Abstract
The mechanisms of DNA transactions driven by long range protein-mediated inter and intra-chromosomal interactions are currently of considerable general interest. Here, we report that site-specific replication termination catalyzed by the dimeric Reb1 protein of Schizosaccharomyces. pombe at its cognate replication termini (Ter) did not occur independently at each site but was modulated by the Reb1-mediated interactions between pairs of Ter sites located either in the same or in different chromosomes. The interactions between two Ter sites in cis, placed in a mutually anti-parallel orientation, caused looping out of the intervening DNA in vitro and enhancement of fork arrest in vivo. A Ter on chromosome 2 interacted pair-wise with two Ter sites located on chromosome1 by chromosome kissing. Mutational inactivation of the major interacting Ter on chromosome1 abolished or significantly reduced fork arrest at the Ter site on chromosome 2, thereby revealing a novel mechanism of control of replication termination.
Keywords: Replication Termination, DNA looping, Chromosome kissing, Reb1 replication terminator protein, programmed fork arrest
Introduction
Action at a distance refers to induction and/or modulation of a DNA transaction (e.g. transcription) catalyzed by a DNA binding protein remote from its primary binding site. The process probably involves movement of chromosome segments that brings the primary binding site (e.g., an enhancer) into contact with a distant, second site (e.g., a promoter) where the biological activity is manifested, either in the same (Choy and Adhya, 1992; Dunn et al., 1984; Hochschild and Ptashne, 1986; Nemeth et al., 2008; Tolhuis et al., 2002) or in a different chromosome (Ling et al., 2006; Spilianakis et al., 2005). All of the aforementioned reports were on transcriptional control. An important and interesting question that invites investigation is whether action at a distance also controls the 3 steps of DNA replication namely initiation, ongoing replication and termination.
We have previously shown that in prokaryotic plasmid systems, DNA looping controls replication initiation both positively (Miron et al., 1992; Mukherjee et al., 1988) and negatively (Zzaman and Bastia, 2005). The possible role of long range protein-DNA interactions in control of replication in eukaryotes previously has not been investigated. In this context, we wished to address the question as to whether two Ter sites located on 2 different chromosomes control programmed fork arrest by long range protein-DNA interactions. The present work addresses this question.
Replication termination mechanism(s) has been best elucidated in prokaryotes in which a replication terminator protein binds to a specific Ter sequence to impede fork movement in a polar mode. The terminator protein-Ter complexes of prokaryotes arrest replicative helicase-catalyzed DNA unwinding in one direction but allow the enzyme approaching from the opposite direction to pass through unimpeded (Bastia et al., 2008; Kaul et al., 1994; Khatri et al., 1989; Lee et al., 1989). A recent review critically discusses the current status of the field including alternative models of fork arrest mechanisms (Kaplan and Bastia, 2009). Physiologically programmed polar fork arrest occurs in eukaryotes in the non-transcribed spacers of rDNA from yeast to man and at certain other locations in the chromosomes, although not every replicon necessarily contains a Ter site (see reviews by Kaplan and Bastia, 2009; Bastia and Mohanty 2006). The existing data favor a model of polar fork arrest that involves not only terminator protein-Ter interaction but also protein-protein interactions between the terminator protein and the DNA unwinding enzyme(s) that drive the forks (Bastia et al., 2008; Mulugu et al., 2001). The detailed mechanism of polar fork arrest in eukaryotes has not yet been elucidated, although indirect evidence suggests that the mechanism not only involves terminator protein-Ter DNA but probably also replisome-terminator protein interactions (Biswas and Bastia, 2008; Eydmann et al., 2008).
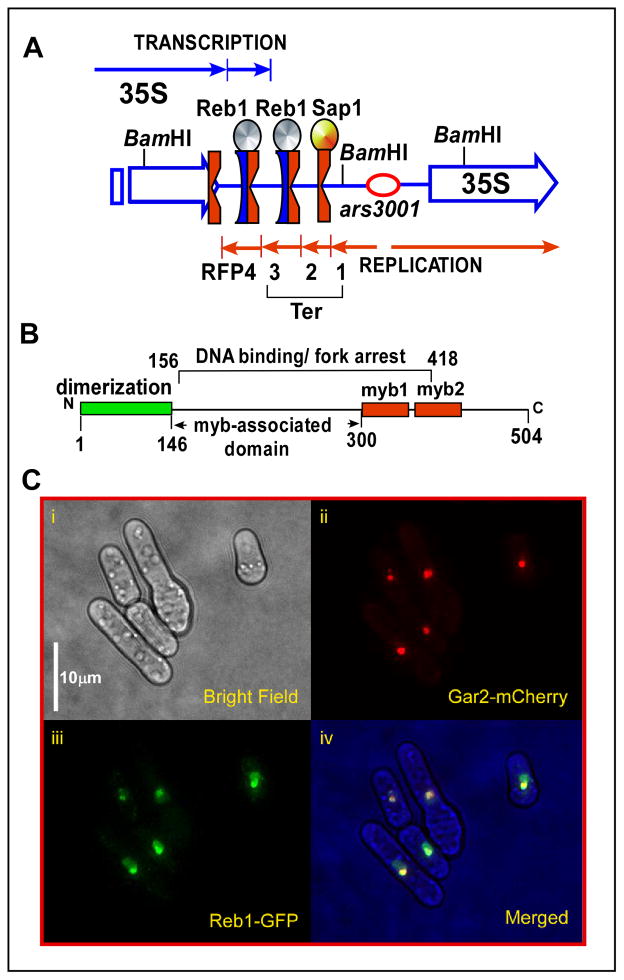
Besides the nontranscribed spacers of rDNA (Bastia and Mohanty, 1996; Bastia and Mohanty, 2006), Ter sites are also present outside these loci, e.g., upstream of the mating type locus Mat1 and at other locations in the 3 chromosomes of S. pombe (e.g., this work). The replication terminus located upstream of the mating type switch locus of S. pombe binds to the protein Rtf1 and promotes mating type switching by preventing a replication fork from traversing the Mat1 locus from the wrong direction (Dalgaard and Klar, 2001). Three replication termini (Ter1-Ter3) and a fork pausing site (FPS4) are present in the nontranscribed spacer region of each rDNA repeat of S. pombe (Krings and Bastia, 2004). The Ter1 binds to Sap1 protein (Krings and Bastia, 2005, 2006; Mejia-Ramirez et al., 2005) that also binds to the switch activating site (SAS1) located near the mating type locus Mat1 (Arcangioli and Klar, 1991), but does not arrest forks at the SAS1 site (Krings and Bastia, 2005). Ter2 and Ter3 of rDNA bind to Reb1, a myb-like protein, which not only promotes polar fork arrest at the binding sites (Krings and Bastia, 2004; Sanchez-Gorostiaga et al., 2004) but also impedes RNA polymerase I-catalyzed transcription approaching the site from the opposite direction (Fig. 1A) (Zhao et al., 1997). The Reb1 protein is dimeric and contains an N-terminal dimerization domain (Fig. 1B) that is dispensable for a basal level of fork arrest at a single Ter site in vivo (Biswas and Bastia, 2008).
Fig. 1. Locations of the Ter sites of S. pombe in rDNA and of Reb1 terminator protein in the nucleoli.
A, schematic representation of the nontranscribed spacer region of rDNA, the 3 Ter sites and the fork pausing site RFP4 are shown, Ter1 binds to Sap1 and Ter2 and Ter3 bind to Reb1 proteins, Ter2 and Ter3 arrest replication forks approaching from one direction (red arrows), and transcription catalyzed by RNA polymerase I from the opposite direction (blue arrows); B, schematic representation of Reb1 protein showing the N-terminal dimerization domain in green and the predicted myb1 domains in red, the myb associated domain; the region of Reb1 from the amino acid residues 156 to 418 is able to arrest replication forks; C, (i) micrographs showing a field of cells in phase contrast, the same field illuminated at a wavelength that shows red fluorescence for Gar2, a nucleolar protein, yellow-green fluorescence emitted by Reb-GFP and a merged image of Gar2-cherry and Reb1-GFP shown in the panels (ii), (iii) and (iv) respectively.
In this work we report on novel results that show that Reb1-dependent Ter sites of S. pombe do not function in isolation but communicate pair wise with other such sites either by DNA looping or by chromosome kissing that regulate the magnitude of physiologically programmed fork arrest.
Results
Reb1 protein localization is pan-nucleolar
A diagram of the replication termini present in the spacer region of rDNA of S. pombe is shown in Fig. 1A in which the red notched surface of Ter denotes the fork-arresting and the blue concave surface, the transcription-blocking face, respectively. We attempted to localize the intracellular address of Reb1 by fluorescence microscopy using fission yeast strains that expressed the nucleolar-specific Gar2M-cherry protein and the wt, dimeric Reb1-GFP or separately an N-terminally truncated monomeric-GFP fusion protein. Representative micrographs of cells in mid log phase (Fig. 1Ci) showed nucleoli painted with red fluorescence characteristic of the Gar2M-cherry fusion protein (Fig. 1Cii). The Reb1-GFP expression was marked by the characteristic yellow-green fluorescence that co-localized with the cherry-red fluorescence of the Gar2M protein. However, significant fluorescence was also detected in the extranucleolar space within the nuclei (Fig 1Ciii and iv). These observations supported the conclusion that Reb1 expression was pan-nucleolar with the average ratio of integrated Reb1-GFP fluorescent intensities in the nucleolus to that in extranucleolar space in the nucleus was ~1.6/1.0 (data not shown).
WT dimeric Reb1 but not the N-terminally truncated monomeric form promoted Ter-Ter interaction in vitro
We have previously shown that WT Reb1 is a dimer and that truncation of the N-terminal 145 amino acids generates a monomeric form that has identical DNA binding affinity for the 17 bp consensus Ter sequence in comparison with the WT dimeric protein (Biswas and Bastia, 2008). We wished to test the hypothesis that the dimeric Reb1 protein caused Ter-Ter interactions and that such interactions enhanced the magnitude of fork arrest at the interacting sites over that of a solo noninteracting Ter (by inducing cooperativity at a distance). We tested this hypothesis by performing DNA ligase-catalyzed circularization enhancement experiments in vitro (Zzaman and Bastia, 2005). The principle of the assay is that if two Ter sites, located close to the ends of a linear DNA and separated by 200 bp or more (> the “persistence length” of the DNA) are brought together by interaction with WT Reb1 protein, such an interaction should significantly increase the rate of DNA end ligation leading to enhanced circularization in comparison with the controls. In the control experiments we measured the rate of DNA circularization of substrates with only a single binding site in the presence of the WT protein, a pair of sites incubated with the monomeric, form of Reb1 (146 Reb1) and the two site substrate ligated without pre-incubation with protein etc.
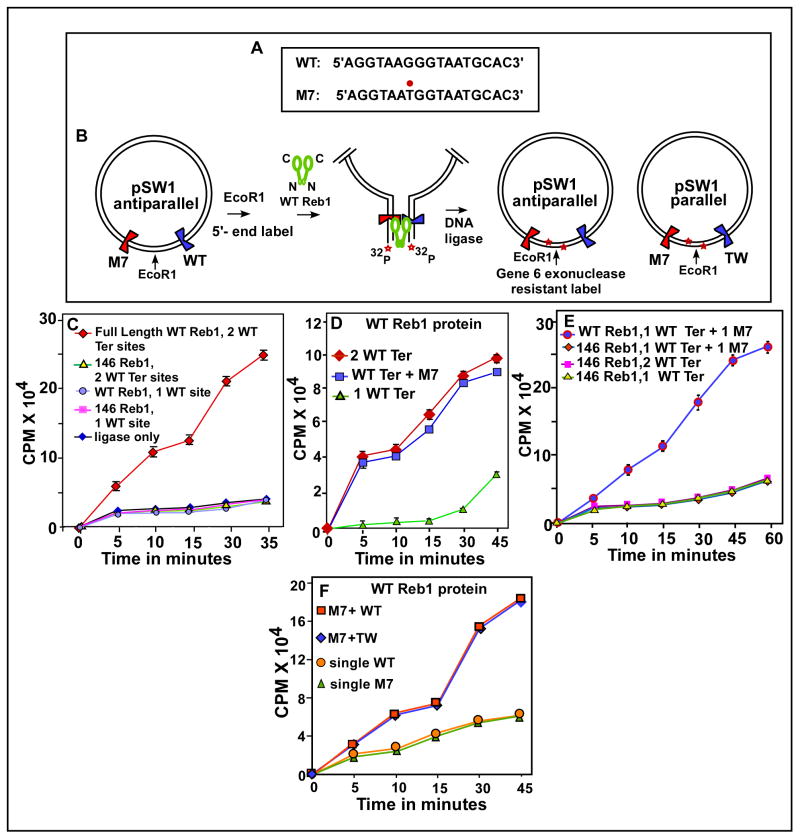
Some of the experiments were carried out with DNA substrates that not only contained the WT Ter sites but also a mutant form called M7 that was generated by a single G to T transversion at the 7th base from the left end of the consensus Ter sequence (Fig. 2A). This mutation is known to cause both a ~90% reduction in the binding affinity of the mutant Ter site for Reb1 protein in vitro and failure to arrest replication forks in vivo (Biswas and Bastia, 2008).
Fig. 2. In vitro Reb1-mediated ligation enhancement assay showing that Reb1 promotes interactions between not only two WT Ter sites but also between a WT and a nonfunctional mutant Ter site M7 in vitro.
A, sequences showing the WT, canonical Ter and the M7 mutant form with the G to T transversion marked with a red dot; B, a schematic representation of the assay method, the plasmid pSW1 contains either two WT Ter sites or a WT and a M7 site placed on either sides of the unique EcoR1 site, DNA looping caused by the binding of the WT Reb1 but not a truncated, monomeric mutant for is manifested by enhancement of DNA circularization of the linearized, labeled DNA that is measured by counting gene 6 exonuclease resistant label; C, Ligation enhancement kinetics showing that two WT sites loop DNA in the presence of the WT Reb1 but not when the truncated form of the protein was supplied, controls show no enhancement of ligation kinetics when a single Ter site was incubated with either the WT or the truncated form of Reb1 or when only ligase was added to the reaction mixture containing the DNA with two WT sites but no reb1 protein; D, ligation kinetics showing that both the substrate containing either two wt sites or a wt site and a M7 site yield approximately equal amounts of enhancement of ligation rate; E, measurement of ligation kinetics showing that a DNA substrate with one WT site and a M7 site show significant enhancement of ligation rate when incubated with the WT Reb1 but not with the mutant monomeric form of Reb1 protein, control experiments using the monomeric protein and DNA substrates with either two or a single WT Ter site did not show more than background levels of ligation enhancement; F, ligation kinetics showing that placing the WT and M7 sites in a mutually parallel orientation does not affect DNA looping.
The DNA substrates were cut with EcoR1, the 5′-ends labeled with γ32P [ATP] and pre-incubated with the WT, dimeric Reb1 or the monomeric 146 Reb1 proteins (Fig. 2B). The DNA protein complexes were then incubated with T4 DNA ligase at 16°C for the indicated periods of time and subsequently digested with T7 gene 6 exonuclease. The reaction products were precipitated with cold 10% TCA and the exonuclease-resistant radioactivity counted after retention on glass fiber filters. The terminal label, after ligation, became internalized and became resistant to gene 6 exonuclease. The enzyme is known to hydrolyze double stranded DNA with 5′ sticky ends in a 5′ to 3′ direction (Zzaman and Bastia 2005). The results showed that a substrate containing 2 WT sites caused significant enhancements of the rate of ligation of the labeled DNA ends in the presence of the WT dimeric but not the monomeric, 146 Reb1 protein (Fig 2C). The WT protein also enhanced the rate of ligation of the substrates with either 2 WT sites or a WT and a M7 site almost to the same extent. In contrast, the rate of ligation of the substrate containing a single WT or an M7 site was not detectably enhanced under identical conditions (Fig. 2D). The monomeric Reb1 protein failed to enhance the rate of ligation of the WT-M7 substrate (Fig. 2E and F). The data taken together supported the conclusion that the dimeric Reb1 but not the monomeric form promoted interactions between either 2 WT sites or a WT site and a M7 site. Both forms of the protein, that had indistinguishable binding affinities for a canonical Ter site, bound very poorly to a solo M7 site in vitro (Biswas and Bastia 2008).
We also investigated whether the magnitude of interaction in vitro between the M7 and the WT sites was independent of the relative orientation of the two sites namely, anti-parallel versus parallel. We constructed two DNA substrates namely pSW1-parallel and pSW1–anti-parallel (Fig. 2B) and measured the kinetics of WT Reb1 mediated enhancement of DNA ligation using EcoR1 digested substrates as described above and discovered that the rates were indistinguishable from each other and significantly higher than that of the control substrates containing either a single WT site or a single M7 site (Fig. 2F). Therefore, the magnitude of Reb1-mediated DNA looping between two Ter sites in vitro was not influenced by their relative orientation.
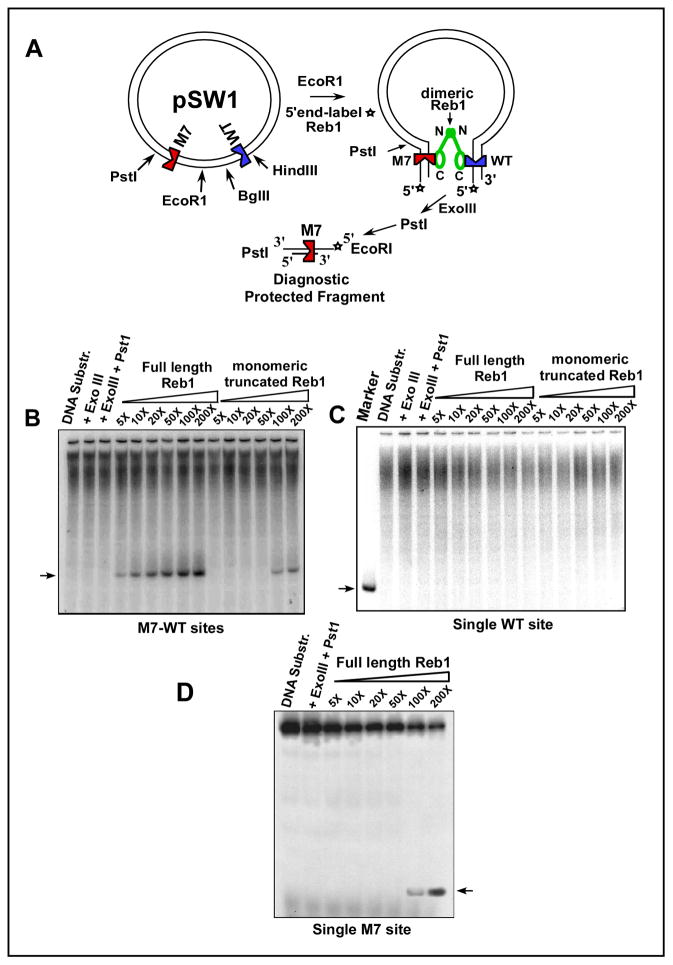
Exonuclease III footprinting confirmed Ter-Ter interaction in vitro
We wished to confirm the aforementioned ligation enhancement results by an independent method. The method is schematically shown in Fig. 3A (Mukherjee et al., 1988; Zzaman and Bastia, 2005). The plasmid DNA substrate pSW1 contained a M7 and a WT Ter site cloned on either sides of the unique EcoR1 site. The M7 site was flanked by a single Pst1 site at its left boundary. The substrate DNA containing the WT and the mutant site or a control substrate containing either a single WT site or a solo M7 site were cut with EcoR1, 5′-end labeled, incubated with either the WT or the truncated form of Reb1protein, digested with exonuclease III (exo III), cut with Pst1 and the DNA resolved in nondenaturing 5% polyacrylamide gels. Exo III is a 3′-5′ exonuclease. Dimeric Reb1 protein-mediated M7-WT site-site interaction would protect the M7 site from resection by exo III digestion, thereby yielding a diagnostic EcoR1-Pst1 protected fragment containing M7. Absence of M7-WT interaction should be revealed by unimpeded resection of DNA by exo III and consequent destruction of the Pst1 site. The autoradiograms of the gels showed that the WT Reb1 protected the Pst1 site in the double M7-WT Ter substrate resulting in the accumulation of the diagnostic EcoR1-Pst1 fragment. The truncated, monomeric Reb1 failed to protect the site. Only at the 2 highest concentration of the monomeric protein, residual protection was observed (Fig. 3B). Neither the WT Reb1 nor the truncated form of the protein protected the Pst1 fragment when only a single WT Ter site was present in the substrate (Fig. 3C). Control experiments with a solo M7 substrate and WT Reb1 protein showed protection of the Pst1 site at the two highest concentrations of the protein (Fig. 3D) and therefore the data in comparison with that in Fig. 3B confirmed that Reb1-mediated looping interaction between the WT with the M7 sites generated cooperativity at a distance. We examined protection from exoIII digestion of a single WT site by the WT or the 146 Reb1 form of the protein and observed similar levels of protection (Fig. S1).
Fig. 3. Autoradiogram of a 6% polyacrylamide nondenaturing gel showing exonuclease III footprints of WT Reb1 supporting Ter-Ter interaction in vitro.
A, schematic representation of the exonuclease III footprinting experiments, the plasmid pSW1 contains a WT Ter site and a mutant M7 site placed on either side of the unique EcoR1 site, protection of the M7 site caused by dimeric Reb1 protein-mediated site-site interaction was measure by the protection of the unique Pst1 site located immediately upstream of the M7 site that generated the ~130bp diagnostic labeled DNA fragment; B, the labeled DNA substrates (25 pmole, each) were incubated with 0.5X (125pmoles), 10X (250 pmoles), 20X, 50X, 100X and 200X wt Reb1 protein and the truncated, monomeric Reb1 of the same range of concentrations, the substrate DNA (25 pmoles) digested with exo III to completion and resolved by electrophoresis. The arrow shows the protected fragment diagnostic of WT Ter – M7 interaction; the truncated Reb1 protein did not generate any protected fragment excepting at the two highest concentration of the protein (100X and 200X) that showed trace amounts of the fragment; C, autoradiogram showing that the control substrate containing a single WT Ter site did not generate any protected fragment when bound to either the WT or the truncated form of Reb1. The location of the protected fragment is marked by an arrow in the “marker” lane; D, autoradiogram showing the extent of protection of the solo M7 region in the presence of WT Reb1 of the same concentration range as in 3B. See also Fig. S1.
WT Reb1 but not the truncated form promoted fork arrest in vivo at a M7 Ter by promoting interaction with a WT Ter
The in vivo relevance of the in vitro Ter-Ter interaction was investigated as follows. We wished to determine whether the dimeric WT Reb1 but not the monomeric form would cause WT-M7 site-site interaction in vivo as manifested in successful replication fork arrest at the M7 site. The experimental plan for monitoring fork arrest in vivo is schematically shown in Fig. 4A. We constructed the plasmid pIRT2-M7-WT-a that contained an ARS-proximal M7 site and a WT site separated by 1.3 kb of DNA (Fig. 4A). The two Ter sites were present in a relative anti-parallel orientation with respect to each other and in the blocking orientation with respect to the fork moving in a counter clockwise direction starting from the ARS. The plasmids were transformed into two S. pombe strains, one expressing the dimeric WT Reb1 and the other, the monomeric form. The replication intermediates were cleaved with PvuII resolved by 2D gel electrophoresis (Brewer and Fangman, 1987), blotted onto nylon membranes and hybridized with 32P-labeled Leu2 probe.
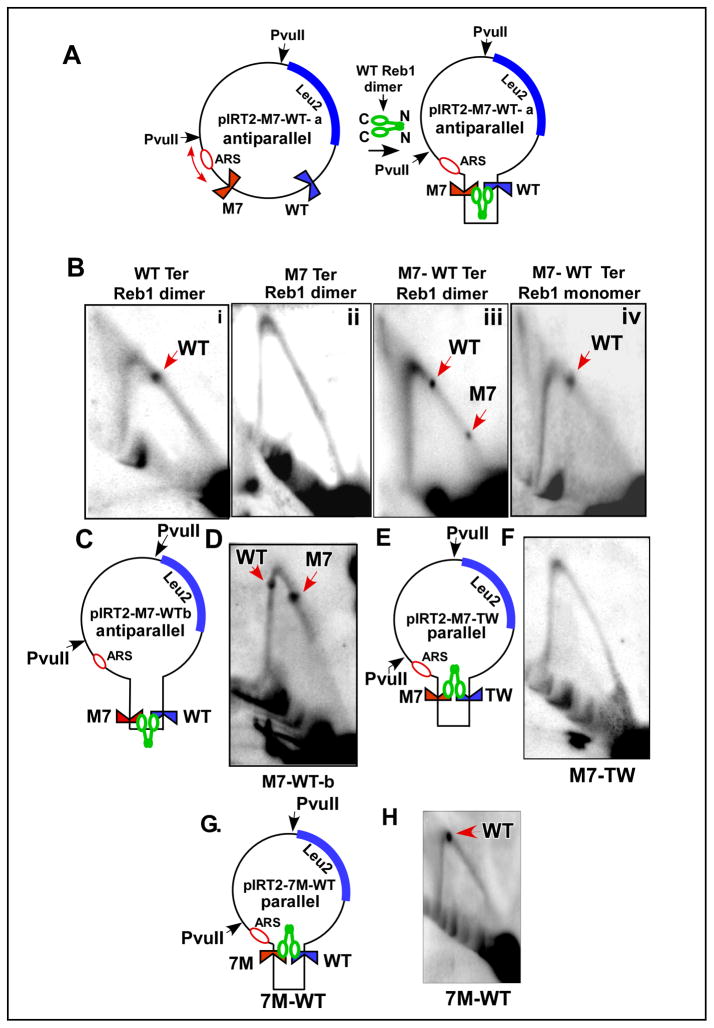
Fig. 4. Representative images of 2D gels of replication intermediates of various DNA substrates.
A, schematic diagram of the plasmid pIRT2 WT-M7 containing an ARS of S. pombe, Leu2 marker, a nonfunctional M7 Ter site and a WT Ter site located 1.3 kb away from the M7 and 2 PvuII sites; the diagram also shows that the dimeric WT Reb1 is expected to promote looping interaction between the M7 and the WT site located in an antiparallel orientation; B, i–iv, images of 2D gels of DNA substrates containing a single WT site, with a single M7 site, containing both the M7 and the WT site in cells expressing the WT Reb1 and the same M7-WT substrate but extracted from cells expressing the truncated, monomeric form of Reb1, respectively; C, diagram of pIRT2-M7-WT-b that contained the two Ter sites separated by 250bp; D, a representative image of a 2D gel of the plasmid shown in C; E, diagram of the pIRT2-M7-TW plasmid with the two Ter sites present in a parallel orientation; F, image of a 2D gel of replication intermediates of the plasmid shown in E; G, diagram of pIRT2-7M-WT plasmid (inverted M7 site) and H, 2D gel pattern of the plasmid shown in G. With the exception of B, iv, in which the replication intermediates were isolated from cells expressing the monomeric Reb1, all of the other samples were extracted from cells expressing the WT reb1 protein. See also Fig. S2.
Representative images of the blots are shown in Fig. 4B. The plasmid DNA replicating in cells expressing WT Reb1 and containing a single WT Ter site, as expected, generated a single termination spot at the expected location on the Y arc (Fig. 4Bi). A solo M7 site in the plasmid, as expected, failed to arrest forks in cells expressing the WT protein (Fig. 4Bii). In contrast, the intermediates of pIRT2-M7-WT-a plasmid that contained a M7 and a WT Ter site revealed two termination spots corresponding to the two Ter sites. The termination spot corresponding to the WT Ter site that was ~2 times more intense than that of the M7 site (Fig. 4Biii). The results supported the interpretation that interaction of the WT Ter with the nonfunctional M7 site converted the latter to a functional replication terminus probably by DNA looping-mediated enhancement of Reb1 binding to M7. In contrast the same plasmid replicating in cells that expressed the monomeric form of Reb1, showed fork arrest at the WT Ter but not at M7 site (Fig. 4Biv). It should be noted that the WT and the M7 sites were present in an anti-parallel orientation with respect to each other in the pIRT2-M7-WT-a plasmid.
In order determine whether the WT and the truncated monomeric protein were both equally expressed in vivo we performed Western blots of host cell lysate (containing the plasmid shown in Fig. 4Biii and Fig. 4Biv), that were resolved in SDS-polyacrylamide gels and developed with polyclonal, monospecific antibodies (Ab) raised against Reb1. We used polyclonal Ab to β-actin to visualize the loading controls. The results showed that both the WT and the truncated proteins were expressed at approximately equal levels (Fig. S2A). We also performed chromatin immunoprecipitations (ChIP) to determine whether both WT and 146 Reb1 bound with similar affinities to Ter3 of rDNA in vivo and the results were in the affirmative (Fig S2B).
Fork arrest required interaction between M7 and WT Ter only in an antiparallel orientation
Anti-parallel refers to pairing of a Ter site A1B1 located in cis with an identical site A2B2 by looping of the intervening DNA in such a way that B1 is aligned against A2 and B2 with A1 in the folded DNA (Fig. 4A). The following experiments were designed to address two questions: (i) to what extent the distance between the M7 and the WT Ter sites determines the magnitude of fork arrest at M7?; (ii) does fork arrest require only Reb1-mediated site-site interactions between the M7 and the WT sites, independently of their relative orientation? The significance of the first question is that it addresses the maximum distance over which Ter-Ter looping might occur. To answer the first question, we constructed the pIRT2 M7-WT-b plasmid derivative in which the distance between the M7 and the WT sites was reduced from 1.3 kb to 250 bp while maintaining their relative anti-parallel orientation (Fig. 4C). We analyzed replication intermediates of the plasmid extracted from cells expressing WT Reb1 protein. A representative image of the 2D gels showed that reduced spacing between the M7 and the WT site significantly enhanced fork arrest at the mutant site in comparison with the WT site as revealed by a stronger termination spot at M7 (~2 fold stronger than the upstream spot; compare Fig. 4D with Fig. 4Biii). The displacement of the WT site to a location just after the inflection point of the Y arc in Fig. 4D was caused by a smaller ~800 bp insert in the plasmid as contrasted with a ~1.3Kb insert in the plasmid shown in Fig. 4Biii. Although the insert was smaller, the Ter sites were located further away from the ARS site in comparison with the plasmid containing the 1.3 Kb insert that accounts for the presence of the two termination spot after the inflection point of the Y arc. The data supported the conclusion that Ter-Ter interaction was significantly reduced if the distance between the two sites was increased from 250bp to 1.3Kb. The limited data tend to suggest that cis interactions between 2 sites separated by as much as 100,000 bp probably would be undetectable in vivo.
In order to address the second question posed above, we constructed the plasmid pIRT2 M7-TW which was a derivative of pIRT-M7-WT-a but contained the WT site in the reversed orientation with respect to the M7 (Fig. 4E). In the absence of any twisting of the intervening DNA, the two Ter sites in this plasmid were expected to interact in a parallel orientation. It should be noted that the M7 and the WT sites in pIRT2 M7-TW were present in the blocking and the nonblocking orientations, respectively, with respect to the counterclockwise moving fork initiated at the ARS. We performed 2D gel analysis of the replication intermediates of the M7-TW plasmid and detected no termination spot (Fig. 4F). The data supported the conclusion that Reb1-mediated pairing between two Ter sites is necessary but not sufficient for induction of fork arrest and that it also requires pairing of the sites in an anti-parallel orientation with respect to each other. We performed the control experiment where the M7 site was reversed with respect to the ARS but the WT site was present in the blocking orientation (Fig. 4G). The 2D gel analysis showed that there was fork arrest at the WT but not at the M7 site (Fig. 4H).
Action at a distance enhanced fork arrest at a natural Ter site in the chromosomal sequence context
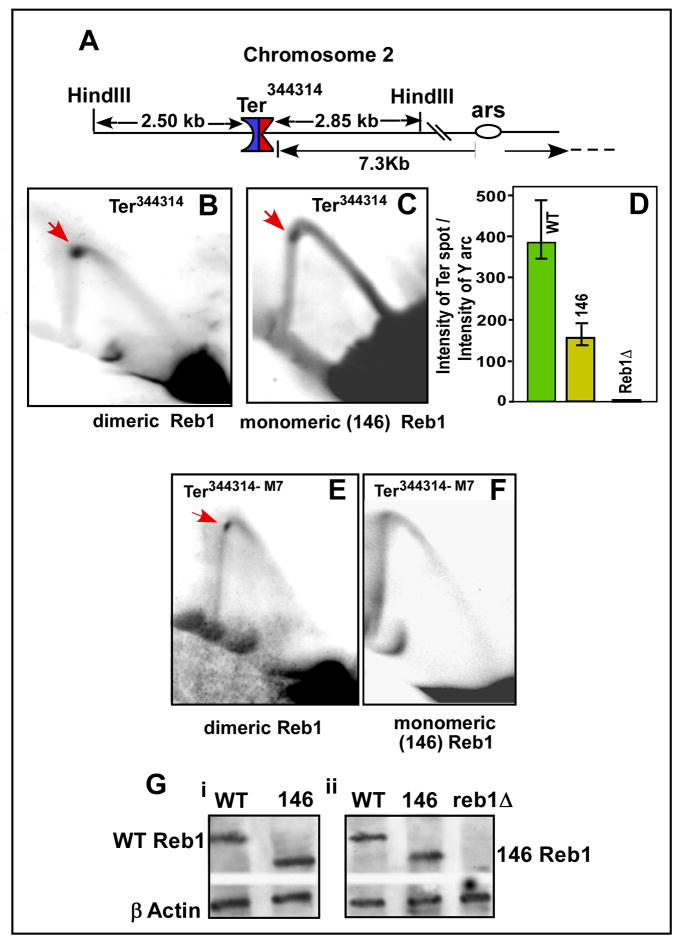
Does Ter-Ter interaction enhance fork arrest at Ter sites present at a natural chromosomal location? In order to address this question, we looked for Ter like sequences present in all 3 chromosomes of S. pombe by a sequence BLAST search using the 17 bp, Reb1-binding core consensus sequence or its close variants as baits. The search revealed several such sequences in all 3 chromosomes. We selected a sequence (Ter344314) present on chromosome 2 at the coordinate 344314 for further work for the following reasons: (i) an inspection of sequences within 100,000 bp on its either side revealed no other recognizable potential Reb1-binding site. This arrangement is expected to prevent cis interaction but should reveal possible trans interactions (chromosome kissing) with another Reb1-binding site and (ii) a potential ARS sequence was located ~7000 bp to the right of the Ter site (Segurado et al., 2003). We expected that it (Ter) might be functional in arresting leftward moving forks initiated from that the ARS site (Fig. 5A). We isolated replication intermediates from cultures harvested at the mid log phase. Comparative FACS profiles of the cultures used at the time of harvest for the preparations of replication intermediates were similar (Fig. S3A). We performed 2D gel analysis of fork movement in the neighborhood of Ter344314 after digesting the chromosomal DNA with HindIII. The results showed that in cells expressing the WT Reb1 protein, forks originating from the ARS and moving to the left were readily arrested at the Ter site (Fig. 5B). In control experiments, in cells that expressed the monomeric form of Reb1, fork arrest was reduced to <50% of that caused by the WT Reb1 protein. As expected, there was no detectable fork arrest at Ter344314 in reb1Δ cells (Fig. 5D). The magnitude of fork arrest was quantified by calculating the ratios of the intensity of the Ter spot over that of the integrated intensities of the Y arc from 4 independent experiments and the data are shown with standard deviation bars (Fig. 5D). All of the 2D gel images used to derive the data in Fig. 5D is shown in Fig. 5B and C and in the supplementary data (Fig. S3B). Western blots showed that the relative levels of expression of WT and 146 Reb1 forms of the protein in vivo corresponding to Fig. 5C and D were approximately equal in strains containing Ter344314(Fig. 5Gi).
Fig. 5. Two dimensional gels of replication intermediates showing that Ter-Ter interactions occur in vivo at natural Ter sites to enhance fork arrest.
A, the location of Ter344314 in chromosome II showing the HinD III sites used to cleave the replication intermediates for 2D gel analysis; B, image of a representative 2D gel, of intermediates purified from cells expressing the WT Reb1, showing fork arrest at the Ter; C, the same as in B excepting that the cells were expressing the monomeric form of Reb1; D, quantification of the replication intermediates with standard deviation bars showing that fork arrest at Ter344314; E, a representative image of a 2D gel of fork arrest at at Ter344314-M7 in cells expressing WT Reb1; F, 2D gel pattern of Ter344314-M7 in cells expressing the truncated monomeric Reb1 protein; note that fork arrest was abolished in cells expressing the monomeric Reb1; G, Western blots showing expression of WT Reb1 and 146 Reb1 in cells containing the normal (Gi) and the M7 form of Ter344314; the blots were developed using polyclonal antibodies raised against Reb1, the loading control is β tubulin. Arrows show termination spots.
We wished to confirm this result further as follows. We have already shown, in the context of a plasmid, that the M7 form of Ter could not arrest forks by itself but required a looping interaction with a WT site to do so (Fig. 4Bii and iii). We reasoned that if a solo Ter344314 promoted efficient fork arrest in vivo, introduction of a M7 mutation into its sequence should abolish its function. On the other hand, if the Ter site was dependent for its activity on interaction with another (WT) site, fork arrest at Ter344314-M7 should still occur in cells expressing WT Reb1 but not in the ones expressing the monomeric, non-looping form of the protein. We introduced the M7 mutation into Ter344314 by site-directed mutagenesis of the sequence cloned into a plasmid and then integrated the Ter344314-M7 mutant sequence into the correct chromosomal location by homologous recombination. The genetic manipulations were done in such a way that the sequence between the mutated Ter and the ARS on the right remained unchanged in comparison with the WT sequence context. We performed 2D gel analyses of fork movement about Ter344314-M7 in cells (i) expressing the WT Reb1, and (ii) the monomeric form of the protein. The images of representative 2D gels showed significant fork arrest at the normal Ter344314-M7 in WT cells (Fig. 5E) However, there was no detectable fork arrest at Ter344314-M7 in cells expressing the truncated, monomeric form of Reb1 (Fig. 5F). The data, derived from 3 independent experiments, supported the interpretation that Ter344314 interacted either in cis or in trans with a site(s) located in the same or in a different chromosome, respectively. We performed Western blots with loading controls to make sure that both the WT and the 146 Reb1 proteins were both well expressed in the cells used in Fig. 5E and F and found that the expression levels of both forms of the protein were very similar (Fig, 5Gii). Which chromosomal sequences interacted with Ter344314? We addressed this question by using the following strategy.
Chromosome kissing occurred between Ter344314 of chromosome 2 and two Ter sites on chromosome 1
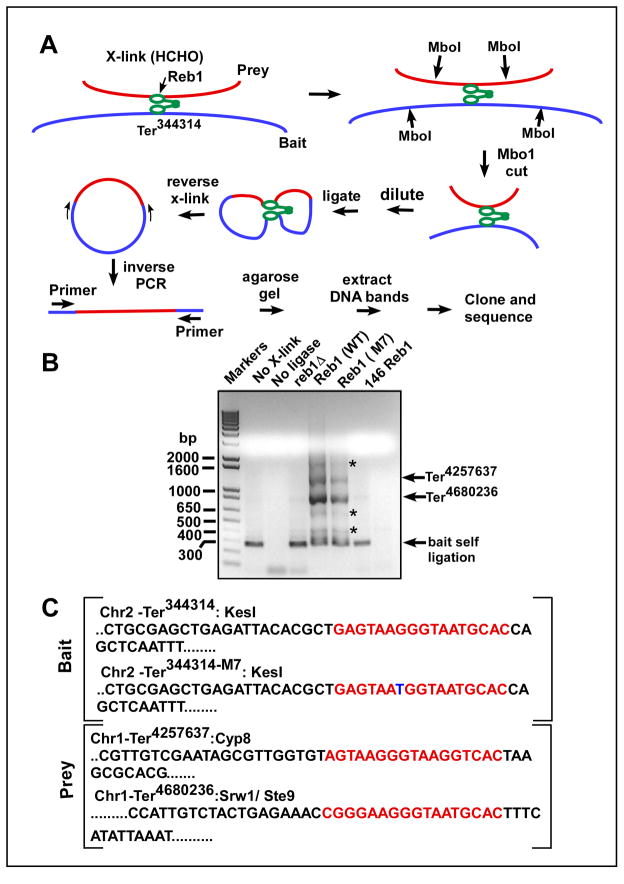
In order to identify sequences interacting with Ter344314, we performed 4C (Circular Chromosome Conformation Capture) analysis by modifications of a published procedure (details shown in supplementary methods), originally designed for mammalian cells (Gondor et al., 2008). Briefly, S. pombe cells expressing WT Reb1 and in control experiment, cells expressing the truncated form were treated with HCHO to crosslink proteins to DNA, the chromatin extracted with SDS to remove non-cross-linked proteins, SDS displaced with Triton X100, the DNA digested with Mbo1 and the samples were diluted and incubated with DNA ligase to promote intra-molecular end joining. Reversal of cross-links and amplification of the specified regions by inverse PCR (as depicted in Fig. 6A) yielded putative interacting sequences. The PCR products were resolved in 1% agarose gels and stained with ethidium bromide. It should be noted that we made no effort to select for Reb1 binding sequences during 4C by a prior immunoprecipitation of the cross-linked chromatin with anti-Reb1-antibodies; this ensured an unbiased selection of the interacting sequences. The data showed that two major DNA bands were reproducibly captured by both bait sequences, i.e., Ter344314 and Ter344314-M7 (Fig. 6B). The third band was a self ligated product that, as expected, was not visible when DNA ligase was omitted. In the absence of cross-linking the two prey bands were not captured. Furthermore, in Reb1Δ cells and in those expressing the truncated form of the protein the two DNA bands were also missing (Fig. 6B). Western blots of cell lysates, with appropriate loading controls were carried out to make sure that failure to capture interacting sequence in cells expressing the 146 Reb1 was not caused by inadequate expression of the truncated protein (Fig. 5Gi).
Fig. 6. Identification of sequences interacting with Ter344314 by 4C analysis.
A, schematic diagram showing the major steps of 4C analysis, the key enzymes/manipulation used at each step are indicated (see the text for further explanation); B, representative agarose gel of the inverse PCR products from a representative 4C analysis that used Ter344314 and Ter344314-M7 as baits, the asterisks show faint minor products that were found to be nonspecific PCR products, the lanes marked Reb1 (WT) and Reb1 (M7) refer to products from cells expressing WT Reb1 protein and which contained the Ter344314 and Ter344314-M7 bait sequences respectively; C, sequences of the bait and the prey sites; the two prey sequences captured by 4C namely Ter4680236 and Ter4257637 are both located in chromosome I, the third major band was self ligated bait sequence which, as expected, is missing from the ligase minus control lane in B (the primer sequences are shown in supplementary data).
The major DNA bands were excised, eluted and either directly sequenced or phosphorylated with polynucleotide kinase, cloned into the SmaI site of pUC19 and several recombinant clones were individually sequenced. The three major DNA bands, from bottom to the top, were identified as self-ligated prey sequence (which, as expected, was missing in the ligase minus control; Fig. 6B), Ter4680236 and Ter4257637, respectively. The Ter4680236 was the most intense (most frequently interacting) band. The two Ter sites, thus captured, were located on chromosome I. The 4C analysis also occasionally showed ~3 faint minor bands (Fig. 6, asterisks). We attempted to identify these by PCR amplification after elution from the gels with the original primer pair but never recovered the same PCR products again. Therefore, we believe that the faint minor bands were probably artifacts of the PCR process caused by spurious binding of the primers to nonspecific sequences.
Chromosome kissing between Ter344314 and Ter 4680236 regulated fork arrest at the former site
We proceeded to obtain more definitive evidence in support of the conclusion that chromosome kissing causes enhanced fork arrest. We inactivated the major interacting Ter4680236 by introducing multiple mutations (Fig. 7A) that altered most of the residue of the consensus site known to contact Reb1 (Biswas and Bastia, 2008). The mutated sequence Ter4680236-M, as expected, did not bind to Reb1 (not shown). We analyzed replication fork arrest at the Ter344314-M7 site in control cells that had the major Ter4680236 site in comparison with those that harbored Ter4680236-M. Both strains expressed WT Reb1 from the natural promoter present at the normal chromosomal location. The cells were harvested under identical conditions in mid log phase and replication intermediates were isolated and analyzed by 2D gels. The images of the gels revealed that fork arrest at Ter344314-M7 was, as expected, readily detectable in control cells with Ter4680236 (Fig. 7Bi). In contrast, fork arrest was reproducibly abolished at Ter344314-M7 in cells containing the mutated Ter4680236-M (Fig. 7Bii). The results unequivocally demonstrate that chromosome kissing between Ter344314-M7 of chromosome 2 and Ter4680236 of chromosome 1 was primarily responsible for optimal fork arrest at the former site.
Fig. 7. 2D gel analysis showing that mutational knockout of Ter4680236 on chromosome 1 abolishes fork arrest at Ter344314-M7 on chromosome 2.
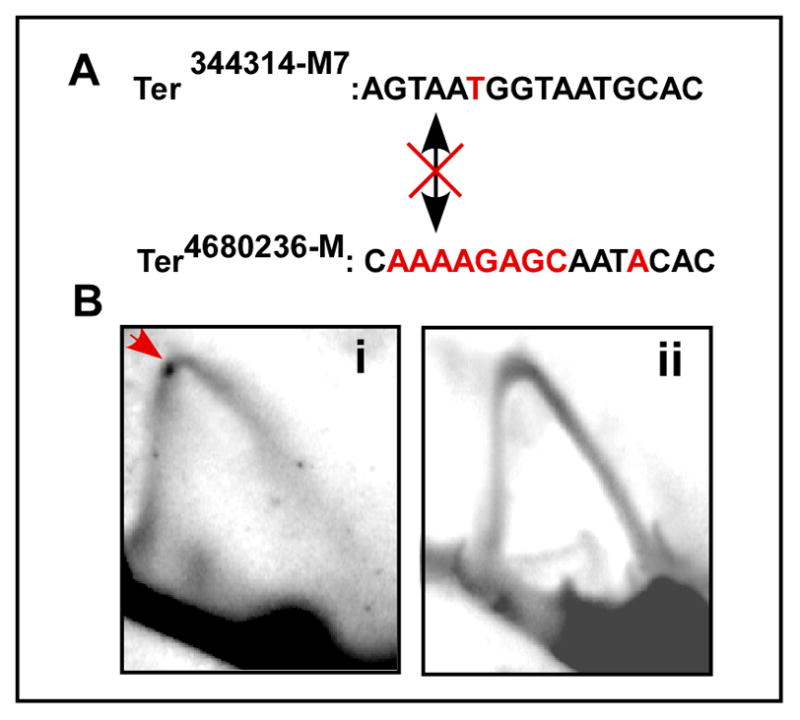
A, sequences of the mutated Ter4580236-M and Ter344314-M7, the altered bases are shown in red; B, images of representative 2D gels showing fork arrest (red arrow) at the control M7 site (Bi) and abolition of the same in cells harboring Ter2680436-M (Bii). The crossed double headed arrow indicates that the two sites do not interact with each other because Ter4580236-M no longer binds to Reb1.
No cis-interacting Ter site of chromosome 2 was recovered by the 4C procedure. The trans interactions by “chromosome kissing” showed remarkable specificity in that the Ter344314 interacted only with the two Ter sites of chromosome 1 under the specified conditions. It is also interesting that although the bulk of Reb1 protein is associated with Ter2 and Ter3 of rDNA repeats present at either ends of chromosome 3, these sites were never captured as interacting sequences using the specified bait sequences. The interaction data pertaining to Ter sites of rDNA will be reported elsewhere.
Discussion
Analyses of the relative spatial orientation of interphase chromosomes by recently developed chromosome conformation capture techniques have revealed extensive intra and inter-chromosomal interactions in yeast and human cells (Dekker et al., 2002; Lieberman-Aiden et al., 2009). While many of the interactions could be caused by random walk, at least some appear to be programmed and biologically significant (Ling et al., 2006; Spilianakis et al., 2005). It should be interesting to test the generality of this mode of control of site-specific replication termination reported in this work by determining whether other replication terminator proteins (e.g., Sap1) of S. pombe also control fork arrest by chromosome kissing. Such work is in progress. Some replication origins fire early whereas others late in the S-phase (Kim and Huberman, 2001). It might be interesting to determine if the activity of each class of the origins is coordinated by physical contacts with other members within each class.
How is cooperativity generated at a distance? It has been suggested that when a weak protein binding site is tethered by DNA to a strong site, the effective concentration of the protein at the weak site becomes greater than that at a solo weak site present at the same location in an identical length of DNA; this difference promotes cooperativity at a distance (Schleif, 1992). Both theoretical modeling and experimental work have suggested that looping can occur over distances up to ~10Mb and chromatin folding influences the site-site interactions. Chromatin apparently folds into a fractal globule in such a way that it possible to unfold and fold any segment of the chromatin without having to break DNA knots (Lieberman-Aiden et al., 2009).
Although, reb1Δ cells can survive under laboratory conditions, the deletion caused some what slower growth rate (data not shown). The Ter344314M7 also had the same slow growth phenotype (data not shown). Reb1 binds to the enhancer/promoter region of Ste9 (Fig. 6B) and activates its transcription (P. Hernandez, personal communications). Ste9 controls pre-start G1 arrest necessary for mating and sporulation, under conditions of nitrogen starvation, by APC-CSte9-mediated degradation of mitotic cyclins (Blanco et al., 2000; Kitamura et al., 1998). Consequently, deletion of Reb1 causes sterility by failure to arrest the cell cycle in pre-start G1. A reb1Δ is synthetically lethal with wee1, the kinase necessary for inactivation of mitotic CdK in G1 (P. Hernandez, personal communications).
It is already known that reb1Δ causes head on collision between replication and transcription units, that precipitates unscheduled fork stalling, which can be potentially destabilizing by causing unscheduled recombination. The fork stalling by collision is clearly manifested in the form of intensification of the descending part of a Y arc as visualized in 2D gels. The effect is amplified when all three Ter sites in the rDNA are rendered nonfunctional by deleting Swi1 (Krings and Bastia, 2004) (G. Krings; 2007 Ph.D. dissertation, MUSC). We postulate that Reb1-dependent chromosome kissing modulates transcriptional activation in at least two ways, by (i) enhancing Reb1 interaction with the transcription complex at the promoter, (ii) by arresting replication forks from interfering with transcriptional activation. In mice, the protein CTCF promotes chromosome kissing that apparently contributes to coordinate expression of genes (Ling et al., 2006). It is possible that Reb1-mediated chromosome kissing in fission yeast also could be contributing to coordinate gene expression.
Although a significant fraction of Reb1 was concentrated in rDNA, presumably bound to each of the two Reb1-binding Ter sites that are repeated in the rDNA array, 4C experiments did not detect any interaction between the Ter sites of rDNA and Ter344314. In reciprocal experiments that used Ter3 of rDNA as bait, no Ter sites from Chromosome I or II were captured as interacting sequences (data not shown). The apparent lack of interaction between Ter sites of rDNA and those located outside the rDNA are perhaps caused by the intra-nucleolar compartmentalization of Reb1-bound Ter sites of rDNA.
Three dimensional optical analyses of inter-phase nuclei suggest that although the chromosomal territories do not seem to move noticeably in response to developmental cues (Hochstrasser and Sedat, 1987), physiologically significant chromosome kissing and DNA looping occurs perhaps via contacts between chromatin loops (de Laat, 2007; Fraser and Bickmore, 2007; Spector, 2003). What drives the chromatin loop movements that promote chromosome kissing? Is it entirely caused by random walk or is it directed by a motor protein? A recent study of transcribing DNA in both budding and fission yeasts reports that the promoter and the terminator sequences of transcribing templates associate with each other forming loops that require interaction with a nucleolar pore protein and a myosin-like protein Mlp1 (Tan-Wong et al., 2009). Further work will be necessary in the future to determine whether Reb1-mediated Ter-Ter interactions also require anchor and motor proteins.
Methods of procedure
Strains and plasmids
These are shown in Table S1 (supplementary data).
Genetic manipulations
Gene deletions and insertions were carried out as described (Grimm et al., 1988).
2D agarose gel electrophoresis
Preparation and separation of replication intermediates (both plasmid and single copy) by two-dimensional (2D) gel electrophoresis were performed following modifications of published procedures (Brewer and Fangman, 1987; Wu and Gilbert, 1995) (see supplement).
Ligation enhancement assay
The assay was carried out essentially as described by Zzaman and Bastia, 2005.
Exonuclease III footprinting
This procedure has been described (Mukherjee et al., 1988). See further details in the online supplementary methods.
Live cell imaging
Cells (FY4460 and FY 4153) were transformed with C-terminally tagged full-length (FL) or 146-N-terminal deletion Reb1 truncation constructs. Cells were grown in pombe minimal media with glutamate (PMG) without uracil and in the presence of 5 μg/ml thiamine to prevent plasmid loss and to repress the nmt1 promoter. A Delta Vision Spectris microscope system (Applied Precision, Issaquah, WA), 60× 1.4 NA objective, was used to determine Reb1 localization. Details are presented in the supplementary methods.
4C (Circular Chromosome Conformation Capture) analysis
This procedure used here was a modification of a published procedure (Gondor et al., 2008). The modifications are described in the online supplementary methods section.
Supplementary Material
Acknowledgments
This work was supported by the grants GM 049264 and GM049264-17S1 to DB. We thank Drs. Louise Pape (University of Wisconsin) for strains and S. Biswas for technical help with one of the experiments. We also thank Dr. Carlos Escalante (VCU, Richmond, VA) for performing bioinformatics analysis of Reb1 and pointing out the presence of SANT-like domains within the myb-associated domain of Reb1, Drs. Tony Carr, Robert Schleif, Fred Winston for useful comments on our manuscript and Dr. Pablo Hernandez for allowing us to cite his work before its publication.
Footnotes
Supplemental data: These contain 3 figures, one table, supplemental methods and references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcangioli B, Klar AJ. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. Embo J. 1991;10:3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D, Mohanty BK. Mechanisms for completing DNA replication. In: DePamphilis M, editor. DNA Replication in Eukaryotic Cells. Plainview, NY: Cold Spring harbor Laboratory Press; 1996. pp. 177–215. [Google Scholar]
- Bastia D, Mohanty BK. Termination of DNA Replication. In: DePamphilis M, editor. DNA replication and human disease. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 155–174. [Google Scholar]
- Bastia D, Zzaman S, Krings G, Saxena M, Peng X, Greenberg MM. Mechanism of replication termination as revealed by Tus-mediated polar arrest of a sliding helicase. Proc Natl Acad Sci USA. 2008;105:12831–12836. doi: 10.1073/pnas.0805898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Bastia D. Mechanistic insights into replication termination as revealed by investigations of the Reb1-Ter3 complex of Schizosaccharomyces pombe. Molecular & Cellular Biology. 2008;28:6844–6857. doi: 10.1128/MCB.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, Sanchez-Diaz A, de Prada JM, Moreno S. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. Embo J. 2000;19:3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Choy HE, Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc Natl Acad Sci U S A. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 2001;15:2060–2068. doi: 10.1101/gad.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W. Long-range DNA contacts: romance in the nucleus? Curr Opin Cell Biol. 2007;19:317–320. doi: 10.1016/j.ceb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dunn TM, Hahn S, Ogden S, Schleif RF. An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984;81:5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydmann T, Sommariva E, Inagawa T, Mian S, Klar AJ, Dalgaard JZ. Rtf1-mediated eukaryotic site-specific replication termination. Genetics. 2008;180:27–39. doi: 10.1534/genetics.108.089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gondor A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nature Protocols. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. J Cell Biol. 1987;104:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL, Bastia D. Mechanism of polar arest of a replication fork. Mol Microbiol. 2009;72:279–285. doi: 10.1111/j.1365-2958.2009.06656.x. [DOI] [PubMed] [Google Scholar]
- Kaul S, Mohanty BK, Sahoo T, Patel I, Khan SA, Bastia D. The replication terminator protein of the gram-positive bacterium Bacillus subtilis functions as a polar contrahelicase in gram-negative Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:11143–11147. doi: 10.1073/pnas.91.23.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri GS, MacAllister T, Sista PR, Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA. Regulation of replication timing in fission yeast. Embo J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Maekawa H, Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol Biol Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2004;101:14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D. Sap1p Binds to Ter1 at the Ribosomal DNA of Schizosaccharomyces pombe and Causes Polar Replication Fork Arrest. J Biol Chem. 2005;280:39135–39142. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- Krings G, Bastia D. Molecular architecture of a eukaryotic DNA replication terminus-terminator protein complex. Molecular & Cellular Biology. 2006;26:8061–8074. doi: 10.1128/MCB.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci U S A. 1989;86:9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Mejia-Ramirez E, Sanchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernandez P. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol. 2005;25:8755–8761. doi: 10.1128/MCB.25.19.8755-8761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A, Mukherjee S, Bastia D. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance [published erratum appears in EMBO J 1992 May;11(5):2002] Embo J. 1992;11:1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Erickson H, Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Mulugu S, Potnis A, Shamsuzzaman, Taylor JK, Bastia D. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc Natl Acad Sci U S A. 2001;98:9569–9574. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008;27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol. 2004;24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;11:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Molecular Cell. 2002;20:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Zhao A, Guo A, Liu Z, Pape L. Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res. 1997;25:904–910. doi: 10.1093/nar/25.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zzaman S, Bastia D. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol Cell. 2005;20:833–843. doi: 10.1016/j.molcel.2005.10.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.