Abstract
6,6,8-Triethyldesmosdumotin B (2) was discovered as a MDR–selective flavonoid with significant in vitro anticancer activity against a multi-drug resistant (MDR) cell line (KB-VIN) but without activity against the parent cells (KB). Additional 2-analogues were synthesized and evaluated to determine the effect of B-ring modifications on MDR-selectivity. Analogues with a B-ring Me (3) or Et (4) group had substantially increased MDR–selectivity. Three new disubstituted analogues, 35, 37 and 49, also had high collateral sensitivity (CS) indices of 273, 250 and 100, respectively. Furthermore, 2–4 also displayed MDR-selectivity in an MDR hepatoma-cell system. While 2–4 showed either no or very weak inhibition of cellular P-glycoprotein (P-gp) activity, they either activated or inhibited the actions of the first generation P-gp inhibitors verapamil or cyclosporin, respectively.
Introduction
Incidences of drug resistance still present major and serious obstacles to the effective chemotherapeutic treatment of cancer, despite many efforts to overcome it.1,2 Resistance to one drug often implies simultaneous resistance to structurally and mechanistically diverse anticancer drugs. This efflux phenotype, called multidrug resistance (MDR),3,4 is in part mediated by the over-expression of plasma membrane transporters, such as P-glycoprotein (P-gp, MDR1 or ABCB1, localized at 7q21.1, a 170 kDa protein),5 MDR-associated proteins (MRP1 or ABCC1, localized at 16p13.1, a 190 kDa protein, and MRP2),6 or breast cancer resistant protein (BCRP or ABCG2, localized at 4q22, a 72 kDa protein).7–9 These three kinds of proteins belong to the superfamily of ATP-binding-cassette (ABC) transporters.10 The emergence of MDR pumps causes cancer drugs to be pumped out of the cell, thus reducing intracellular drug concentrations below cytotoxic levels. Because P-gp has broad substrate specificity, tumor cells that over-express P-gp show resistance to many classical and newer molecular-targeted antitumor drugs. The development of agents targeted toward MDR1 or MRP1 is greatly needed in order to improve anticancer chemotherapeutic strategies. MDR1/P-gp is a well-characterized efflux system and a major mediator of MDR.11–14 Intrinsic or acquired over-expression of P-gp dramatically reduces drug response, at times to less than one-quarter, causing poor clinical outcomes following chemotherapy.15
To date, the major pharmacological approaches to overcome MDR have focused on inhibition of the pump function and/or down-regulation of pump over-expression.16–19 Numerous pump inhibitors (or modulators) have been found, and are generally classified as first, second, or third generation chemosensitizers. Many second and third generation inhibitors, such as valspodar, zosuquidar, and tariquidar, are more potent and less toxic than first generation compounds like verapamil (VERAP) or cyclosporine A (CSA). Although some of these new generation compounds are still in clinical trials, the benefits of chemosensitization remain disappointing, in part because the modulator causes undesirable changes in drug pharmacokinetics.20
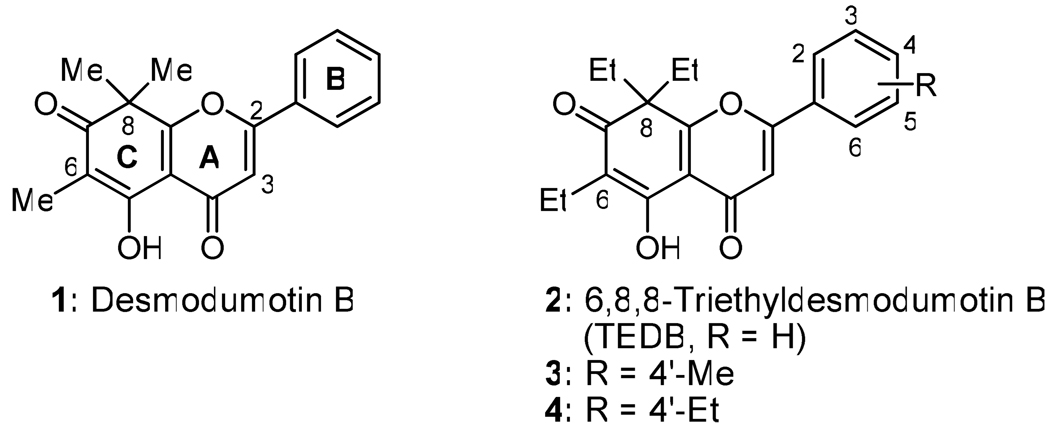
Flavonoids are the most widespread natural compounds produced in plants, and they exhibit diverse, important biological activities, including antioxidant, anticarcinogenic, antiinflammatory, antiproliferative, antiangiogenic and antiestrogenic effects.21 Many reports have demonstrated that flavonoids can also interact with ABC transporters and some act as P-gp modulators.22–25 We found that the flavonoid desmosdumotin B (1, Figure 1) exerted unique in vitro anticancer activity against a P-gp expressing MDR tumor cell line (KB-VIN, ED50 = 2.0 µg/mL), resulting in a >20 fold index of selectivity over the drug sensitive KB parent cell line.26 We also discovered that 6,8,8-triethyldesmosdumotin B (TEDB, 2) and its 4′-methyl (3) and 4′-ethyl (4) analogues were significantly more potent and showed MDR-selectivity of >250.27 In contrast, the related 6,8,8-tripropyl as well as 6,8-diethyl compounds were less optimized for MDR-selectivity.
Figure 1.
The hypersensitivity of drug-resistance was first reported using Escherichia coli in 1952, and the phenomenon was termed “collateral sensitivity” (CS).28 A significant CS agent should show at least two-fold greater activity against the MDR line than the parental line. Recently, Hall et al published a review concerning CS, and argued that exploiting CS agents, which especially kill MDR cancer cells, is an exciting challenge and opportunity for new drug discovery as well as clinical development.29 A thiosemicarbazone derivative (NSC73306) was discovered through the US National Cancer Institute (NCI) anticancer drug screen as a drug lead for targeting MDR tumor cell populations, and several derivatives showed improved MDR-selectivity.30,31 While it was toxic toward a diverse panel of Pgp-expressing tumor cell lines, the CS index was only 7.3-fold at best. Thus, the MDR-selectivity of 2 and its known active analogues is substantially better, but many questions remain about these unique flavonoids. In this report, we address some critical issues by looking at effects on P-gp function, in vitro anticancer activity using a second drug-selected (hepatoma) MDR-cell system, and general mode and mechanism of cytotoxicity. We also explored structure-activity relationship (SAR) results for this unique compound class with major focus on the modification of B-ring substituents.
Chemistry
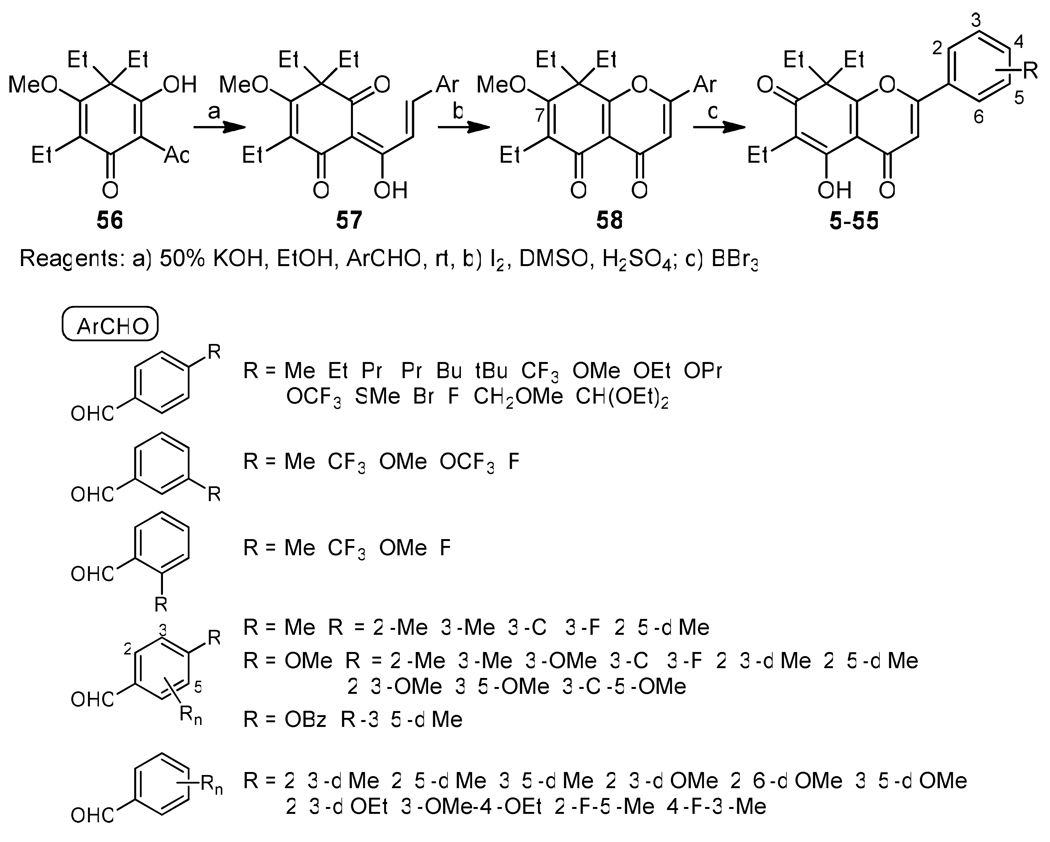
Generally, analogues 5–55 were synthesized through a three-step sequence, Claisen-Schmidt condensation of 5626 with a substituted benzaldehyde (ArCHO), cyclization to form the pyranone A-ring, and C-7 demethylation with BBr3 as shown in Scheme 1. Because the C-7 methoxy group was removed faster than a phenyl B-ring methoxy group, treatment with BBr3 was carried out at a low temperature to retain the latter group. Analogues 10 and 44–46, which contain hydroxy groups, were obtained from the related methoxy (for 10, 44, and 45) or benzyloxy (for 46) phenylaldehydes. The methyl and benzyl ethers were cleaved at the final stage to provide the alcohols. 4′-Bromomethylphenyl analogue 18 was produced by BBr3 treatment when 4′-(methoxymethyl)benzaldehyde, derived from commercially available [4-(dimethoxymethyl)phenyl]methanol, was used as the ArCHO. Analogues 2–4 were synthesized previously.
Scheme 1.
Syntheses of TEDB Analogues
Results and Discussion
All synthesized 2-analogues were evaluated in vitro against two human tumor cell lines, the KB-VIN cell line, an MDR P-gp expressing cloned subline stepwise selected using vincristine, and its parental non-MDR KB cell line. The cytotoxic activity data including KB/KB-VIN sensitivity are listed in Tables 1 (mono-substituted phenyl B-ring) and 2 (multi-substituted phenyl B-ring). Except for 10, 26, and 53, which showed no MDR selectivity, all 2-analogues demonstrated greater cytotoxicity against KB-VIN than KB, resulting in CS indices of over 2.0. Regarding the mono-substituted B-ring analogues (Table 1), 4′-Me (3) and 4′-Et (4) compounds inhibited KB-VIN cell growth most strongly with ED50 values of 0.08 and 0.07 uM, and CS indices of 460 and 320, respectively. Other compounds with significant KB-VIN inhibition (ED50 <2.0 uM) had the following rank order: 11 (4′-OMe, ED50 = 0.68 uM), 6 (4′-iPr, ED50 = 1.08 uM), 22 (3′-OMe, ED50 = 1.22 uM), 15 (4′-SMe, ED50 = 1.38 uM), and 17 (4′-F, ED50 = 1.85 uM). Among of 4’-substitued analogues, 2–19, compounds with a longer side chain (over three carbons), OH, CHO, and CH2Br at the 4′-position or 2′-CF3 lost potency against KB-VIN and the order of substituents by decreasing selectivity was as follows: Me, Et > H ≫ SMe, OMe > F > OEt > OPr > CF3, etc.
Table 1.
Activity of 3–29 (mono-substituted phenyl B-ring) against KB and KB-VIN
| R | ED50 (µM)a | Selectivity | |||||
|---|---|---|---|---|---|---|---|
| 2’ | 3’ | 4’ | 5’ | KB | KB-VIN | KB/KB-VIN | |
| 2 | >236.7 | 1.07 | >221 | ||||
| 3 | Me | 39.2 | 0.08 | 460 | |||
| 4 | Et | 21.9 | 0.07 | 320 | |||
| 5 | Pr | >53 | 12.16 | >4 | |||
| 6 | iPr | 8.13 | 1.08 | 3 | |||
| 7 | Bu | 110.7 | 36.95 | 3.0 | |||
| 8 | tBu | 29.3 | 17.4 | 2 | |||
| 9 | CF3 | 35.7 | 10.35 | 3.5 | |||
| 10 | OH | 20.6 | 20.6 | 1 | |||
| 11 | OMe | 17.7 | 0.68 | 26 | |||
| 12 | OEt | >52.4 | 7.54 | >7 | |||
| 13 | OPr | >50.5 | 29.17 | >2 | |||
| 14 | OCF3 | 36.49 | 9.95 | 4 | |||
| 15 | SMe | >52.1 | 1.38 | >38 | |||
| 16 | Br | >48.1 | 15.70 | 3 | |||
| 17 | F | 33.0 | 1.85 | 18 | |||
| 18 | CH2Br | >46.5 | 27.91 | >2 | |||
| 19 | CH2OMe | >52.4 | 3.40 | 15 | |||
| 20 | Me | 49.7 | 3.69 | 14 | |||
| 21 | CF3 | 35.7 | 5.42 | 6.6 | |||
| 22 | OMe | 13.3 | 1.22 | 11 | |||
| 23 | OCF3 | >47.4 | 9.95 | >5 | |||
| 24 | F | >56.2 | 3.09 | >18 | |||
| 25 | Me | 40.9 | 2.41 | 17 | |||
| 26 | CF3 | 45.6 | 43.10 | 1 | |||
| 27 | OMe | 40.8 | 3.26 | 13 | |||
| 28 | F | >56.2 | 3.93 | >14 | |||
Cytotoxicity as ED50 values for each cell line, the concentration of compound that caused 50% reduction in absorbance at 562 nm relative to untreated cells using the sulforhodamine B assay.
Compounds 20–28 carry a single substitution on the C2′- or C3′-position of the benzene ring. All of them, except 26, exhibited some degree of selectivity; however, they showed lower activity against KB-VIN compared to the parent unsubstituted compound.
Regarding multi-substituted B-ring analogues (Table 2), except for 54, all analogues showed potent cytotoxicity against KB-VIN. Among them, 35, 37, and 49 had notable CS indices of 273, 250, and 100, respectively. Multiple substitutions on the benzene ring of compounds 29–55 demonstrated that, in general, increased selectivity and activity towards KB-VIN is afforded by small hydrophobic groups in C4’- and C3’-positions. The decreasing rank order for selectivity was as follows: 3-Me-4-OMe (35); 3’-F-4’-OMe (37) > 3’,5’-diMe (49) > 3’,4’-diMe (30) > 3’-Me-4’-F (55) > 2’,3’-diMe-4’-OMe (39), > 3’-Cl-4’-Me (31) = 3’-Cl-4’-OMe (38) > 2’-F-5’-Me (54) > 3’,5’-diOMe (52) > 3’-F-4’-Me (32) > 2’,3’-diOMe (50), etc. These results indicate that substitutions on C3′ and C4′ significantly contribute to the KB/KB-VIN sensitivity. Especially, C3′-substitution was the most important for the activity towards KB-VIN (see, for instance, the change in activity for the pairs of compounds 29 and 30, 34 and 35, 39 and 40, 48 and 49). It is unclear to what extent the methoxy and halogen groups acting as hydrogen bond acceptors contribute to the observed changes.
Table 2.
Activity of 30–56 (multi-substituted phenyl B-ring) against KB and KB-VIN
| R | ED50 (µM)a | Selectivity | |||||
|---|---|---|---|---|---|---|---|
| 2’ | 3’ | 4’ | 5’ | KB | KB-VIN | KB/KB-VIN | |
| 29 | Me | Me | 41.2 | 8.52 | 5 | ||
| 30 | Me | Me | 35.5 | 0.71 | 50 | ||
| 31 | Cl | Me | 39.0 | 2.15 | 18 | ||
| 32 | F | Me | 28.1 | 2.95 | 10 | ||
| 33 | Me | Me | Me | 8.2 | 4.37 | 2 | |
| 34 | Me | OMe | 20.9 | 2.62 | 8 | ||
| 35 | Me | OMe | 28.5 | 0.10 | 273 | ||
| 36 | OMe | OMe | 14.7 | 1.00 | 15 | ||
| 37 | F | OMe | 26.7 | 0.10 | 250 | ||
| 38 | Cl | OMe | 24.9 | 1.37 | 18 | ||
| 39 | Me | Me | OMe | 5.43 | 0.28 | 20 | |
| 40 | Me | OMe | Me | 36.6 | 7.58 | 5 | |
| 41 | OMe | OMe | OMe | 9.4 | 1.57 | 6 | |
| 42 | OMe | OMe | OMe | 46.7 | 5.6 | 8 | |
| 43 | Cl | OMe | OMe | 21.8 | 8.40 | 3 | |
| 44 | OH | OH | 10.0 | 5.41 | 2 | ||
| 45 | Me | Me | OH | 6.5 | 2.75 | 2 | |
| 46 | Me | OH | Me | 18.3 | 8.90 | 2 | |
| 47 | Me | Me | 3.1 | 0.85 | 4 | ||
| 48 | Me | Me | 14.5 | 3.41 | 4 | ||
| 49 | Me | Me | 45.5 | 0.45 | 100 | ||
| 50 | OMe | OMe | 35.2 | 4.02 | 9 | ||
| 51 | OMe | OMe | 8.8 | 2.51 | 4 | ||
| 52 | OMe | OMe | 45.2 | 3.27 | 14 | ||
| 53 | OEt | OEt | 44.6 | 31.69 | 1.4 | ||
| 54 | F | Me | 47.3 | 3.24 | 15 | ||
| 55 | Me | F | 40.5 | 1.62 | 24 | ||
Cytotoxicity as ED50 values for each cell line, the concentration of compound that caused 50% reduction in absorbance at 562 nm relative to untreated cells using the sulforhodamine B assay.
The previously synthesized 2–4 were also evaluated in vitro against a human hepatoma MDR cell system, and the data are shown in Table 3. Hep3B-VIN cells are P-gp-expressing and selected using vincristine from the parent non-MDR Hep3B cell line. The CS indices measured for 2–4 were 30, 21, and 8.5, respectively, which are lower than values against KB-VIN cells. Interestingly, the order of potency was changed to 3 (GI50 0.35 µM) > 4 (GI50 0.72 µM) > 2 (GI50 1.57 µM), compared with that against KB-VIN [4 (ED50 0.068 µM) = 3 (ED50 0.085 µM) > 2 (ED50 1.07 µM)] (Tables 1 and 3). However, it is significant that all three compounds displayed selective in vitro anticancer action against the hepatoma-derived MDR cell line, because this finding shows that the activity is not peculiar to a single MDR-cell line.
Table 3.
Activity of 2–4 against Hep3B and Hep3B-VIN
| GI50 (µM) | |||
|---|---|---|---|
| Hep3B | Hep3B-VIN | Hep3B/Hep3B-VIN | |
| 2 | 47.57 | 1.57 | 30 |
| 3 | 7.31 | 0.35 | 21 |
| 4 | 6.09 | 0.72 | 8.5 |
| paclitaxel | 0.0004 | 16.17 | - |
| vincristine | 0.0029 | 6.86 | - |
The effects of 2–4 on P-gp function in KB VIN (MDR) cells was also investigated as shown in Figure 2. Drug pumping activity and inhibition of P-gp were measured using the standard calcein-AM loading assay as described under methods. Figure 2A shows the effects of treatment 2, 3 or 4 compared with those of first generation competitive inhibitors VERAP and CSA. The data show that the latter two prototype P-gp inhibitors stimulated calcein-AM influx, while compounds 3 and 4 inhibited the pump but only very weakly over the concentration range tested (as compared to the action of VERAP and CSA shown in Figure 2A), and compound 2 was inactive. These results indicated that 2 had no detectable effect on P-gp function in KB-VIN cells, while 3 and 4 were weak inhibitors, but only at concentrations much higher than those that induced CS. The effects of co-treatment with 2, 3, or 4 and a fixed concentration of an active inhibitor (2.5 µg/ml VERAP or 4 µg/mL CSA) are shown in Figures 2B–D respectively. Interestingly, inhibition of P-gp activity was influenced by co-treatment with each MDR-selective compound and the magnitude and type of interaction depended on the identity of compound and P-glycoprotein inhibitor, respectively. All three compounds stimulated calcein-AM influx upon co-treatment with VERAP, but had the opposite effect when co-treated with CSA. The activation of VERAP inhibition of Pg-p was clearly concentration-dependent over the range tested, while the responses were dose-limiting and more complicated for CSA with compound 3 or 4. Compound 2 possessed the weakest activity, and compounds 3 and 4 were substantially more potent (compare accumulation of intracellular fluorescence between Figures 2B–D). A direct correlation between P-gp functional effects (Figures 2B–D) and in vitro anticancer activity against KB-VIN cells (Table 1) was not observed, although the rank-order of the three compounds’ activity in both assays is apparent. In conclusion, compounds 2, 3 and 4 are not potent P-gp inhibitors (Figure 2A), although they can clearly influence interaction of the enzyme with substrates/inhibitors in MDR cells.
Figure 2.
Effect on P-gp function in KB-VIN cells
As shown in Figure 3, compound 2 was evaluated against KB-VIN cells together with non-toxic concentrations of P-gp modulators VERAP or the allosteric inhibitor flupenthixol. Co-treatment of 2 with VERAP or flupenthixol (Z-isomer) partially reversed the in vitro anticancer activity of 2, showing that the MDR-selectivity was dependent in-part on P-gp function and consistent with the effect on P-gp activity measured using the co-incubation treatment protocol. Similar results showing partial reversal of 3 and 4 MDR-selectivity were observed (data not shown). Further analysis using suitable probe analogues will be needed to determine if the agents interact directly with P-gp and to map possible interaction domains.
Figure 3.
Chemoprotection of 2 by MDR-1 modulators in KB-VIN cells. Co-treatment with non-toxic concentrations of VERAP or flupenthixol partially reversed in vitro anticancer activity of 2 against MDR-1 cell line (measured by use of the anionic protein dye sulforhodomine-B (SRB). These results show that a competitive substrate-type inhibitor (VERAP) and an allosteric-type pump inhibitor (Flupent) can negate MDR-selective cytotoxicity.
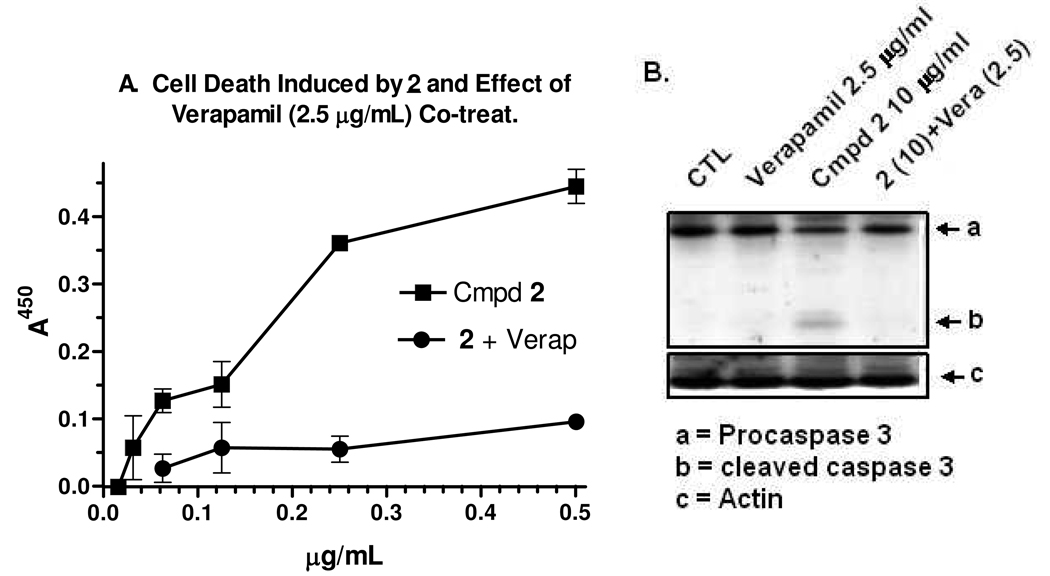
Cell death induced by 2 and effect of co-treatment with VERAP are shown in Figure 4. The effect of 2 on apoptosis in KB-VIN cells was measured using a Cell Death Detection ELISAPLUS kit (Roche Diagnostics, Mannheim, Germany). As shown in Figure 4A, one-day treatment induced a dose-dependent increase in adsorbance associated with histone-associated-DNA-fragments in the cytoplasm. The half-maximal effect occurred between 1–2 ug/ml of 2, consistent with in vitro anticancer activity (Table 1). Co-treatment with VERAP reversed the 2-induced DNA fragmentation. At one day post-treatment, immuno-blot analysis clearly showed pro-caspase-3 cleavage, which again was prevented by VERAP co-treatment (Fig 4B). These data demonstrate that 2 can induce classical apoptosis in KB-VIN cells, and P-gp activity is implicated and required due to the antagonistic action of the pump inhibitor.
Figure 4.
The effect of 2 on apoptosis in KB-VIN cells was measured using a Cell Death Detection ELISAPLUS kit (Roche Diagnostics, Mannheim, Germany).
Whether these effects are mediated via direct interaction with the P-gp enzyme (perhaps at an allosteric site resulting in conformational change) or via indirect action (by affecting membrane properties and permeation of substrate/inhibitors) remain to be elucidated. However, the current data are consistent with 2–4 being novel P-gp actives and suggest that elucidation of the relationship between the MDR phenotype and the potent and selective in vitro anticancer action is warranted and will be worthwhile.
Conclusions
In summary, all analogues demonstrated greater cytotoxicity against KB-VIN than KB, resulting in CS indices of over 2.0, except for 10, 26, and 53. Among newly synthesized analogues, 35, 37, and 49 displayed potent in vitro anticancer activity against KB-VIN, with CS-ratios of 273, 250 and 100, respectively. These results indicated that substitutions on C3′ and C4′ significantly contribute to the KB/KB-VIN sensitivity.
Previously synthesized 2–4 also showed MDR selectivity against the drug-resistant human hepatoma cell line Hep3B-VIN. While analogues 2–4 did not inhibit or only weakly inhibited drug efflux P-gp pump activity, they either activated or inhibited the drug efflux pump activity of prototype P-gp inhibitors VERAP and CSA, respectively. The flavonoid compounds clearly influenced interaction of the enzyme with substrates/inhibitors in MDR cells, although they did not inhibit P-gp. The MDR selectivity was reversed by co-treatment with diverse P-gp inhibitors, such as VERAP or flupenthixol, suggesting dependence on efflux pump activity, and 2-treatment induced apoptosis in KB-VIN cells.
Because of the unique bioactivity of this flavonoid series against P-gp over-expressing tumor cell lines, we plan to explore detailed mechanism of action and preclinical application studies on their potential as new cancer drugs.
Experimental Section
Chemistry. General
All chemicals and solvents were used as purchased. All melting points were measured on a Fisher-Johns melting point apparatus without correction. 1H and 13C NMR spectra were recorded on a Varian Gemini 2000 (300 MHz) NMR spectrometer with TMS as the internal standard. All chemical shifts are reported in ppm. NMR spectra were referenced to the residual solvent peak; chemical shifts δ in ppm; apparent scalar coupling constants J in Hz. Mass spectroscopic data were obtained on a TRIO 1000 mass spectrometer. Analytical thin-layer chromatography (TLC) was carried out on Merck precoated aluminum silica gel sheets (Kieselgel 60 F-254). All target compounds were characterized by 1H-NMR, MS, and elemental analyses. The purities of >95% were determined by 1H-NMR and elemental analyses.
General synthetic procedures for 1-analogues
The aryl-substituted intermediate compound (57) was dissolved in 0.1% conc H2SO4 in DMSO, then I2 (0.1 eq. mol) was added and the reaction mixture heated at 90–95 °C for 1 h. The reaction mixture was quenched with ice-cold aq 10% Na2S2O3 and extracted with EtOAc. The extract was washed with brine, dried over Na2SO4 and concentrated in vacuo. The residue was chromatographed on silica gel eluting with EtOAc–hexane (1:4 to 1:2, v/v) to afford the 7-methoxy substituted analogue (58), which was dissolved in anhydrous CH2Cl2. The mixture was cooled to −78 °C. BBr3 (3 eq. mol, 1.0 M solution in CH2Cl2) was added to the solution, which was warmed to 0 °C spontaneously and stirred until the starting material was consumed. After addition of water, the reaction mixture was extracted three times with CH2Cl2. The extracts were combined, washed with brine, dried over Na2SO4 and concentrated in vacuo. The residue was chromatographed on silica gel eluting with EtOAc–hexane (1:4) to obtain the target compound (5–55). The physical data of representative compounds, 5 for 4′-substituted analog, 20 for 3′-substituted analog, 27 for 2′-substituted analog, 29 for 2′,4′-disubstituted analog, 35 for 3′,4′-disubstituted analog, and 49 for 3′,5′-disubstituted analog are described below. Data for other compounds are in the supporting information.
4′-Propyl-6,8,8-triethyldesmosdumotin B (5)
Pale yellow prisms, mp 169–170 °C (EtOAc-Hexane). 1H NMR (300 MHz, CDCl3): δ 13.14 (s, 1H, 5-OH), 7.72 (d, 2H, J = 8.2 Hz, Ar-H), 7.36 (d, 2H, J = 8.2 Hz, Ar-H), 6.87 (s, 1H, 3-H), 2.69 (t, 2H, J = 7.3 Hz, 4′-CH2CH2CH3), 2.45 (q, 2H, J = 7.3 Hz, 6-CH2CH3), 2.33-2.19 (m, 2H, 8-CH2CH3), 2.07–1.92 (m, 2H, 8-CH2CH3), 1.76-1.62 (m, 2H, 4′-CH2CH2CH3), 1.04 (t, 3H, J = 7.4 Hz, 6-CH2CH3), 0.97 (t, 3H, J = 7.3 Hz, 4′-CH2CH2CH3), 0.67 (t, 6H, J = 7.3 Hz, 8-CH2CH3). MS (ESI+) m/z: 381 (M++1). Anal. (C24H28O4) C, H, O.
3′-Methyl-6,8,8-triethyldesmosdumotin B (20)
Pale yellow prisms, mp 109–110 °C (EtOAc-hexane). 1H NMR (300 MHz, CDCl3): δ 13.09 (s, 1H, 5-OH), 7.64-7.56 (m, 2H, Ar-H), 7.49-7.39 (m, 1H, Ar-H), 6.89 (s, 1H, 3-H), 2.48 (s, 3H, 3′-CH3), 2.46 (q, 2H, J = 7.4 Hz, 6-CH2CH3), 2.34-2.20 (m, 2H, 8-CH2CH3), 2.08-1.93 (m, 2H, 8-CH2CH3), 1.04 (t, 3H, J = 7.4 Hz, 6-CH2CH3), 0.67 (t, 6H, J = 7.4 Hz, 8-CH2CH3). MS (ESI+) m/z: 353 (M++1). Anal. (C22H24O4) C, H, O.
2′-Methoxy-6,8,8-triethyldesmosdumotin B (27)
Pale yellow prisms, mp 139–140 °C (EtOAc-hexane). 1H NMR (300 MHz, CDCl3): δ 13.24 (s, 1H, 5-OH), 7.70 (dd, 1H, J = 8.0, 1.6 Hz, 6′-H), 7.55 (ddd, 1H, J = 8.5, 7.4, 1.6 Hz, 4′-H), 7.23 (s, 1H, 3-H), 7.13 (dd, 1H, J = 8.0, 7.4 Hz, 5′-H), 7.08 (d, 1H, J = 8.5 Hz, 3′-H), 2.46 (q, 2H, J = 7.4 Hz, 6-CH2CH3), 2.30-2.16 (m, 2H, 8-CH2CH3), 2.04-1.90 (m, 2H, 8-CH2CH3), 1.04 (t, 3H, J = 7.4 Hz, 6-CH2CH3), 0.67 (t, 6H, J = 7.4 Hz, 8-CH2CH3). MS (ESI+) m/z: 369 (M++1). Anal. (C22H24O5) C, H, O.
2′,4′-Dimethyl-6,8,8-triethyldesmosdumotin B (29)
Pale yellow prisms, mp 129–130 °C (EtOAc-hexane). 1H NMR (300 MHz, CDCl3): δ 13.14 (s, 1H, 5-OH), 7.35 (d, 1H, J = 8.5 Hz, 6´-H), 7.21-7.14 (m, 2H, 3′ and 5′-H), 6.59 (s, 1H, 3-H), 2.1–2.38 (m, 2H, 6-CH2CH3), 2.43 (s, 3H, 2′or 3′-CH3), 2.41 (s, 3H, 2′ or 3′-CH3), 2.28-2.12 (m, 2H, 8-CH2CH3), 1.98-1.82 (m, 2H, 8-CH2CH3), 1.04 (t, 3H, J = 7.4 Hz, 6-CH2CH3), 0.65 (t, 6H, J = 7.4 Hz, 8-CH2CH3). MS (ESI+) m/z: 389 (M++Na). Anal. (C23H26O4) C, H, O.
4′-Methoxy-3′-methyl-6,8,8-triethyldesmosdumotin B (35)
Pale yellow prisms, mp 169–170 °C (EtOAc-hexane). 1H NMR (300 MHz, CDCl3): δ 13.26 (s, 1H, 5-OH), 7.65 (dd, 1H, J = 8.2, 2.5 Hz, 6′-H), 7.55 (d, 1H, J = 2.5 Hz, 2′-H), 6.96 (d, 1H, J = 8.2 Hz, 5′-H), 6.79 (s, 1H, 3-H), 3.93 (s, 3H, 4′-OCH3), 2.45 (q, 2H, J = 7.3 Hz, 6-CH2CH3), 2.30 (s, 3H, 3′-CH3), 2.33-2.18 (m, 2H, 6- or 8-CH2CH3), 2.06-1.92 (m, 2H, 6- or 8-CH2CH3), 1.04 (t, 3H, J = 7.3 Hz, 6-CH2CH3), 0.67 (t, 3H, J = 7.3 Hz, 8-CH2CH3). MS (ESI+) m/z: 383 (M++1). Anal. (C23H26O5) C, H, O.
3′,5′-Dimethyl-6,8,8-triethyldesmosdumotin B (49)
Pale yellow prisms, mp 138–139 °C (EtOAc-hexane). 1H NMR (300 MHz, CDCl3): δ 13.13 (s, 1H, 5-OH), 7.38 (br s, 2H, Ar-H), 7.24 (br s, 1H, Ar-H), 6.87 (s, 1H, 3-H), 3.95 and 3.89 (s, 3H each, 2′ and 3′-OCH3), 2.45 (q, 2H, J = 7.3 Hz, 6-CH2CH3), 2.43 (s, 6H, 3′ and 5′-CH3), 2.32-2.20 (m, 2H, 8-CH2CH3), 2.06-1.92 (m, 2H, 8-CH2CH3), 1.04 (t, 3H, J = 7.4 Hz, 6-CH2CH3), 0.67 (t, 6H, J = 7.3 Hz, 8-CH2CH3). MS (ESI+) m/z: 399 (M++Na). Anal. (C23H26O4) C, H, O.
Cytotoxic Activity Assay
All stock cultures were grown in T-25 flasks. Freshly trypsinized cell suspensions were seeded in 96-well microtiter plates at densities of 1500–7500 cells per well with compounds added from DMSO-diluted stock. After three days in culture, attached cells were fixed with cold 50% trichloroacetic acid and then stained with 0.4% sulforhodamine B. The absorbency at 562 nm was measured using a microplate reader after solubilizing the bound dye. The mean ED50 is the concentration of agent that reduces cell growth by 50% under the experimental conditions and is the average from at least three independent determinations that were reproducible and statistically significant. For the VERAP reversal experiments, cells were co-treated with VERAP (1 µg/mL). Control experiments showed this concentration had no effect on the replication of KB-VIN cells. The following human tumor cell lines were used in the assay: KB (nasopharyngeal carcinoma) and KB-VIN (vincristine-resistant KB subline). All cell lines were obtained from Lineberger Cancer Center (UNC-CH) or from ATCC (rockville, MD), except KB-VIN, which was a generous gift of Professor Y.-C. Cheng, Yale University. Cells were cultured in RPMI-1640 medium supplemented with 25mM HEPES, 0.25% sodium bicarbonate, 10% fetal bovine serum, and 100 µg/mL kanamycin.
Detection of P-gp activity (calcein-AM loading assay)
Calcein-AM is a probe substrate of P-gp and is converted into a fluorescent derivative via cellular enzymes. MDR cells give low intrinsic fluorescent signals with this probe substrate, unless P-gp is inhibited and then relative fluorescence will increase (see Figure 2-A for effects of classical P-gp inhibitors). For the assay, 10,000 KB-VIN (MDR) cells per well were seeded into 96-well plates with RPMI-1640 medium containing 5% FBS and incubated in a humidified incubator for adhesion and growth. After 24 h incubation, the indicated concentrations of test compounds were added into wells for 30 min at 37°C. Medium was aspirated then replaced with fresh medium containing 1 µM calcein-AM and indicated compounds. After continuing culture for 30 min at 37 °C in the dark, medium was removed and the plates were washed gently and quickly with cold isotonic (PBS) buffer in the dark. Cells were lysed using hypotonic Tris-HCl buffer, and fluorescence was detected using an ELISA reader (Ex. 494 nm, Em. 517 nm.). Composite data obtained from two or more independent experiments were analyzed using Prizm™ (Graphpad Software, San Diego, CA).
Supplementary Material
Acknowledgement
This study was supported by grant CA-17625 from the National Cancer Institute, NIH, awarded to K. H. L and by a grant from the University Research Council, awarded to K.N.G.
Abbreviations
- ABC
ATP-binding-cassette
- ArCHO
substituted benzaldehyde
- BCRP
breast cancer resistant protein
- CS
collateral sensitivity
- CSA
cyclosporine A
- MDR
multidrug resistance/resistant
- NCI
National Cancer Institute
- P-gp
P-glycoprotein
- SAR
structure-activity relationship
- TEDB
6,8,8-triethyldesmosdumotin B
- VERAP
verapamil
Footnotes
Supporting Information Available. Elemental analysis data for compounds 5–55. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell. Mol. Life Sci. 2008;65:3145–3167. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Tomas R. Multidrug resistance: retrospect and prospects in anti-cancer drug treatment. Curr. Med. Chem. 2006;13:1859–1876. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- 3.Fojo T, Bates S. Strategies for reversing drug resistance. Oncogene. 2003;22:7512–7523. doi: 10.1038/sj.onc.1206951. [DOI] [PubMed] [Google Scholar]
- 4.Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci. 2003;94:15–21. doi: 10.1111/j.1349-7006.2003.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endicott JA, Ling V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annu. Rev. Biochem. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- 6.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 7.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATPbinding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998;58:5337–5339. [PubMed] [Google Scholar]
- 9.Miyake K, Mickley L, Litman T, Zhan Z, Robey R, Cristensen B, Brangi M, Greenberger L, Dean M, Fojo T, Bates SE. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: Demonstration of homology to ABC transport genes. Cancer Res. 1999;59:8–13. [PubMed] [Google Scholar]
- 10.Recent review for ABC transporters: Eckford PDW, Sharom FJ. ABC efflux pump-based resistance to chemotherapy drugs. Chem. Rev. 2009;109:2989–3011. doi: 10.1021/cr9000226. Sharom FJ. ABC multidrug transporters: Structure, function and role in chemoresistance. Future Medicine. 2008;9:105–127. doi: 10.2217/14622416.9.1.105.
- 11.Crowley E, Callaghan R. Multidrug efflux pumps: drug binding-gates or cavity? FEBS. 2010;277:530–539. doi: 10.1111/j.1742-4658.2009.07484.x. [DOI] [PubMed] [Google Scholar]
- 12.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura Y, Morita S, Matsuo M, Ueda K. Mechanism of multidrug recognition by MDR1/ABCB1. Cancer Sci. 2007;98:1303–1310. doi: 10.1111/j.1349-7006.2007.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 15.Modok S, Mellor HR, Callaghan R. Modulation of multidrug resistance efflux pump activity to overcome chemoresistance in cancer. Curr. Opin. Pharm. 2006;6:350–354. doi: 10.1016/j.coph.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Takara K, Sakaeda T, Okumura K. An update on overcoming MDR1-mediated multidrug resistance in cancer chemotherapy. Curr. Pharm. Design. 2006;12:273–286. doi: 10.2174/138161206775201965. [DOI] [PubMed] [Google Scholar]
- 17.Avendaño C, Menéndez JC. Recent advance in multidrug resistance modulators. Med. Chem. Rev. 2004;1:419–444. [Google Scholar]
- 18.Kawase M, Motohashi N. New multidrug resistance reversal agents. Curr. Drug Targets. 2003;4:31–43. doi: 10.2174/1389450033347064. [DOI] [PubMed] [Google Scholar]
- 19.Lehne G. P-glycoprotein as a drug target in the treatment of multidrug resistant cancer. Curr. Drug Targets. 2000;1:85–99. doi: 10.2174/1389450003349443. [DOI] [PubMed] [Google Scholar]
- 20.Coley HM. Overcoming multidrug resistance in cancer: clinical studies of P-glycoprotein inhibitors. Methods Mol. Biol. 2010;549:341–358. doi: 10.1007/978-1-60761-416-6_15. [DOI] [PubMed] [Google Scholar]
- 21.Havsteen BH. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002;96:67–202. doi: 10.1016/s0163-7258(02)00298-x. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez AI, Real R, Pérez M, Mendoza G, Prieto JG, Merino G. Modulation of the activity of ABC transporters (P-glycoprotein, MRP2, BCRP) by flavonoids and drug response. J. Pharm. Sci. 2010;99:598–617. doi: 10.1002/jps.21851. [DOI] [PubMed] [Google Scholar]
- 23.Bansal T, Jaggil M, Khar RK, Talegaonkar S. Emerging significance of flavonoids as P-glycoprotein inhibitors in cancer chemotherapy. J. Pharm. Pharmaceut. Sci. 2009;12:46–78. doi: 10.18433/j3rc77. [DOI] [PubMed] [Google Scholar]
- 24.Morris ME, Zhang S. Flavonoid-drug interactions: effects of flavonoids on ABC transporters. Life Sciences. 2006;78:2116–2130. doi: 10.1016/j.lfs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Boumendjel A, Di Pietro A, Dumontet C, Barron D. Recent advances in the discovery of flavonoids and analogs with high-affinity binding to P-glycoprotein responsible for cancer cell multidrug resistance. Med. Res. Rev. 2002;22:512–529. doi: 10.1002/med.10015. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa-Goto K, Bastow KF, Wu J-H, Tokuda H, Lee KH. Total synthesis and bioactivity of unique flavone desmosdumotin B and its analogs. Bioorg. Med. Chem. Lett. 2005;15:3016–3019. doi: 10.1016/j.bmcl.2005.04.070. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa-Goto K, Bastow KF, Cgen TH, Morris-Natschke SL, Lee KH. Antitumor agents 260. New desmosdumotin B analogues with inproved in vitro anticancer activity. J. Med. Chem. 2009;51:3297–3303. doi: 10.1021/jm701208v. [DOI] [PubMed] [Google Scholar]
- 28.Szybalski W, Bryson V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 1952;64:489–499. doi: 10.1128/jb.64.4.489-499.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hall MD, Handley MD, Gottesman MM. Is resistance useless? Multidrug resistance and collateral sensitivity. Trends in Pharmacological Sciences. 2009;30:546–556. doi: 10.1016/j.tips.2009.07.003. (Note: The structure of desmosdumotin analog 11 in Figure 3 is missing a triethyl group on A-ring; it should be the same as 3 in this article)
- 30.Ludwig JA, Szakács G, Martin SE, Chu BF, Cardarelli C, Sauna ZE, Caplen NJ, Fales HM, Ambudkar SV, Weinstein JN, Gottesman MM. Selective toxicity of NSC73306 in MDR1-positive cells as a new strategy to circumvent multidrug resistance in cancer. Cancer Res. 2006;66:4808–4815. doi: 10.1158/0008-5472.CAN-05-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall MD, Salam NK, Hellawell JL, Fales HM, Kensler CB, Ludwig JA, Szakács G, Hibbs DE, Gottesman MM. Synthesis, activity, and pharmacophore development for isatin-β-thiosemicarbazones with selective activity toward multidrug-resistant cells. J. Med. Chem. 2009;52:3191–3204. doi: 10.1021/jm800861c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.