Abstract
Although it is known that most HIV-1 infections worldwide result from exposure to virus in semen, it has not yet been established whether transmitted strains originate as RNA virions in seminal plasma or as integrated proviral DNA in infected seminal leukocytes. We present phylogenetic evidence that among six transmitting pairs of men who have sex with men, blood plasma virus in the recipient is consistently more closely related to the seminal plasma virus in the source. All sequences were subtype B, and the env C2V3 of transmitted variants tended to have higher mean isoelectric points, contain potential N-linked glycosylation sites, and favor CCR5 co-receptor usage. A statistically robust phylogenetically corrected analysis did not detect genetic signatures reliably associated with transmission, but further investigation of larger samples of transmitting pairs holds promise for determining which structural and genetic features of viral genomes are associated with transmission.
INTRODUCTION
Although individuals may be exposed to HIV through contact with virus in many bodily fluids, most HIV infections worldwide result from sexual transmission of virus harbored in semen (1). Infection after sexual exposure has been associated with the viral loads in blood (2) and seminal plasma (3), stage of infection of the source partner (4), concurrent sexually transmitted infections (STIs) (5), sexual positioning (6), and other behavioral and biological factors (7, 8). Viral genetics likely influence the transmissibility of HIV from semen, as certain sequence features may confer selective advantage for occupying the male genital tract or effectively establishing infection in susceptible individuals (9, 10).
Semen is a complex mixture of cellular and plasma components (11) that harbors both HIV RNA virions within seminal plasma and proviral HIV DNA within infected seminal cells (12). It remains unclear whether cell-free virions, cell-associated proviruses, or both are the source reservoir for sexually transmitted HIV (13). One possible mechanism of transmission is that a seminal lymphocyte or macrophage carrying replication-competent provirus (cell-associated HIV DNA) releases its viral cargo after entering the recipient partner. This hypothesis is supported by documented cases of HIV transmission from a source partner receiving suppressive antiretroviral therapy (14) and also by finding multiple related viral variants in the blood of acutely infected individuals (15, 16). An alternative hypothesis is that cell-free virus, measured as HIV RNA in seminal plasma, is the source of transmitted virus, which is supported by the monoclonal acute infections found in most studies (17, 18) and the positive correlation between blood plasma HIV RNA concentrations and transmission risk (2, 3). Finally, both mechanisms might occur naturally (13), with one route predominating for undetermined reasons.
Separately evolving subpopulations of HIV are frequently established in diverse tissues, including the lung (19), central nervous system, spleen (20), and male genital tract (9, 21), after infection. This anatomic compartmentalization is a result of selection by local factors such as differential immune responses, drug concentrations, and tissue tropism (22). Conserved, compartment-specific genetic motifs of the env-coding region have been found in viral subpopulations from the semen (9) and the blood of acutely infected individuals (18, 23). Putative genetic correlates of sexual transmission in the gene coding for envelope glycoprotein (env) have also been reported (10, 24, 25); however, there is little concordance between the studies, which suggests that such correlates are too complex to be evinced from currently available samples. Here, we report on the genetic and structural elements of the C2V3 region of env extracted from the blood and semen of subjects belonging to a transmission cohort of recently infected men who have sex with men (MSM). We show HIV RNA viral subpopulations in the blood of recipient partners to be more similar to the HIV RNA virus in seminal plasma than to the seminal cell–associated HIV DNA virus from source partners’ semen, thus providing ample support for seminal plasma as the origin of transmitted virus among MSM.
RESULTS
Cell-free seminal plasma virus is the origin of transmitted HIV
We determined the estimated date of infection (EDI), herpes simplex virus type 2 (HSV-2) serostatus, infection status of bacterial STIs (syphilis, gonorrhea, or Chlamydia), blood plasma HIV viral loads, source partner seminal plasma HIV viral loads, and CD4+ cell counts for six transmission pairs, each consisting of an HIV-infected source and recipient sex partner (Table 1). HIV RNA was extracted and quantified from the blood of both partners and from the seminal plasma of source partners, and HIV DNA was extracted from the source partner’s infected seminal cells (see Supplementary Material). Epidemiological linkage analysis confirmed HIV transmission between the identified source and recipient for each pair. Recipient partners provided biological samples an average of 72 days after their EDI, and all reported having engaged in receptive anal intercourse with their source partners. Source partners provided samples of urine for STI testing and samples of blood and semen an average of 88 and 94 days, respectively, after their recipient partners’ EDI (Table 1). In four pairs, source partners had been infected with HIV for less than 6 months at the time of HIV transmission, and the remaining two source partners (A and F) were chronically infected. One individual was the source partner for three identified recipient partners: two (B and C) ~100 days after his own EDI and the third (A) ~900 days later. None of the source partners had a concomitant bacterial STI at study enrollment, but half (n = 3) of the recipient partners were infected by gonorrhea. Four source partners were HSV-2 seropositive, and four recipient partners were HSV-2 seronegative, resulting in four HSV-2 serodiscordant pairs. Rates of STI, HSV-2 serostatus, and respective means of the source and recipient partners’ ages (33 and 39 years), CD4 cell counts (439 and 673 cells per microliter), and viral loads of HIV-1 RNA in the blood (5.35 and 5.15 log HIV RNA copies per milliliter) were not significantly different (P > 0.05, exact and Wilcoxon rank tests); however, statistical power was limited by the small sample size.
Table 1.
Comparisons within and between partner pairs. Average is mean for ages and laboratory values, except as indicated below. S, source partner; R, recipient partner; SEDI, source partner’s EDI; REDI, recipient partner’s EDI; SEDI-REDI, number of days between the SEDI and the REDI (average reported is median); GC, gonococcus; BPVL, blood plasma viral load; SPVL, seminal plasma viral load (applies to source partners only); REDI-BPVL, number of days between the REDI and collection of blood sample (average reported is median); REDI-SPVL, number of days between the REDI and collection of semen sample (average reported is median); NA, not applicable.
| Trait | Transmission pairs | Group | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pair | A | B | C | D | E | F | Averages | |||||||
| Partner | S | R | S | R | S | R | S | R | S | R | S | R | S | R |
| Age (years) | 37 | 42 | 35 | 42 | 35 | 47 | 39 | 36 | 23 | 39 | 26 | 30 | 33 | 39 |
| HSV-2 seropositive | Yes | Yes | Yes | No | Yes | No | No | No | Yes | No | No | Yes | 66% | 33% |
| Bacterial STI | No | No | No | GC | No | GC | No | No | No | GC | No | No | 0% | 50% |
| CD4 (cells/μl) | 352 | 614 | 377 | 647 | 377 | 930 | 568 | 506 | 730 | 380 | 230 | 962 | 439 | 673 |
| BPVL (log copies/ml) | 4.96 | 4.92 | 5.76 | 6.08 | 5.76 | 7.19 | 6.30 | 4.84 | 4.12 | 4.05 | 5.20 | 3.80 | 5.35 | 5.15 |
| REDI-BPVL (days) | 92 | 83 | 52 | 44 | 29 | 21 | 128 | 128 | 123 | 73 | 83 | 84 | 88 | 72 |
| SPVL (log copies/ml) | 3.83 | 4.15 | 4.15 | 4.36 | 2.56 | 4.84 | 3.98 | |||||||
| REDI-SPVL (days) | 92 | 19 | 42 | 143 | 130 | 96 | 94 | |||||||
| SEDI-REDI (days) | 1059 | 93 | 116 | 67 | 22 | Chronic | NA | |||||||
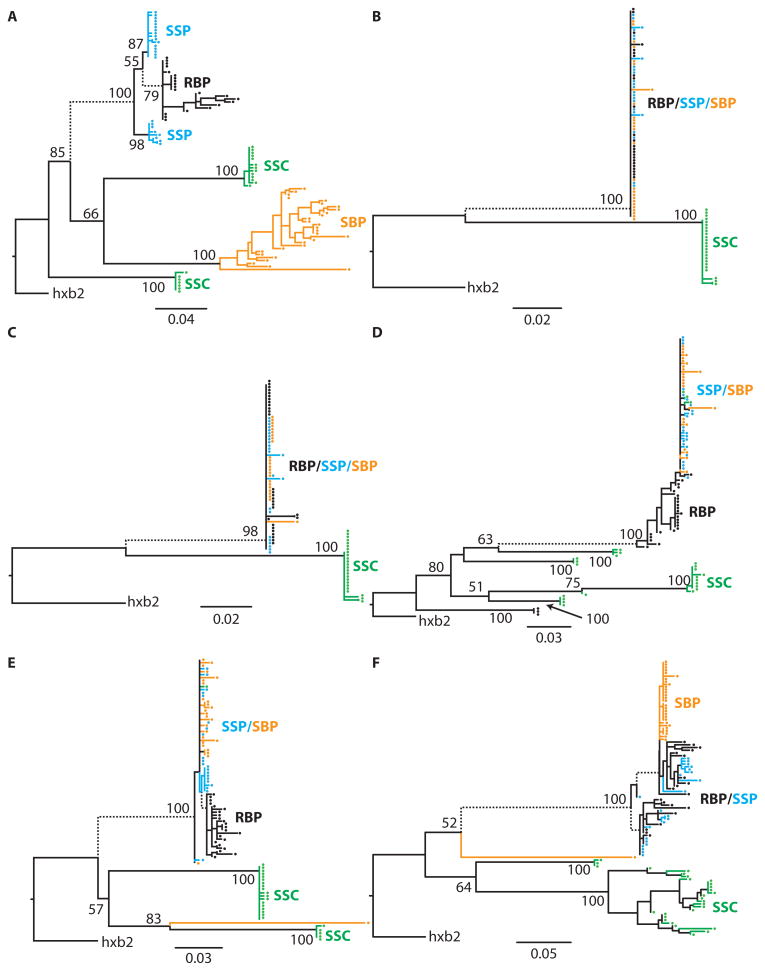
Blood-derived HIV RNA from source and recipient partners and semen-derived HIV RNA and HIV DNA from source partners were analyzed using at least 20 sequences from each sample type. Sequences of the C2V3 region of HIV-1 env were generated either by single-genome amplification of HIV RNA extracted from blood or by clonal amplification of HIV RNA from seminal plasma and HIV DNA from seminal cells (see Supplementary Material) using previously described techniques (23). The origin of transmitted virus and degree of differentiation of viral subpopulations within source partners was inferred using phylogenetic and population genetic methods (see Materials and Methods). All sequences were of subtype B and did not show evidence for superinfection within hosts. For all six pairs, viral sequences derived from the recipient partner’s blood plasma clustered with the sequences derived from his source partner’s seminal plasma rather than those from his seminal cells (Fig. 1). In all cases, most (91%) of source seminal cell–associated HIV DNA sequences clustered independently (100% bootstrap support; Fig. 1) of HIV RNA sequences from the source partner’s seminal and blood plasma. on Thus, HIV-1 transmitted during sexual exposure between MSM likely originates in seminal plasma rather than in seminal cells. High concentrations of genetic differentiation between seminal cell–associated HIV DNA subpopulations and the HIV RNA subpopulations sampled from both seminal plasma (mean Fst = 0.79 ± 0.07; P < 0.001 for all pairs) and blood plasma (mean Fst = 0.80 ± 0.07; P < 0.001 for all pairs) were inferred (table S1). A minority (9%) of seminal cell–associated HIV DNA sequences clustered with HIV RNA sequences from seminal plasma for two of the six transmission pairs (Fig. 1, D and E). These minority HIV DNA variants were probably extracted from seminal cells in which HIV RNA had been recently integrated from contemporaneous viral subpopulations in blood or seminal plasma, whereas most of the sampled HIV DNA variants likely represent archived virus (26). Additionally, the estimated time to the most recent common ancestor (TMRCA) for recipient blood and source seminal plasma HIV-1 RNA sequences was less than that for recipient blood plasma and source seminal cell HIV-1 DNA sequences (table S2), further indicating that the transmitted viral subpopulations were not derived from archived provirus (26).
Fig. 1.
(A to F) Phylogenetic analysis of source and recipient viral sequences. Maximum likelihood phylogenies of viral subpopulations in anatomic compartments for each transmission pair (A to F, Table 1). RBP, recipient HIV RNA in blood plasma; SBP, source HIV RNA in blood plasma; SSP, source HIV RNA in seminal plasma; SSC, source seminal cell–associated virus.
The degree of compartmentalization differs between acute and chronic infection
Differentiation of the three viral subpopulations was evident within all source individuals to varying degrees (Fig. 1). Viral subpopulations sampled from blood plasma and seminal plasma within source patients were admixed for transmission pairs B to E (Fig. 1 and table S1) but not for transmission pairs A and F, where the source partners were chronically infected. Three of the transmission pairs involved a source partner who transmitted HIV to recipient partners around 100 days (B and C) and again 900 days (A) after he acquired HIV. The HIV RNA subpopulations from the blood plasma of these recipient partners clustered with contemporaneous subpopulations of HIV RNA from the source partner’s seminal plasma rather than HIV DNA subpopulations from his seminal cells (Fig. 1 and fig. S1). In the earlier-transmission pairs (B and C), the source partner’s HIV RNA subpopulations sampled from blood and seminal plasma clustered closely together and were genetically indistinguishable (Fst = 0.034 and 0.010; P = 0.203). In the later-transmission pair (A), however, the source partner’s blood HIV RNA population was significantly differentiated from both the HIV DNA population in his seminal cells (Fst = 0.727; P < 0.001) and the HIV RNA population sampled from his seminal plasma at the later time point (Fst = 0.842; P < 0.001) (Fig. 1 and fig. S1). Estimating the TMRCA for transmission pair A proved challenging, as the inferred posterior distributions of the TMRCA were bimodal, highlighting the lack of signal needed to reliably estimate TMRCA in these relatively homogeneous viral sequences (table S2 and fig. S2K). These findings indicate compartmentalization of source blood and seminal plasma HIV RNA subpopulations at the time of the later transmission (A), further supporting HIV RNA subpopulations in seminal plasma as the origin of sexually transmitted virus between MSM.
Transmitted and nontransmitted viruses show structural but not selective differences
The sequenced C2V3 region of cell-free (transmitted) virus had, on average, a higher mean isoelectric point (median, 9.7; range, 8.76–10.95 versus median, 8.89; range, 6.5–10.49; one-sided P < 0.001, Mann-Whitney test), contained more potential N-linked glycosylation sites (median, 7.15; range, 5.36–7.70 per 100 codons versus median, 6.48; range, 4.46–7.70 per 100 codons; one-sided P < 0.001, Mann-Whitney test), and had a greater proportion of CCR5-tropic strains than the cell-associated (nontransmitted) virus (100% and 92%, respectively). However, because clonally derived sequences are unlikely to constitute independent samples from the viral population, the P values reported above are likely to overestimate the statistical significance of differences. All sequences were, on average, subject to purifying selection for all transmission pairs. Six sites were inferred to be positive selection in two of the six transmission pairs (A and F; table S3 and fig. S3), but these sites were not shared across transmission pairs (fig. S3). For all but one of the transmission pairs (C), the genetic algorithm approach (27) for adaptively mapping selective regimes to lineages failed to reject a model in which selection was homogeneous across the phylogeny. In the case of transmission pair C, however, two selective regimes were detected: one accounting for purifying selection (dN/dS = 0.725) and the other for strong positive selection (dN/dS = infinity). This result might be artifactual given the overall low diversity in this transmission pair (samples were obtained less than 30 days after the EDI; Table 1), thus limiting reliability in the estimate of the synonymous mutation rate in some lineages. We also attempted to associate specific sequence motifs with transmitted viral variants while accounting for the shared ancestry of viruses sampled from transmission pairs. We reconstructed unobserved ancestral viral strains using maximum likelihood and mapped substitutions to transmission and pretransmission branches (see Supplementary Material; Fig. 1). Only one codon (position 329 of HXB2 env) shared a substitution between two transmission pairs (Q329R and Q329K; table S4A) along a transmission branch, and one codon (position 293 of HXB2 env) had substitutions in five of six cases along a pretransmission branch (table S4B). In a sample of six transmission pairs with three recipient partners sharing a common source partner, it is not surprising that no shared transmission motifs were detected. The robust identification of signature motifs in the context of HIV transmission remains an open statistical problem, exacerbated by the similarity of sequences due to shared ancestry (rather than positive selection) and the need to account for founder effects when attempting to identify conserved motifs (28).
DISCUSSION
The hypothesis that sexually transmitted HIV originates in the seminal plasma of the source partner is the most likely mechanism of transmission according to our sequence analyses, but we should consider other possible mechanisms. First, it is possible that all six HIV transmissions occurred during anal sex, with the insertive (recipient) partner having been exposed to HIV present in the blood of the receptive (source) partner (for example, through tears in his rectal mucosa, instead of virus in his semen). If we further assume that viral RNA subpopulations in blood and seminal plasma of each of the source partners are closely related (and distinct from the cell-associated seminal HIV DNA), then phylogenetic analyses may be congruent with the patterns seen in Fig. 1. However, at least in one transmission pair (pair A), a clear distinction between blood- and semen-derived HIV RNA sequences was detected in the source partner, and the transmitted virus was more similar to the latter. The probability of this scenario is further reduced by the observations (6) that the rates of HIV transmission during anal sex are much lower when the source partner is the receptive partner rather than the insertive partner, and that the receptive partner is more likely to be exposed to virus in semen than virus in blood. These data also suggest that the HIV RNA population in the blood could be the source of the HIV RNA population in semen, and this finding may be important in the role of antiretroviral therapy to reduce the infectiousness of a potential source partner. Second, discrepant viral loads in blood and seminal biological samples necessitated the use of different sequence amplification techniques (single-genome amplification for blood and clonal amplification for semen), which could have biased the composition of each sample. However, we used the same set of polymerase chain reaction (PCR) primers for all samples to reduce amplification bias, and such a bias would be expected to generate phylogenetic clustering based on the amplification technique (that is, clonally amplified semen sequences would cluster together and separately from the single-genome amplified blood sequences), which was not observed (Fig. 1). Third, the sample of sequences of HIV DNA generated from seminal cells could be biased toward the subpopulation of infected seminal cells containing archived and mostly replication-incompetent provirus (26). Although we cannot eliminate the possibility that unsampled, low-frequency, cell-associated HIV DNA founded the transmitted viral population, it is unlikely to have occurred in all transmission pairs, especially given the high degree of genetic similarity and phylogenetic clustering between the HIV RNA subpopulations sampled from the blood plasma of recipient partners and the seminal plasma of source partners. Fourth, observations reported in this study are based on the C2V3-coding region of env, and alternative results may have been obtained if other coding regions were investigated. This region of env, however, has been widely used in previous studies concerning HIV transmissions, and its choice allows for the identification of important viral characteristics including cellular tropism and infectivity after sexual exposure (9, 10, 18, 23, 24). Fifth, the relatively small number of transmission pairs studied limits the broad generalization of these findings to all MSM exposures; therefore, larger investigations are needed to confirm our findings with a high degree of statistical confidence.
Our results provide the most compelling experimental confirmation for the hypothesis that that cell-free HIV RNA in seminal plasma, and not cell-associated HIV DNA in seminal cells, is the origin of sexually transmitted virus between MSM. Because of the clear importance of identifying genetic correlates of transmissibility for the purposes of guiding preventive efforts, we examined sequence attributes that have previously been associated with transmission (23–25). Whereas mean isoelectric points, numbers of putative N-linked glycosylation sites, and co-receptor tropism differed between the transmitted and the nontransmitted viral subpopulations, we were unable to identify specific motifs or substitutions associated with the sexual transmission of HIV from semen among six transmission pairs of MSM. Such motifs are probably too subtle to deduce because of sample size limitations inherent in this and other studies of HIV transmission (23–25). However, although larger studies are warranted, this study identifies HIV RNA populations in the seminal plasma as an attractive target for biological efforts to interrupt transmission.
MATERIALS AND METHODS
Sampling
Participants with acute HIV infection recruited their antiretroviral-naïve recent sex partners for this study. All were interviewed for sexual histories, examined, and administered HIV counseling. Samples of urine, blood, and semen were obtained. After 48 hours of abstinence, semen was collected by masturbation without lubricant. Time between transmission and collection of specimens was estimated using the recipient’s EDI (29) and date of sample collection. Blood plasma was aliquoted, frozen, and stored at −80°C. Viral transport medium (2 ml of 80% RPMI 1640, 9% fetal bovine serum, 9% penicillin-streptomycin, 2% nystatin) was added to samples at collection. Seminal plasma was separated from seminal cells by centrifugation at 700g for 12 min within 2 hours of collection and stored at −80°C and −150°C, respectively (30). Neisseria gonorrhoeae and Chlamydia trachomatis infection was assessed in urine samples (LabCorp). Syphilis infection was assessed by rapid plasma reagin titers. Total CD4+ lymphocyte counts were measured (LabCorp). HSV-2 serostatus was determined by HSV-2–specific enzyme immunoassay (ARUP Laboratories) with confirmation by HSV-2 Western blotting (University of Washington, Seattle). HIV RNA was extracted and quantified from 500 μl of blood plasma (Amplicor HIV-1 Monitor Test, Roche Molecular Systems Inc.) and from 500 μl of seminal plasma (qc-HIV Assay, GenProbe Inc.) according to the manufacturers’ protocols. HIV DNA was extracted from seminal cells (DNeasy Blood & Tissue kit, Qiagen Inc.) per the manufacturer’s protocols.
Sequencing
ViroSeq v.2.0 (Applied Biosystems) was used to sequence HIV-1 pol from blood per the manufacturer’s instructions. HIV-1 env C2V3 sequences were generated from blood plasma virus (HIV RNA, sources and recipients) and semen virus (seminal cell HIV DNA and plasma HIV RNA, sources only). Briefly, HIV RNA extracted from blood plasma was reverse-transcribed (with RETROscript kit, Applied Bio-systems) into complementary DNA (cDNA), and nested PCR of env C2V3 was performed (31, 32) for single-genome amplification and sequencing as previously described (23). Because template from semen was insufficient for single-genome amplification, HIV DNA from seminal cells and HIV cDNA from seminal plasma were cloned and sequenced (using the same primers as above) with the TOPO-TA Cloning System (Invitrogen), as previously described (33). Sequences were manually checked with Sequencher 4.1 (Gene Codes), screened for laboratory contamination with HIV BLAST (34), aligned with MUSCLE (35), and manually edited with Geneious Pro 4.6 (Biomatters Ltd.) to preserve reading frame.
Sequence characteristics
Sequence characteristics were as follows: co-receptor utilization determined with WebPSSM (36), potential N-linked glycosylation sites identified with a custom script within HyPhy (37), HIV-1 subtype determined as described by Kosakovsky Pond et al. (38), shared transmission motifs not due to common ancestry estimated by reconstructing ancestors using maximum likelihood (39, 40), and mapping substitutions either to the transmission branch (most recent common ancestor of recipient viruses) or to a pretransmission branch (most recent common ancestor of the recipient viruses and most closely related source virus).
Phylogenetic analysis
Epidemiological linkage confirmation was performed on pol sequences from each partner (33). Codons associated with drug resistance were removed to evaluate transmission linkage independent of resistance mutations (41). Linkage results were not disclosed to participants. Maximum likelihood phylogenetic trees were inferred from env C2V3 sequences for each transmission pair using PhyML (42) under a GTR+Γ model (43, 44) and support estimated with 1000 bootstraps. Gene flow between compartments (blood, seminal plasma, and seminal cell) within source individuals and between source compartments and recipient blood plasma was estimated with Fst (45), and statistical significance was assessed with 1000 permutations. An uncorrelated molecular clock (46) in BEAST v1.4.8 (47) was used to estimate TMRCA between sequences obtained from the recipients’ blood HIV RNA and three source compartments (blood HIV RNA, seminal plasma HIV RNA, and seminal cell HIV DNA). Models of DNA substitution and coalescent demographics were compared with Bayes factors. Because the fit of demographic and substitution models was indistinguishable (table S5), GTR+Γ with constant population size was used. Transmission pairs with low diversity (Fig. 1, B and C) were combined with a third recipient (Fig. 1A) because they shared a source (fig. S1). Pair F was excluded because the EDI could not be reliably determined. Bayesian Markov chain Monte Carlo (MCMC) analysis used uninformative priors and 24 (multiple recipients) or 10 (single recipients) independent MCMC chains of 10 million generations (burn-in of 1 million). MCMC parameters were chosen to ensure convergence and sufficient effective sample sizes (>200) for estimated parameters.
Selection
We investigated selection by means of codon models (48, 49). Fixed effects likelihood (FEL) methods identified purifying and diversifying selection across the phylogeny (FEL) and along internal branches (iFEL). A genetic algorithm was used to map selection classes to lineages (27). Model selection was used to select a DNA substitution model, and evidence for recombination was evaluated with GARD (50), and analyses were implemented with Datamonkey (51).
Supplementary Material
www.sciencetranslationalmedicine.org/cgi/content/full/2/18/18re1/DC1
Table S1. Genetic differentiation between compartments.
Table S2. Estimated divergence times of compartments.
Table S3. Number of purifying (dN/dS < 1) and positive (dN/dS > 1) selection sites.
Table S4. Inferred substitutions along transmission branches for pairs A, E, and F (panel A) and along pretransmission branches for all pairs A to K (panel B).
Table S5. Marginal likelihoods, estimated as the harmonic mean of the sampled likelihoods in Bayesian MCMC analysis.
Fig. S1. Maximum likelihood phylogenetic tree of viral sequences from a source who infected multiple partners at two independent time points.
Fig. S2. Posterior distributions of parameters sampled during Bayesian MCMC analyses.
Fig. S3. Site-specific nonsynonymous (dN) and synonymous (dS) rates of substitution.
Acknowledgments
Funding: NIH grants MH083552, AI077304, AI69432, AI38858, AI43638, AI074621, AI43752, AI29164, AI47745, MH62512, AI047745, and AI57167; National Science Foundation grant NSF-0714991; University of California, San Diego, Center for AIDS Research grant AI36214; Centers for Disease Control and Prevention Contract 200-2002-00656; and San Diego Veterans Affairs Healthcare System.
Footnotes
Author contributions: D.M.B., M.K.L., and P.M.C. assisted in the collection, analysis, and interpretation of data and the writing of the report. W.D. assisted in the analysis and interpretation of data and the writing of the report. S.L.K.P. assisted in the study design, the analysis and interpretation of data, and the writing of the report. S.J.L. and D.D.R. assisted in the study design, the collection and interpretation of data, and the writing of the report. D.M.S. conceived the study design and assisted in the collection, analysis, and interpretation of data and the writing of the report.
Competing interests: D.D.R. has consulted for Chimerix, Gen-probe, Merck, Bristol Myers Squibb, Gilead, Idenix, Monogram Biosciences, Tobira, Myriad, Biota, and Theraclone. D.M.S. has received grant support from Pfizer. The other authors have no competing interests. Accession numbers: Sequences have been deposited in GenBank with accession numbers GU597090 to GU597321.
REFERENCES AND NOTES
- 1.UNAIDS. AIDS epidemic update: December 2007. 2007 [Google Scholar]
- 2.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 3.Butler DM, Smith DM, Cachay ER, Hightower GK, Nugent CT, Richman DD, Little SJ. Herpes simplex virus 2 serostatus and viral loads of HIV-1 in blood and semen as risk factors for HIV transmission among men who have sex with men. AIDS. 2008;22:1667–1671. doi: 10.1097/QAD.0b013e32830bfed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, Kiwanuka N, Kigozi G, Kiddugavu M, Lutalo T, Nalugoda F, Wabwire-Mangen F, Meehan MP, Quinn TC. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 5.Cohen MS. Sexually transmitted diseases enhance HIV transmission: No longer a hypothesis. Lancet. 1998;351(Suppl. 3):5–7. doi: 10.1016/s0140-6736(98)90002-2. [DOI] [PubMed] [Google Scholar]
- 6.Vittinghoff E, Douglas J, Judson F, McKirnan D, MacQueen K, Buchbinder SP. Per-contact risk of human immunodeficiency virus transmission between male sexual partners. Am J Epidemiol. 1999;150:306–311. doi: 10.1093/oxfordjournals.aje.a010003. [DOI] [PubMed] [Google Scholar]
- 7.Boily MC, Baggaley RF, Wang L, Masse B, White RG, Hayes RJ, Alary M. Heterosexual risk of HIV-1 infection per sexual act: Systematic review and meta-analysis of observational studies. Lancet Infect Dis. 2009;9:118–129. doi: 10.1016/S1473-3099(09)70021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta K, Klasse PJ. How do viral and host factors modulate the sexual transmission of HIV? Can transmission be blocked? PLoS Med. 2006;3:e79. doi: 10.1371/journal.pmed.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai SK, Good B, Kosakovsky Pond SL, Wong JK, Strain MC, Richman DD, Smith DM. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol. 2005;79:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, Huang W, Quinn TC. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owen DH, Katz DF. A review of the physical and chemical properties of human semen and the formulation of a semen simulant. J Androl. 2005;26:459–469. doi: 10.2164/jandrol.04104. [DOI] [PubMed] [Google Scholar]
- 12.Levy JA. The transmission of AIDS: The case of the infected cell. JAMA. 1988;259:3037–3038. [PubMed] [Google Scholar]
- 13.Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, Cooper DA, Ho DD. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: Evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stürmer M, Doerr HW, Berger A, Gute P. Is transmission of HIV-1 in non-viraemic serodis-cordant couples possible? Antivir Ther. 2008;13:729–732. [PubMed] [Google Scholar]
- 15.Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JA, Kitrinos KM, Hicks CB, Eron JJ, Jr, Swanstrom R. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu T, Wang N, Carr A, Wolinsky S, Ho DD. Evidence for coinfection by multiple strains of human immunodeficiency virus type 1 subtype B in an acute seroconvertor. J Virol. 1995;69:1324–1327. doi: 10.1128/jvi.69.2.1324-1327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh A, Besson G, Mobasher A, Collman RG. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J Virol. 1999;73:6680–6690. doi: 10.1128/jvi.73.8.6680-6690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: Evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiessling AA, Fitzgerald LM, Zhang D, Chhay H, Brettler D, Eyre RC, Steinberg J, McGowan K, Byrn RA. Human immunodeficiency virus in semen arises from a genetically distinct virus reservoir. AIDS Res Hum Retroviruses. 1998;14(Suppl. 1):S33–S41. [PubMed] [Google Scholar]
- 22.Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, Mittler JE, Mullins JI. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immuno-deficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 25.Frost SD, Liu Y, Kosakovsky Pond SL, Chappey C, Wrin T, Petropoulos CJ, Little SJ, Richman DD. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Böni J, Hirschel B, Weber R, Trkola A, Gunthard HF. Swiss HIV Cohort Study, HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA. 2008;105:16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosakovsky Pond SL, Frost SD. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol. 2005;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 29.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of HIV viremia and antibody sero-conversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 30.Smith DM, Kingery JD, Wong JK, Ignacio CC, Richman DD, Little SJ. The prostate as a reservoir for HIV-1. AIDS. 2004;18:1600–1602. doi: 10.1097/01.aids.0000131364.60081.01. [DOI] [PubMed] [Google Scholar]
- 31.Koelsch KK, Smith DM, Little SJ, Ignacio CC, Macaranas TR, Brown AJ, Petropoulos CJ, Richman DD, Wong JK. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS. 2003;17:F11–F16. doi: 10.1097/00002030-200305020-00001. [DOI] [PubMed] [Google Scholar]
- 32.Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow MM, Eron JJ, Jr, Fiscus SA, Cohen MS, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: Underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DM, Wong JK, Shao H, Hightower GK, Mai SH, Moreno JM, Ignacio CC, Frost SD, Richman DD, Little SJ. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: Implications for secondary transmission. J Infect Dis. 2007;196:356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 34.Los Alamos National Laboratory. www.lanl.gov.
- 35.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen MA, Li FS, van’t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosakovsky Pond SL, Frost SD, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 38.Kosakovsky Pond SL, Posada D, Stawiski E, Chappey C, Poon AF, Hughes G, Fearnhill E, Gravenor MB, Leigh Brown AJ, Frost SD. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sankoff D. Minimal mutation trees of sequences. SIAM J Appl Math. 1975;28:35–42. [Google Scholar]
- 40.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafer RW, Rhee SY, Pillay D, Miller V, Sandstrom P, Schapiro JM, Kuritzkes DR, Bennett D. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 43.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 44.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 45.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anisimova M, Kosiol C. Investigating protein-coding sequence evolution with probabilistic codon substitution models. Mol Biol Evol. 2009;26:255–271. doi: 10.1093/molbev/msn232. [DOI] [PubMed] [Google Scholar]
- 49.Delport W, Scheffler K, Seoighe C. Models of coding sequence evolution. Brief Bioinform. 2009;10:97–109. doi: 10.1093/bib/bbn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Frost SD. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencetranslationalmedicine.org/cgi/content/full/2/18/18re1/DC1
Table S1. Genetic differentiation between compartments.
Table S2. Estimated divergence times of compartments.
Table S3. Number of purifying (dN/dS < 1) and positive (dN/dS > 1) selection sites.
Table S4. Inferred substitutions along transmission branches for pairs A, E, and F (panel A) and along pretransmission branches for all pairs A to K (panel B).
Table S5. Marginal likelihoods, estimated as the harmonic mean of the sampled likelihoods in Bayesian MCMC analysis.
Fig. S1. Maximum likelihood phylogenetic tree of viral sequences from a source who infected multiple partners at two independent time points.
Fig. S2. Posterior distributions of parameters sampled during Bayesian MCMC analyses.
Fig. S3. Site-specific nonsynonymous (dN) and synonymous (dS) rates of substitution.