 activation using the small molecule nutlin [Maimets et al., 2008]. Treatment with nutlin rapidly induced expression of p21 and HDM2, two direct p53 target genes, and resulted in an increased percentage of cells in G1 and an increase in markers of differentiation [Maimets et al., 2008]. Furthermore, in this study, activation of p21 was accompanied by phosphorylation of CDK2 and subsequent degradation of Cyclin A and Cyclin E, which correlated with p53 de-phosphorylation [Maimets et al., 2008]. Treatment of hESCs with sodium butyrate, an inducer of differentiation, activated p21, but not p53, resulting in expression of differentiation markers similar to that seen with nutlin treatment, suggesting that the effects of p53 activation are mediated by p21 [Maimets et al., 2008]. These data again point to a tight correlation between proliferation and differentiation in hESCs.
c-Myc is both a target of E2F and a regulator of the expression of Cyclin E and CDK2 [Bartek and Lukas, 2001; Neganova and Lako, 2008]. Overexpression of c-Myc in hESCs triggers both apoptosis and differentiation [Sumi et al., 2007]. This phenotype coincides with an increase in p53 and p21 protein levels; c-Myc-induced differentiation occurred independently of p53 upregulation, as assayed in knock-down cells, but the role of p21 was not tested in this experiment [Sumi et al., 2007]. Also lacking is the distribution of the cell cycle in response to c-Myc overexpression. One could postulate that an increase in p21 after overexpression of c-Myc would delay or arrest the cells in the G1 phase, resulting in the differentiation of c-Myc-expressing hESCs. Future experiments could test this possibility, and if correct, it would add to the growing evidence that cell cycle regulation is tightly controlled and critically linked to self-renewal ability in hESC populations.
Finally, manipulation of the telomerase reverse transcriptase, TERT, in hESCs has shown its importance in maintaining pluripotency and in cell cycle regulation. Overexpression of TERT in hESCs resulted in increased proliferation and suppression of differentiation, with an increased percentage of cells in S phase, at the expense of G1 [Yang et al., 2008]. This was accompanied by an increase in protein levels of Cyclin D1, transcription of CDC6, an E2F target gene, and an increase in RB phosphorylation [Yang et al., 2008]. Knockdown of TERT resulted in the opposite effect; decreased proliferation with an increase in the percentage of cells in G1, down-regulation of Cyclin D1 and CDC6, accompanied by loss of pluripotency and an increase in markers of differentiation [Yang et al., 2008]. These data suggest that TERT may play a critical role in regulating the cell cycle and pluripotency of hESCs.
An increasing number of cell cycle regulators have been shown to be regulated themselves by microRNAs, including at the G1/S transition [Card et al., 2008; Sengupta et al., 2009; Wang et al., 2008]. Some of these experiments suggest that p57, p21, p130, and Cyclin D1 are all regulated by microRNAs and may play a critical role in cell cycle progression in hESCs [Card et al., 2008; Sengupta et al., 2009; Wang et al., 2008]. However, each of the microRNAs studied has multiple targets and, in the absence of functional experiments, it is difficult to identify which target has a more prominent role in hESCs.
Thus far, the data reviewed has shown that a delay or arrest of the cell cycle, particularly in G1, results in an increase of differentiation in hESCs. This is strong evidence that hESCs are susceptible to differentiation signals when G1 is lengthened. Therefore, tight control of a rapid cell cycle is critical to the self-renewal of hESCs. However, the precise epistatic relationship between the control of self-renewal and differentiation as regulated by the RB pathway remains unclear. Important functional studies of RB itself are lacking in hESCs, and it is possible that RB has critical functions in hESCs aside from cell cycle control. Finally, there has been little evidence for the role of p107 and p130, the two family members of RB. Future studies should examine the specific functions of RB, p107, and p130 in hESCs.
Future directions
The studies reviewed here provide substantial evidence that proper regulation of the G1/S transition of the cell cycle is critical for hESCs. These studies have also begun to point to a specific role for certain members of the RB pathway in hESCs. However, it is clear that these experiments are just touching on the complexity of cell cycle regulation in these cells and that a number of follow-up studies will be required to better understand how hESCs achieve indefinite proliferation while retaining the ability to produce differentiated progeny. One first important aspect of these experiments will be to study the expression and mechanisms of action of the RB pathway in several independently derived lines of hESCs, to ensure that the observations made are not due to the specific genetic make-up of one or a few cell lines or to specific culture conditions.
A second area of future investigation is to continue exploring the differences between hESCs and ESCs from other species, including mESCs (Figure 2). For instance, it has been suggested that mESCs and hESCs represent two different stages of embryo development [Brons et al., 2007; Tesar et al., 2007]. This raises the question whether the cell cycle differences between mESCs and hESCs are due to the fact that these cells arise from two distinct developmental stages. These cell cycle differences may also eventually inform us about the minimal and maximal length of the G1 phase that is compatible with maintenance of self-renewal potential and rapid proliferation in ESCs.
Another key area to continue to investigate in future studies is the connection between the cell cycle machinery and the network of factors that are essential to maintain the undifferentiated phenotype of ESCs and induce the reprogramming of cells into pluripotent cells (iPSCs), including core factors such as Oct4, Sox2, c-Myc, Klf4 and Nanog [Hanna et al., 2008; Yamanaka, 2009]. An obvious link between the RB pathway and these stemness factors is c-Myc, a transcription factor that works both downstream and in parallel of the RB pathway [Santoni-Rugiu et al., 2000; Sears and Nevins, 2002]. E2F was also found to be a transcription factor whose targets are significantly enriched in a network of proteins associated with pluripotency in murine pluripotent stem cells, a category that includes embryonic stem cells [Muller et al., 2008]. Recent evidence also suggests that E2F activity may act as a regulatory co-factor for Oct4 on the promoter of Oct4 target genes and that ORC1L, a direct E2F target involved in DNA replication, belongs to the core Oct4 regulatory network, suggesting that E2F and Oct4 activities are linked (Figure 3) [Chavez et al., 2009]. Both Sox2 and Nanog may display some binding activity on the E2F3 promoter, although these data from high throughput chromatin immunoprecipitation experiments need to be functionally validated [Boyer et al., 2005]. Nanog may also directly regulate cell cycle progression by directly binding to CDK6 and CDC25A, two regulators of the G1/S [Zhang et al., 2009]. Finally, very little is known on how the cell cycle regulators of the RB pathway may impact the reprogramming of cells into iPSCs, but it is very likely that accelerating or slowing the cell cycle may change the reprogramming efficiency. For instance, decreased p53 levels have been shown to increase cellular reprogramming [Zhao et al., 2008]. It is likely that many more connections exist between the master regulators of cell cycle and stemness, including via the regulation of microRNAs [Gunaratne, 2009], and this will be an area of active investigation in the next few years. These studies will have an impact not only on our basic knowledge of embryonic development and stem cell biology but certainly also on our understanding of the mechanisms of tumorigenesis. Some evidence points to a direct connection of the RB pathway to self-renewal; other evidence has simply shown that some of the cell cycle regulators are critical for maintaining the rapid cell cycle, and a disturbance of this can result in loss of self-renewal. Future research in the areas described here will undoubtedly gain us knowledge that will be critical for the use of hESCs and iPSCs in therapeutic applications.
Figure 3. Functional interactions between the RB pathway and the core regulatory network maintaining stemness in hESCs.
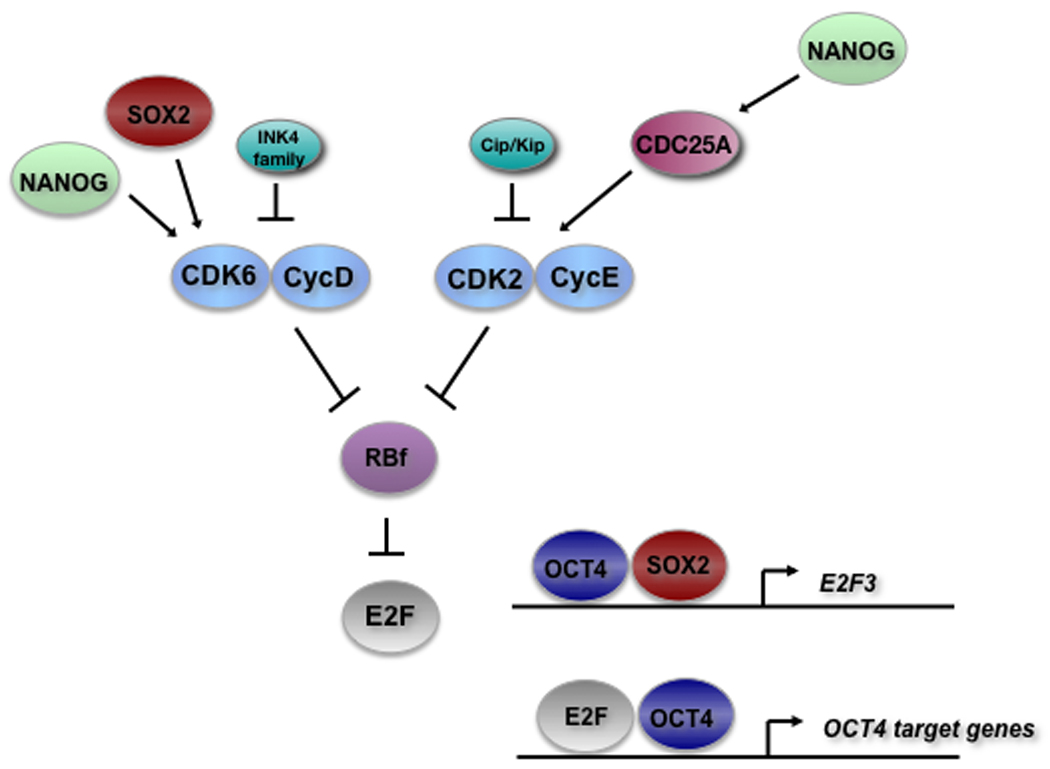
Emerging evidence suggests that members of the RB pathway are regulated by and function with factors involved in the maintenance of self-renewal and stemness in hESCs. See text for references.
ACKNOWLEDGMENTS
The authors are very grateful to Eric Chiao, Luis Sinberger Batista, and Stacey Wirt for critical reading of the manuscript. We sincerely apologize to all those colleagues whose important work is not cited because of space limitations. Research on embryonic stem cells in the Sage lab is sponsored by the California Institute for Regenerative Medicine (CIRM – contract grant number: RS1-00298-1). Julien Sage is a Leukemia and Lymphoma Society Scholar and a Morgridge Scholar. Jamie Conklin is supported by the Tobacco Related Disease Program (TRDRP) of California.
REFERENCES
- Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Blomen VA, Boonstra J. Cell fate determination during G1 phase progression. Cell Mol Life Sci. 2007;64:3084–3104. doi: 10.1007/s00018-007-7271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- Chavez L, Bais AS, Vingron M, Lehrach H, Adjaye J, Herwig R. In silico identification of a core regulatory network of OCT4 in human embryonic stem cells using an integrated approach. BMC Genomics. 2009;10:314. doi: 10.1186/1471-2164-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych MA, Sicinski P. Cell cycle in mouse development. Oncogene. 2005;24:2877–2898. doi: 10.1038/sj.onc.1208608. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco G, Comes F, Simone C. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene. 2006;25:5244–5249. doi: 10.1038/sj.onc.1209623. [DOI] [PubMed] [Google Scholar]
- Egozi D, Shapira M, Paor G, Ben-Izhak O, Skorecki K, Hershko DD. Regulation of the cell cycle inhibitor p27 and its ubiquitin ligase Skp2 in differentiation of human embryonic stem cells. FASEB J. 2007;21:2807–2817. doi: 10.1096/fj.06-7758com. [DOI] [PubMed] [Google Scholar]
- Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a) Oncogene. 2004;23:491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- Filipczyk AA, Laslett AL, Mummery C, Pera MF. Differentiation is coupled to changes in the cell cycle regulatory apparatus of human embryonic stem cells. Stem Cell Res. 2007;1:45–60. doi: 10.1016/j.scr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Fluckiger AC, Marcy G, Marchand M, Negre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24:547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S, Wood JG, Livingston DM. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci U S A. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220(NPAT) in human embryonic stem cells. J Cell Physiol. 2007;213:9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, Rao MS. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- Gunaratne PH. Embryonic Stem Cell MicroRNAs: Defining Factors in Induced Pluripotent (iPS) and Cancer (CSC) Stem Cells? Curr Stem Cell Res Ther. 2009 doi: 10.2174/157488809789057400. [DOI] [PubMed] [Google Scholar]
- Hanna J, Carey BW, Jaenisch R. Reprogramming of somatic cell identity. Cold Spring Harb Symp Quant Biol. 2008;73:147–155. doi: 10.1101/sqb.2008.73.025. [DOI] [PubMed] [Google Scholar]
- Jagadeesh S, Banerjee PP. Telomerase reverse transcriptase regulates the expression of a key cell cycle regulator, cyclin D1. Biochem Biophys Res Commun. 2006;347:774–780. doi: 10.1016/j.bbrc.2006.06.172. [DOI] [PubMed] [Google Scholar]
- Kalaszczynska I, Geng Y, Iino T, Mizuno S, Choi Y, Kondratiuk I, Silver DP, Wolgemuth DJ, Akashi K, Sicinski P. Cyclin a is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008 doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, Pandit SK, Khanizadeh M, Weinstein M, Leone G, de Bruin A. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong MX, Smith KP, Stein GS. Human embryonic stem cell registries: value, challenges and opportunities. J Cell Biochem. 2008;105:625–632. doi: 10.1002/jcb.21872. [DOI] [PubMed] [Google Scholar]
- Macleod KF. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer. 2008;8:769–781. doi: 10.1038/nrc2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimets T, Neganova I, Armstrong L, Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27:5277–5287. doi: 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
- Mitra P, Ghule PN, van der Deen M, Medina R, Xie RL, Holmes WF, Ye X, Nakayama KI, Harper JW, Stein JL, Stein GS, van Wijnen AJ. CDK inhibitors selectively diminish cell cycle controlled activation of the histone H4 gene promoter by p220NPAT and HiNF-P. J Cell Physiol. 2009;219:438–448. doi: 10.1002/jcp.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Luo Y, Khrebtukova I, Brandenberger R, Zhou D, Thies RS, Vasicek T, Young H, Lebkowski J, Carpenter MK, Rao MS. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 2004;13:694–715. doi: 10.1089/scd.2004.13.694. [DOI] [PubMed] [Google Scholar]
- Moon NS, Dyson N. E2F7 and E2F8 keep the E2F family in balance. Dev Cell. 2008;14:1–3. doi: 10.1016/j.devcel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, Schmidt NO, Loring JF. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Lengerke C. Patient-specific pluripotent stem cells: promises and challenges. Nat Rev Endocrinol. 2009;5:195–203. doi: 10.1038/nrendo.2009.18. [DOI] [PubMed] [Google Scholar]
- Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova I, Zhang X, Atkinson S, Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 2009;28:20–30. doi: 10.1038/onc.2008.358. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Pera MF, Reubinoff B, Trounson A. Human embryonic stem cells. J Cell Sci. 2000;113(Pt 1):5–10. doi: 10.1242/jcs.113.1.5. [DOI] [PubMed] [Google Scholar]
- Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Involvement of Myc activity in a G(1)/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol. 2000;20:3497–3509. doi: 10.1128/mcb.20.10.3497-3509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, von Zglinicki T, Lako M. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, Jirmanova L, Vitelli L, Samarut J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol. 2002;185:27–33. doi: 10.1385/1-59259-241-4:27. [DOI] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–11620. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b Controls the G1/S Checkpoint Gene p57 in Human Embryonic Stem Cells. Stem Cells. 2009;27:1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Apoptosis and differentiation of human embryonic stem cells induced by sustained activation of c-Myc. Oncogene. 2007;26:5564–5576. doi: 10.1038/sj.onc.1210353. [DOI] [PubMed] [Google Scholar]
- Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007;102:1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2) Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Yang C, Przyborski S, Cooke MJ, Zhang X, Stewart R, Anyfantis G, Atkinson SP, Saretzki G, Armstrong L, Lako M. A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells. 2008;26:850–863. doi: 10.1634/stemcells.2007-0677. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, Hughes O, Strachan T, Stojkovic M, Hinds PW, Armstrong L, Lako M. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol. 2009;184:67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zheng L, Flesken-Nikitin A, Chen PL, Lee WH. Deficiency of Retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res. 2002;62:2498–2502. [PubMed] [Google Scholar]


