Abstract
The binding of Hedgehog to its receptor Patched causes de-repression of Smoothened resulting in the activation of the Hedgehog pathway. Here, we show that Smo activation is dependent on the levels of phospholipid, Phosphatidyl Inositol-4 Phosphate (PI4P). Loss of STT4 kinase required for the generation of PI4P exhibits hh-loss of function phenotypes while loss of Sac1 phosphatase required for the degradation of PI4P results in hh-gain of function phenotypes in multiple setting during Drosophila development. Furthermore, loss of Ptc function which results in the activation of Hedgehog pathway also causes an increase in PI4P levels. Sac1 functions downstream of STT4 and Ptc in the regulation of Smo membrane localization and Hh pathway activation. Taken together, our results suggest a model in which Ptc directly or indirectly functions to suppress the accumulation of PI4P. Binding of Hh to Ptc derepresses the levels of PI4P, which in turn promotes Smo activation.
Introduction
The Hedgehog signaling pathway, discovered through genetic analysis during Drosophila development is highly conserved across evolution and functions in a diverse array of developmental decisions (Jiang and Hui, 2008). The embryonic functions of the Hh pathway are recapitulated in the adult where it is required for the maintenance of stem cell fate and in tissue repair (Beachy et al., 2004). Consistent with its diverse role, loss of Hh signaling results in developmental defects and its over-activation has been implicated in the etiology of cancer (Jiang and Hui, 2008).
Activation of the signaling cascade occurs upon binding of Hedgehog (Hh) to its receptor, the 12 trans-membrane protein Patched (Ptc), which differs from conventional receptors for intercellular signals in that it functions as a pathway inhibitor (Hooper and Scott, 1989; Jiang and Hui, 2008; Stone et al., 1996). Upon ligand binding, Ptc is inactivated, which in turn leads to a relief of repression of Smoothened (Smo), a 7 trans-membrane protein similar to G-coupled receptors, that transduces the signal inside the cell (Alcedo et al., 1996; van den Heuvel and Ingham, 1996). The specific details of the intra-cellular relay mechanism activated by Smo differs somewhat between vertebrates and flies but culminates in the activation of a transcriptional effector, Cubitis Interruptus (Ci) in Drosophila and a group of Gli proteins in vertebrates (Ruiz i Altaba et al., 2002). In the absence of the Hh signal, the full length (155kd) Ci protein is processed by the proteasome-ubiquitin pathway to generate a 75 kd product that functions as a repressor of Hh pathway target genes. Upon activation of the signal, the proteasome-ubiquitin mediated processing of Ci is blocked, full length Ci is stabilized and migrates to the nucleus functioning as an activator of Hh target genes. Ptc, the receptor for Hh, and also the negative regulator of the pathway, is transcriptionally up-regulated by Ci, setting up a feedback loop to control the amplitude and duration of the signal.
A major unresolved issue in the Hedgehog signal transduction pathway is the mechanism by which Ptc inhibits Smo to block the activation of the pathway and constitutes a primary focus of this study. Biochemical analysis suggests that Smo can exist either in an active or an inactive state and that Ptc downregulates Smo function by stabilizing the inactive state of Smo. This process appears to be catalytic rather than through stoichiometric interactions since Ptc inhibits Smo even when the latter is present in molar excess (Taipale et al., 2002). As a further complication, in spite of multiple binding studies, Ptc has never been found to be physically associated with Smo (Denef et al., 2000; Taipale et al., 2002). Genetic studies in flies suggest that Ptc regulates the membrane localization of Smo and in ptc mutants, Smo is localized constitutively to the membrane, activating the pathway. Likewise, in vertebrate model systems, localization of Smo to the primary cilium of the cell is essential for Hh pathway activation and Ptc inhibits its transport to these primary cilia (Huangfu et al., 2003; Rohatgi et al., 2007). Finally, the Ptc–Smo interaction appears to be one of the most important steps in the Hh pathway because it is one of the most frequently disrupted steps in Hh pathway-related human cancers (Rohatgi and Scott, 2007).
Apart from Ptc mediated relief of repression, Smo activation has also been proposed to require the binding of a small molecule, rather than a protein ligand. Cyclopamine, a plant alkaloid that is teratogenic in sheep, binds Smo and inhibits its function. Likewise, studies in tissue culture cells suggest that sterol like molecules can bind Smo and also the Ptc protein has a sterol sensor domain, a 180 amino acid module present in proteins involved in sterol metabolism and vesicular transport (Eaton, 2008). Finally, sequence homology studies suggest that Ptc protein shows similarity to the Bacterial Resistance, Nodulation Division family of proteins that function as homotrimeric small molecule pumps (Taipale et al., 2002). In this scenario, Ptc is proposed to function as a pump to change the concentration of a small molecule involved in Smo activation. The nature of the proposed small molecule and its relationship to the Ptc/Smo interaction remains unresolved.
Phosphoinositiols (PIs) are lipid constituents of the plasma and organelle membranes of all cells and occur as a collection of seven different phosphorylated versions. The interconversion between different PI moieties is regulated by multiple kinases and phosphatases (Blero et al., 2007; Skwarek and Boulianne, 2009). Different versions of PI show specific localization to membranes of different sub-cellular compartments and regulate cytoskeletal organization, signal transduction and membrane and protein trafficking (Skwarek and Boulianne, 2009). In yeast, the generation of PI4P, the first step in the synthesis of all PIs, is regulated by two genes, PIK1 and STT4, which are required for cell viability and are highly conserved across evolution(Audhya et al., 2000; Flanagan et al., 1993; Yoshida et al., 1994). These two genes function in a non-redundant manner since over-expression of STT4 in PIK1 mutant background does not rescue its phenotype and suggests that STT4 and PIK1 generate distinct non-overlapping pools of PI4P (Foti et al., 2001). Localization studies further suggest the PIK1 is primarily present in the nucleus and the Golgi (Audhya et al., 2000; Walch-Solimena and Novick, 1999) while STT4 is primarily localized to the cytoplasm. In yeast, the cytoplasmic STT4 is recruited to the plasma membrane by trans-membrane proteins for localized synthesis of PI4P on the membrane and is required for PKC-1 dependent activation of the MAP kinase cascade (Audhya and Emr, 2002). Inhibition of the mammalian STT4 homolog, PI4III kinase α, delocalizes PH domain-based reporters from the plasma membrane, suggesting its function in providing PI4P at the cell surface is evolutionarily conserved with yeast (Balla et al., 2005). A PIK1 homolog, PI4III kinase β, is also present in mammals but studies suggest it functions primarily to produce PI4P in the Golgi, where it is used to regulate trafficking to the cell surface (Godi et al., 2004; Wong et al., 1997). To a great extent, however, the precise degree to which STT4 and PIK1 function has been conserved and apportioned among their mammalian orthologs remains unclear
In yeast, suppressor of actin-1 (sac1) phosphatase has been shown to dephosphorylate PI4P to PI and its loss results in a 8-10 fold increase in the levels of PI4P (Foti et al., 2001). Sac1 is localized to the Golgi and the Endoplasmic Reticulum and is highly conserved in its sequence between Drosophila, mice and humans. In Drosophila, sac1 was first identified as a lethal mutation with embryonic defects showing a puckering phenotype similar to that seen upon an increase of Jun Kinase signaling (Wei et al., 2003). Loss of sac1 function causes an increase in Jun Kinase activity during dorsal closure and results in ectopic expression of decapentaplegic (dpp), the signal for the TGF-β pathway.
In this paper, we show that loss of sac1 function causes ectopic activation of Hedgehog signaling. Our studies further show that this increased signaling upon loss of sac1 is due to PI4P accumulation and occurs at the level of Ptc and Smo interaction. Consistent with this observation, our results show that loss of sac1 and the gene encoding the kinase required for PI4P production generate Hh gain and loss of function phenotype, respectively, in multiple settings during Drosophila development. We propose a model in which control of lipid metabolism by Ptc plays a novel and critical role in transducing the Hh pathway signal.
Results
Activation of Jun Kinase (JNK) Wg and Dpp in sac1 mutant clones during imaginal disc development
The previously studied function of Drosophila sac1 relates to its role in the activation of JNK during embryonic dorsal closure (Wei et al., 2003). Consistent with these studies, sac1 mutant clones generated in eye imaginal discs also show increased JNK signaling exemplified by a significant increase in the level of pJUN staining (Fig 1A) as well as by the increased expression of a JNK reporter gene (Fig 1B-C). Activation of the JUN kinase pathway in imaginal discs has been shown to cause cell death mediated by activation of Caspase-3 (Igaki et al., 2002). Indeed, Caspase-3 is activated in sac1 mutant clones (Fig 1D) and the resulting apoptotic signal is suppressed by mutations in the genes encoding JNK (basket, bsk) or and in the gene encoding the upstream kinase that activates the JNKKKK (misshapen, msn) (Fig 1E-F). Over-expression of anti-apoptotic genes such as DIAP-1 also block cell death in sac1 mutant tissue (Fig 1G). This causally links Caspase activation in sac1 clones to the activation of the JNK pathway. A second major target of JNK signaling is wingless (wg) (Ryoo et al., 2004), and we found that Wg protein expression is also increased in sac1 mutant clones and that this ectopic Wg expression is also suppressed in msn/msn, sac1/ sac1 double mutant clones (Fig 1H-I).
Figure 1. Activation of Jun kinase (JNK) cascade and Caspase-3 in sac1 mutant clones.
All tissues are third instar eye discs, posterior is to the left.
(A) In a disc containing sac1L2F/ sac1L2Fmutant clones, Anti-phospho-JNK staining (red) is up-regulated in mutant tissue (non-green).
(B) Control, mock clones in which both green and non-green tissue are wild type, show normal weak wild-type expression of msn-lacZ reporter (red) in cells posterior to the furrow (marked by arrowhead).
(C) In a disc containing sac1L2F/ sac1L2F mutant clones (non-green), msn-lacZ reporter (red) is over- expressed in the mutant tissue.
(D) In a disc containing sac1L2F / sac1L2F mutant clone (non-green), Caspase-3 (red) is activated in the mutant tissue.
(E) In a disc containing double mutant sac1L2F/ sac1L2F and bsk1/ bsk1 clones (non-colored) sacIL2F clones with a single copy loss of bsk (blue) and wild-type cells (green), Caspase-3 (red) expression is fully suppressed in cells doubly mutant for sacI/bsk and partially suppressed in sacI-only mutant clones (blue) which are heterozygous for bsk (compare with D). bsk1 is a null allele of JUN kinase in Drosophila.
(F) In a disc containing double mutant sac1L2F ,msn172/ sac1L2F ,msn172 clones (non-green), Caspase-3 (red) expression is suppressed in the mutant tissue (compare with D).
(G) DIAP1 was over expressed using the Ay-Gal4 system (see materials and methods) in a disc containing sac1 mutant clones (non-green). Cells mutant for sac1L2F / sac1L2F which also over-express DIAP1 show a reduction in Caspase-3 (red) activation (compare with D).
(H) Wg (red) is over-expressed in mutant tissue in a disc containing sac1L2F/ sac1L2F mutant clones (non-green).
(I) Wg (red) expression is suppressed in the mutant tissue in a disc containing sac1L2F / sac1L2F, msn172/ msn172 double mutant clones (non-green) (compare with H).
In addition to these phenotypes, that largely confirm previous findings at other developmental stages where sac1 function was studied, we found that sac1 mutant clones also show a dramatic increase in the expression of the gene encoding the BMP ortholog in Drosophila, decapentaplegic (dpp) (Fig 2A-B’). Unlike Wg and Caspase-3, increased dpp expression is independent of JNK signaling as it is maintained in msn, sac1 double mutant clones (Fig 2C-C’). As dpp is a target of Hedgehog (Hh) in the eye imaginal disc, we investigated if sac1 is associated with increased output from Hh signaling.
Figure 2. Activation of Hedgehog signaling pathway in sac1 mutant clones is independent of JNK signaling.
(A) dpp-lacZ reporter (red) is expressed in cells at the morphogenetic furrow (arrow) in a wild-type disc with mock clones (both green and non-green tissue are wild type).
(B-B’) In a disc containing sac1L2F/ sac1L2F mutant clones (non-green), dpp-lacZ reporter (red) is over- expressed in mutant tissue.
(C-C’) In a disc containing double mutant sac1L2F / sac1L2F, msn172/ msn172 clones (non-green), dpp-lacZ reporter (red) continues to be over-expressed and is similar to that seen in sac1L2F / sac1L2F mutant clones (compare with B-B’).
(D) Wild-type eye disc stained with Ci antibody. Ci (red) is expressed in a band of cells anterior to the morphogenetic furrow (arrow).
(E-E’) Ci (red) is increased in the mutant tissue in a disc containing sac1L2F / sac1L2F mutant clones (non-green).
(F-F’) In a disc containing double mutant sac1L2F / sac1L2F, msn172/ msn172 clones (non-green), Ci (red) continues to be over-expressed and is similar to that seen in sac1L2F / sac1L2F mutant clones (compare with E-E’)
(G) Wild-type eye disc stained with anti-Smo antibody. Smo (red) is only expressed in cells posterior to the morphogenetic furrow (arrow).
(H-H’) In a disc containing sac1L2F/ sac1L2F clones (non-green), Smo (red) expression is enhanced in mutant cells even anterior to the furrow.
(I-I’) In a disc containing double mutant sac1L2F / sac1L2F, msn172/ msn172 clones (non-green), Smo (red) is over-expressed and is similar to that seen in sac1L2F / sac1L2F mutant clones (compare with H-H’).
(J) Wild-type eye disc stained with anti-Ptc antibody. Ptc (red) is very weakly expressed in cells of the eye disc.
(K-K’) In a disc containing sac1L2F/ sac1L2F clones (non-green), Ptc (red) is over-expressed in mutant cells.
(L-L’) In a disc containing double mutant sac1L2F / sac1L2F, msn172/ msn172 clones (non-green), Ptc (red) continues to be over-expressed similar to that seen in sac1L2F / sac1L2F mutant clones (compare with K-K’).
In the third instar eye disc, Ci, the transcriptional effector of Hh signaling is activated in a narrow stripe of cells immediately anterior to the morphogenetic furrow which marks the advancing front of the differentiation wave (Fig 2D). In sac1 mutant clones, expression of activated Ci is dramatically increased both ahead and behind the furrow (Fig 2E-E’) and is independent of JNK signaling as it is maintained in msn, sac1 double mutant clones (Fig 2F-F’). Additionally, loss of sac1 function causes high levels of membrane localized Smo to accumulate in the mutant tissue (Fig 2G-H’) and this phenotype is also independent of JNK signaling as it is maintained in msn, sac1 double mutant clones (Fig 2I-I’). Ptc is a downstream target of Hh signaling and is also expressed at higher levels in sac1 mutant clones (Fig 2J-K’) and its expression is also maintained in sac1, msn double mutant combination (Fig 2L-L’). Thus, transducers of Hh pathway and its downstream targets dpp and ptc are ectopically activated in sac1 mutant tissue in a JNK independent manner. Furthermore, membrane associated receptors and ligands of other signaling pathways such as Notch and Delta whose localization and trafficking has been reported to require PI function (Skwarek and Boulianne, 2009) show no increase upon loss of sac1 function (Fig S1A-F). Likewise, localization of proteins associated with plasma membrane such as Crumbs, PDGF/VEGF receptor (PVR) and Armadillo, is unaffected in sac1 mutant clones (Fig S1G-O). These observations suggests that the loss of sac1 function does not result in general defects in protein transport to the membranes but specifically increases output from Hh signaling.
Interestingly, the expression of Hh protein remains unaltered in sac1 mutant clones (Fig 3A-A’). Also, accumulation of ectopic Smo in sac1 is not altered in hhts genetic background under conditions non-permissive for hhts function (Fig 3D-D’). Thus, the Hh pathway is activated in sac1 mutant tissue downstream of the ligand binding event.
Figure 3. Activation of downstream components of Hedgehog signaling pathway in sac1 mutant clones does not require Hedgehog.
(A-A’) In a disc containing sac1L2F/ sac1L2F mutant clones (non-green). Hedgehog expression (red) is similar to that seen in adjacent wild-type (green) cells. Higher magnification of the selected area containing both wild type (green) and mutant (non-green) cells expressing similar levels of Hh is shown in inset.
(B) Control, third instar eye disc from hhts2 / hhts2 homozygous larva grown at a permissive temperature (18°C) and stained with anti-Smo antibody showing wild-type expression of Smo in cells posterior to the morphogenetic furrow.
(C) Eye disc from hhts2 / hhts2 homozygous larva following a 48 hour shift to non-permissive temperature (29°C). Smo expression is virtually eliminated under these conditions.
(D-D’) Third instar eye disc containing sac1L2F/ sac1L2Fmutant clones in a hhts2 / hhts2 mutant background under non-permissive temperature (48 hours at 29° C). Smo expression (D’) is lost in tissue singly mutant for hhts2 / hhts2 (green) as in C, but is maintained at high levels in cells doubly mutant for sac1L2F/ sac1L2F hhts2 / hhts2 (arrows).
(E) Wild-type expression of Smo (red) in third instar salivary gland cells showing very weak staining.
(F) In this control, antp-Gal4 which is expressed in all cells of the salivary gland was used to drive UAS-hh and stained with anti-Smo antibody. Ectopic expression of Hedgehog in these cells causes an accumulation of membrane localized Smo (red).
(G) antp-Gal4; UAS-sac1RNAi salivary glands stained with anti-Smo antibody. Smo (red) is membrane localized even in the absence of Hh protein.
(H-H’) Clone of cells in the salivary gland expressing sac1RNAi (green) to reduce sacI levels was generated using the Ay-Gal4 system (see materials and methods for details). A cell autonomous activation of dpp-lacZ (red), a read out for the Hh pathway, is detected in cells with reduced levels of sacI function (green).
(I-I’) Clone of cells in the salivary gland co-expressing sacIRNAi and smoRNAi (green) to doubly knock down sac1 and smo using the Ay-Gal4 system (see materials and methods) function in the background of dpp-lacZ. Ectopic dpp-lacZ activation seen upon loss of sacI function is suppressed upon simultaneous loss of both sacI and smo (compare with H-H’).
Sac1 is required for the membrane localization of Smo
In wild-type cells, Smo is largely localized to vesicles and translocates to the plasma membrane upon activation by Hh (Denef et al., 2000) (Jia et al., 2004) (Zhu et al., 2003). We further investigated the membrane localization of Smo in a sac1 mutant background using the previously described Drosophila salivary gland model (Zhu et al., 2003). In this system, all components of Hh signaling cascade, except the Hh protein, are normally expressed. Ectopic expression of Hh in these cells using antp-Gal4 as a salivary gland specific driver causes membrane localization of Smo and activation of the pathway (Fig 3E-F). In the antp-Gal4 UAS-sac1RNAi combination where sac1 function is attenuated in the salivary gland, Smo is re-localized to the cell membrane even in the absence of Hh (Fig 3G). Furthermore, this membrane localization of Smo promoted by the loss of sac1 function is sufficient to cause expression of the Hedgehog pathway target, dpp-lacZ (Fig 3H-H’) and is suppressed when sac1RNAi and smoRNAi are co-expressed in the same cells (Fig 3I-I’). These results clearly establish, in a system where Hh is not normally expressed, that loss of sac1 alone is sufficient to promote both the membrane localization of Smo and its ability to activate down-stream components of the pathway.
Increased levels of PI4P in sac1 mutant tissue
Biochemical analysis in S. cerevisiae has established that sac1 encodes a lipid phosphatase whose primary substrate is PI4P (Foti et al., 2001). We used antibodies against PI4P to detect this lipid in eye discs containing sac1 clones and found that the mutant tissue shows a dramatic increase in PI4P levels as compared to adjacent wild-type cells (Fig 4A-A’). This increase in the levels of PI4P upon loss of sac1 function is not due to perturbations in the levels of other PIs such as PI(4,5)P and PI(3)P since their levels in sac1 mutant tissue is similar to that seen in adjacent wild-type tissue (not shown). The synthesis and inter-conversion of PIs in cells is regulated by a balance in the activities of lipid phosphatases and kinases. When we used RNAi to down-regulate the STT4 kinase in sac1L2F clones in the eye disc, PI4P levels were significantly reduced (Fig 4B-B’). Likewise, in the wing imaginal disc, a reduction of sacI function in the dorsal compartment of the wing disc using ap-Gal4, UAS-sacIRNAi combination, resulted in an increase in PI4P levels (Fig 4C-C’), a simultaneous loss of STT4 kinase in this genetic background also caused a significant reduction in PI4P levels (Fig 4D-D’). In contrast, a simultaneous loss of four wheel drive (fwd), the ortholog of yeast PIK1 (Brill et al., 2000) in fwd, sac1 double mutant clones in the eye disc as well as a co-expression of UAS-sacIRNAi and UAS-fwdRNAi in the dorsal compartment of the wing disc does not result in suppression of PI4P phenotype of sacI (Fig S2). This establishes Drosophila STT4 as a major kinase required for PI4P generation in the context of Hh signaling. To further determine the epistatic relationship between the Sac1 phosphatase / STT4 kinase pair and the canonical components of Hh signaling, we used the wing disc as a model since appropriate region-specific drivers are available for this tissue.
Figure 4. Mutation in sac1 leads to increased PI4P.
(A-B’) Third instar eye discs.
(A -A’) In a third eye disc containing, PI4P (red) levels are significantly up-regulated in sac1L2F/ sac1L2F mutant clone (non-green).
(B-B’) In sac1L2F / sac1L2F mutant clones that have also lost STT4 function, PI4P (red) levels are significantly reduced as compared to loss of sacI function alone (compare with A -A’).
(C-D’) Third instar wing discs. The wing pouch is demarcated by a white dotted line. sacI function was reduced in the dorsal compartment of the wing disc (that expresses Apterous, see schematic in Fig. 5A) by a combination of ap-Gal4, UAS-GFP; UAS-sac1RNAi (green).
(C-C’) PI4P (red) levels are significantly increased in the green dorsal compartment (C’) compared to the adjacent non-green tissue that corresponds to the ventral compartment.
(D-D’) A simultaneous loss of STT4 kinase and sacI in the dorsal compartment of the wing disc (green) causes PI4P (red) levels to be significantly reduced as compared to loss of sacI function alone (compare with C-C’).
In the third instar wing imaginal disc, Hh is expressed in cells of the posterior compartment and activates the signaling cascade in the anterior compartment in a graded fashion (Fig 5A). Ci, the transcriptional effector of Hh signaling, is expressed in all cells of the anterior compartment while the transcription of Dpp, which requires high levels of Hh signaling, is seen in a narrow stripe of cells adjacent to the A/P boundary (Fig 5B). Smo, in contrast, accumulates in all cells of the posterior compartment (Fig 5B). Reduction of sac1 function using ap-gal4, a dorsal compartment specific driver in the wing disc, using the combination ap-gal4, UAS-sac1RNAi results in an expansion of the region in which Smo protein accumulates to include the entire dorsal compartment including the anterior half (Fig 5C, C’). This increase in Smo localization on the membrane is not due to an increase in the transcription of smo as quantitative PCR comparison between ap-gal4, UAS-sac1RNAi and its sibling control show no difference in the level of smo transcripts (data not shown). Therefore, as in the eye disc, loss of sac1 function causes increased membrane localization of Smo. Reduction of the STT4 kinase function in the dorsal compartment using ap-gal4, UAS-STT4RNAi causes reduced Smo protein accumulation in the posterior half of the dorsal compartment (Fig 5D-D’). A simultaneous reduction of STT4 and sac1 function was achieved in the dorsal compartment of the wing imaginal disc using the combined genotype ap-gal4, UAS-sac1RNAi, UAS-STT4 RNAi. This causes a reduction in Smo protein accumulation in the posterior half of the dorsal compartment (Fig 5E-E’), a phenotype identical to that seen upon loss of STT4 function (Fig 5D-D’). These results strongly argue for a requirement of the STT4 kinase that gives rise to PI4P in the membrane localization of Smo and the activation of the pathway. Finally, loss of STT4 function in the dorsal compartment of the wing disc causes a strong reduction in the expression of Ptc, a Hh target gene (Fig 5F-G’) and to a lesser extent the expression of activated Ci in the dorsal-anterior compartment indicating an attenuation of Hh signaling (Fig S3A-B).
Figure 5. sac1 phosphatase and stt4 PI4 Kinase cause elevated membrane levels of Smoothened protein.
The wing pouch is marked by a dotted line. Red channel only shown in gray scale in C’, D’, E’, F’ and G’ for clarity.
(A) A schematic representation of the third instar wing disc. The wing pouch is marked by white dotted line. The A/P compartment boundary (A/P) demarcates the anterior and posterior compartments. The D/V compartment (D/V) boundary demarcates the dorsal and the ventral compartments. ap-Gal4 is expressed throughout the dorsal compartment (shown in blue). Smo is expressed in the posterior compartment of the pouch (red dots)
(B) In wild-type, Smo (red) is expressed in the posterior compartment while the Hh target reporter dpp-lacZ (green) is activated along the A/P compartment boundary.
(C-C’) Knockdown of sac1 in the dorsal compartment of the wing disc using the combination ap-Gal4, UAS-GFP; UAS-sac1RNAi (green cells), causes Smo (red) membrane localization to expand to the dorsal-anterior compartment of the wing (arrow) (compare with B). Red channel only shown in gray scale for clarity (C’).
(D-D’) Knockdown of stt4 kinase in the dorsal compartment of the wing disc using the combination ap-Gal4, UAS-stt4RNAi UAS-GFP (green). Smo (red) membrane localization is reduced in the dorsal posterior compartment (arrow). Red channel only shown in gray scale for clarity (D’)
(E-E’) Double knockdown of sac1 and stt4 in the dorsal compartment of the wing disc using ap-Gal4, UAS-GFP; UAS-stt4RNAi; UAS-sac1RNAi (green) genetic combinations. Smo (red) accumulation in the dorsal anterior compartment (arrow) and dorsal posterior compartment (arrowhead) is significantly reduced. Red channel only shown in gray scale for clarity (E’).
(F-F’) As a control, wild-type Ptc (red) is expressed along the A/P boundary at similar levels in cells of the dorsal (green) and ventral (non-green) regions of the wing pouch.
(G-G’) Knockdown of stt4 kinase in the dorsal (green) compartment of the wing disc using the genetic combination ap-Gal4, UAS-stt4RNAiUAS-GFP causes significant reduction in Ptc (red) expression.
(H-H’) Over-expression of Ptc in the dorsal compartment of the wing disc using the combination ap-gal4, UAS-ptc-YFP (green). Smo (red) membrane accumulation is significantly reduced in the dorsal posterior compartment (arrow). Note that the fully functional Ptc-YFP fusion can be directly visualized in the green channel. Red channel only shown in gray scale for clarity (H’)
(I-I’) Over expression of Ptc in a sac1 knockdown background in the dorsal compartment of the wing disc using ap-gal4 (green). Smo (red) membrane localization in the posterior dorsal compartment is reduced (compare with C and C’).
(J-J’) sac1 knockdown in the dorsal compartment of the wing disc using ap-Gal4, UAS-GFP; UAS-sac1RNAi (green) in the background of dpp-lacZ. Ectopic activation of dpp-lacZ (red) is seen the dorsal anterior compartment (compare with 5B). It is currently unclear why the expression of dpp-lacZ is highest near the A/P boundary and lower near the distal edges of the pouch.
(K-K’) Double knockdown of sac1 and stt4 in the dorsal compartment of the wing disc using the combination ap-Gal4, UAS-GFP/ UAS-stt4RNAi; UAS-sac1RNAi (green). Ectopic dpp-lacZ as a read out for activation of the Hh pathway seen upon loss of sacI alone is suppressed (compare with J-J’).
The Ptc protein plays perhaps the most prominent role in the regulation of Smo membrane localization during Hh signaling (Jiang and Hui, 2008; Zhu et al., 2003). For example, over-expression of Ptc in the dorsal compartment of the wing disc using the genetic combination ap-gal4, UAS-ptc causes a loss of membrane associated Smo in the posterior half of the dorsal compartment (Fig 5H-H’). We show that over-expression of Ptc can also override the Smo membrane localization phenotype and PI4P accumulation seen due to down regulation of sacI (Fig 5I-I’, Fig S3C-F). Thus, ptc functions along with sac1 in the regulation of membrane localization of Smo.
In wild-type third instar wing disc, dpp-lacZ expression is dependent on high levels of Hh signaling and is seen as a stripe of cells in the anterior compartment along the A/P boundary (Fig 5B). This expression is expanded in the dorsal-anterior compartment when sac1 function is reduced in the dorsal compartment of the wing disc (Fig 5J-J’) indicating increased Hh signaling in cells which normally respond to Hh. However, a simultaneous loss of STT4 and sac1 in the dorsal compartment of the wing disc prevents this expansion of dpp-lacZ expression and causes a suppression of the sac1 phenotype (Fig 5K-K’). In this combination, lower sac1 levels would increase PI4P levels, but the STT4 kinase is required for the generation of PI4P in the first place. Thus, the simultaneous block of STT4 and sac1 will cause a reduction in PI4P leading to a reduction in dpp-lacZ expression, a readout for Hh signaling. These results link phospholipid metabolism with the ability of Smo to signal and activate Hh signaling. To directly test if PI4P levels increase upon activation of Hh signaling, we once again used the salivary gland as a model. In wild type, salivary gland cells contain low PI4P that increases dramatically when sac1 is downregulated (Fig 6A-A”). Likewise, activation of Hh signaling by removing ptc in flip-out clones using the Ay-gal4 system in the salivary gland (see materials and methods) using the combination Ay-Gal4 UAS-ptcRNAi which causes a loss of ptc expression in cells expressing Gal4 (marked by GFP) causes a cell-autonomous increase in its PI4P content (Fig 6B-B”). Similarly, and increase in PI4P content is seen when ptc function is attenuated in the dorsal compartment of the wing imaginal disc using the combination, ap-gal4, UAS-ptcRNAi or when mutant clones of ptc are analyzed in the eye imaginal disc (Fig S3G-H). The increase in PI4P levels upon loss of ptc function is dependent on STT4 function as simultaneous loss of ptc and STT4 function in the combination ap-gal4, UAS-ptcRNAi and UAS-SST4RNAi results in the suppression of the phenotype (Fig S3I-L). This establishes a causal relationship between the strength of the Hh signal and the level PI4P. Additionally, the change in level of PI4P is not a feedback from a posttranscriptional target of Hh signaling since over expression of activated Ci in the dorsal compartment does not result in an increase in PI4P levels nor does a simultaneous loss of ptc and smo in the dorsal compartment of the wing disc using the combination ap-gal4, UAS-ptcRNAi, UAS-smoRNAi suppress the increased levels of PI4P (Fig S3M-P).
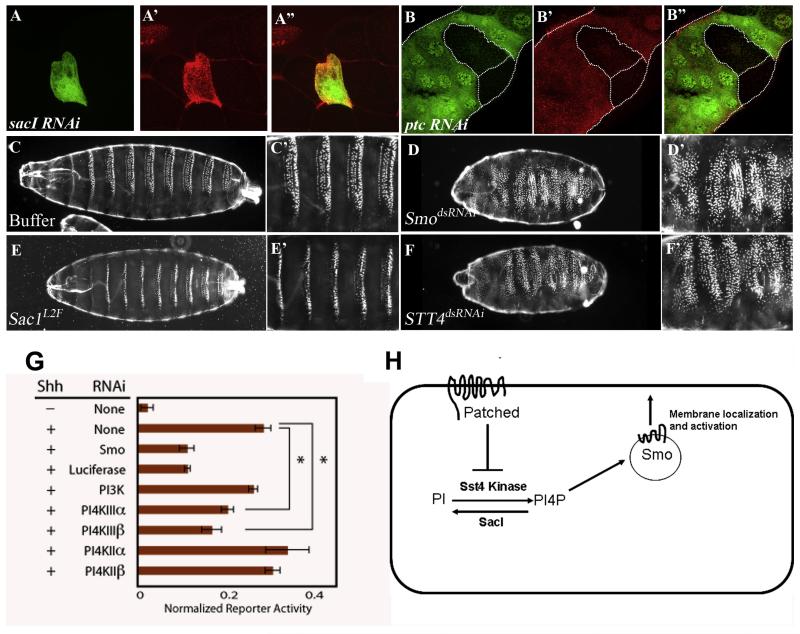
Figure 6. STT4 kinase and SacI phosphatase function in Hh signaling.
(A-A”) Loss of sacI function in positively marked flip-out clones (A, green) generated using the Ay-Gal4 UAS-sacI RNAi combination in salivary gland shows elevated levels of PI4P (A’, red). The merged panel is shown in (A”).
(B-B”) Loss of ptc function in positively marked flip-out clones (B, green) generated using the Ay-Gal4, UAS-ptcRNAi combination in salivary gland shows elevated levels of PI4P (B’, red). The merged panel is shown in (B”). The outline of the salivary gland and wild-type cells are marked with white dotted line.
(C-C’) Organization of denticle belts in wild-type Drosophila embryo microinjected with buffer. A higher magnification of panel (C’) emphasizes normal polarity of denticle belts.
(D-D’) Denticle belt preparation from embryo microinjected with smodsRNAi. The denticle belt pattern is disrupted with the broadening of each belt and a loss in its polarity (magnified image in D’).
(E-E’) sac1L2F /sac1L2F homozygous embryo showing reduction in denticle belt specification, a phenotype similar to that seen in weak ptc mutant embryos.
(F-F’) Denticle preparation of microinjected embryo with STT4dsRNAi showing expansion of denticle belt specification similar to that seen in smodsRNAi (compare with D-D’)
(G) PI4III kinase RNAi inhibits Hh pathway activation in mammalian cells.
Diced siRNA pools were made against PI4III kinase α and PI4III kinase β, the mammalian STT4 and PIK1 homologs, respectively.4 RNAi against other PI4 kinases and PI3 kinase were also tested, along with RNAi against Smo. Hh reporter cells were treated with the indicated RNAi, then grown to confluency and switched into low serum growth medium containing Shh. Following 24 h Shh treatment, cells were lysed and assayed for firefly luciferase-based reporter induction relative to a constitutive Renilla luciferase. Data is reported as the mean of three replicates +/− one standard deviation. *p<0.01, Student’s t-test (two-tailed).
(H) A model. The genetic results presented here are consistent with a model in which Ptc inhibits PI4P formation and this causes retention of Smo away from the plasma membrane. Hh binding to Ptc would activate STT4 kinase and the increase of PI4P will cause a non-stoichiometric amount of Smo to be transported to the cell membrane.
STT4 and Sac1 are conserved components of the Hedgehog signaling pathway
Results presented thus far suggest that STT4 functions as a positive modulator of Hh signaling while Sac1 is required to attenuate this signal. To test if loss of STT4 and sac1 in the embryo results in the signature hedgehog segment polarity phenotypes, we microinjected hairpin versions of STT4 RNA (as no loss of function alleles are available) and for sac1, we isolated homozygous loss of function sac1L2F/ sac1L2F mutants and monitored their denticles belt phenotypes. In wild-type late stage embryos there are 14 denticles belts with a distinct segmental polarity pattern (Fig 6C-C’). As positive controls, microinjection of a short hairpin version of Smo RNA exhibits over-specification of denticle belts and disruption of polarity (Fig 6D-D’) and Ptc mutant embryos show reduction in the denticle belts (Hooper and Scott, 1989). sac1L2F homozygous mutant embryos show a reduction in the width of the denticle belts, a phenotype similar to that seen in ptc, indicative of increased Hh signaling (Fig 6E-E’), while loss of STT4 results in over-specification of denticle belts (Fig 6F-F’), a phenotype similar to that seen in hh and smo mutant embryos (compare Fig 6F-F’ with D-D’), further establishing that STT4 functions as a positive modulator of Hh signaling. Due to the nature of feedback loop between Hh and Wg at the segmental boundary, this effect of sac1/STT4 on Hh is likely to trigger a change in Wg levels as well.
Many aspects of Hedgehog signal transduction, though not all are evolutionarily conserved between Drosophila and mammals. We explored the possibility that PI4P signaling modulates Hh signal transduction in mammalian cells. The mouse genome encodes an ortholog of STT4, called PIKIII kinase α, as established by domain architecture and catalytic domain primary sequence alignment. A second mouse kinase, PI4III kinase β, has a domain architecture that appears to be more closely related to PIK1 (Balla and Balla, 2006). The effect of kinase subunit RNA interference was tested in Shh-LIGHT2 cells, a mouse fibroblast cell line engineered to possess a Gli-dependent luciferase-based reporter system (Taipale et al., 2000). Treatment of these cells with Shh gives robust (>10-fold) increases in reporter activity. In a control experiment, Shh-LIGHT2 cells were treated with RNAi that inactivates smo RNA. Loss of Smo function reduced Shh-induced reporter activation, demonstrating the effectiveness of this system in measuring effects on Hh pathway transduction (Fig. 6G). Next, we tested RNAi treatments designed to reduce kinase functions. RNAi against PI3 kinase or PI4II kinases did not affect Shh-driven reporter induction to a significant extent. In contrast RNAi against the mammalian STT4 homolog, PI4III kinase α, strongly reduced Shh-stimulated reporter activity (Fig. 6G). RNAi against the mammalian PIK1 homolog, PI4PIII kinase β, also affected reporter induction in these cells. The RNAi sequences used were each highly specific for the kinase target, so the results are unlikely to be explained by cross-reactivity. Taken together, these results suggest a non-redundant role for PI4III kinases in mammalian Hh pathway transduction. Note that RNAi against the mammalian ortholog of Sac1 did not affect reporter activity, either in basal or Shh-stimulated conditions (data not shown). However, Shh-LIGHT2 cells are relatively insensitive to Hh pathway de-repression caused by the removal of negative regulators such as Suppressor of Fused (data not shown).
Discussion
A major regulatory step in the modulation of Hedgehog signaling occurs at the level of the two multi-pass transmembrane proteins, Patched and Smoothened. Genetic and biochemical studies suggest that the ligand, Hh, binds Ptc and functions in its inactivation (Taipale et al., 2002) (Denef et al., 2000). This inhibitory step is critical for the activation of Smo which transduces the signal intracelluarly to promote Hh target gene activation. The importance of this regulatory step is further underscored by the observation that the Ptc/Smo interaction is the most commonly disrupted step in cancers caused upon aberrant Hh signaling (Rohatgi and Scott, 2007).
In this paper, we show that phospholipid metabolism plays an important role in the modulation of Hh signaling at the level of Ptc/Smo interaction. In particular, our results show that an increase in the level of PI4P by the inactivation of Sac1 phosphatase leads to Smo protein re-localization to the membrane and an increase in Hh signaling in multiple tissues during Drosophila development. Furthermore the kinase (STT4), which is required for the generation of PI4P, is also required for the proper transduction of Hh signaling as indicated by its effects on Hh target gene expression. PI4P accumulation in the cell is a hallmark of sac1 mutations, and is also seen upon loss of ptc activity. Furthermore, in sac1 mutant tissue we find both increased membrane localization of Smo and accumulation of PI4P while reduction in the PI4P kinase function leads to a hh-like loss of function phenotype. These results establish that phospholipid metabolism provides a critical regulatory input in the modulation of Hh signaling.
Recent studies have proposed that Smo activation requires an input from a non-protein small molecule. Cholesterol and its derivatives (oxysterols) are likely candidates for the small molecules required directly or indirectly for Ptc inhibition or Smo activation since they also promote the translocation of Smo to the cilium (Dwyer et al., 2007). As oxysterols are known to bind to vesicular transport proteins that also interact with phospholipids (Xu et al., 2001), further studies on possible cooperation between these two lipid types could further shed light the mechanism of Smo activation.
Inactivation of Smo by Ptc occurs in a catalytic fashion (Taipale et al., 2002) in that a small number of Ptc molecules can inactivate many more Smo molecules. Our results provide an explanation for this non-stoichiometric inhibitory mechanism. The finding that inactivation of Ptc increases PI4P suggests that Ptc normally functions in keeping PI4P levels low within a cell. This could be achieved either by the down-regulation of the STT4 kinase or by the up-regulation of the Sac1 phosphatase. It is less likely that Ptc modulates Sac1 activity since in vivo localization studies in multiple models system have shown that Sac1 is predominantly localized to the Golgi and due to proximity arguments alone, it seems a more likely possibility that Ptc modulates PI4P levels by down-regulating the lipid kinase. In this model, during normal Hh signaling, binding of Hh to Ptc will relieve repression of the kinase by Ptc and cause an increase in PI4P. As with all genetic analysis in Drosophila, our results do not imply direct protein interactions; currently unknown transduction components could exist and future biochemical analyses will reveal which if any of the interactions is direct. However, our genetic analysis does allow us to propose how an increase in the levels of this lipid can activate Hh signaling. Studies from both flies and vertebrate model system have suggested that the localization of Smo protein to the plasma membrane is essential for the activation of the pathway and studies in multiple model systems have shown that PI4P function is essential in the vesicular transport of cargo proteins from the Golgi to the plasma membrane (Skwarek and Boulianne, 2009). We therefore propose that Hh binding to Ptc releases inhibition of a lipid kinase such as STT4, resulting in high PI4P levels. This aids vesicular transport of Smo to the membrane and causes its activation. A schematic representing the genetic model that is consistent with past and present data is shown in Fig. 6H. Our results using Shh-responsive mouse fibroblasts indicate that mammalian Hh signal transduction is dependent upon the activity of the murine STT4 ortholog, PI4III kinase α. Previous localization studies suggest PI4III kinase α contributes to plasma membrane PI4P pools, an observation consistent with a conserved role for PI4P metabolites in the control of Smo by mammalian Ptc1 (Balla et al., 2005; Wong et al., 1997). The observation that RNAi against the mammalian PIK1 homolog, PI4III kinase β, also reduces Hh signal transduction could suggest it has diverged in function between flies and mammals. Alternatively, PI4P pools could be exchanged more readily between membrane-bound subcellular compartments and the cell surface in mammalian cells, making the removal of either of the PI4III kinases affect global availability of PI4P derivatives. In mammalian cells, Smo activation is associated with translocation of the molecule to the primary cilium, a ubiquitous microtubule-based cell surface protrusion (Corbit et al., 2005; Huangfu et al., 2003). Given that Drosophila cells appear to lack primary cilia, it will be of interest to determine whether PI4III kinase activity is required for Smo translocation.
Supplementary Material
Acknowledgements
This project was initiated as an undergraduate research project in the HHMI Professors program. A.Y., A.F., K.N are undergraduate research students in the UCLA Undergraduate Research Consortium in Functional Genomics which is supported by a Howard Hughes Medical Institute Professor’s Award to U.B. We would like to acknowledge the contribution of Emil Kohan, and Ji-Eun Lee, to early parts of this project. We thank members of the Banerjee lab for helpful suggestions during the course of this work. R.N and G.C are instructors in the UCLA Undergraduate Research Consortium in Functional Genomics. T.H was supported by a Stanford Bio-X fellowship. M. P. S is an Investigator of the Howard Hughes Medical Institute. We would like to thank J. Treisman, H. Wei, K. Moses, P. Ingham, M. Seeger A.J. Zhu, K. Basler and the stock centers of Bloomington, Szeged, NIG (Tokyo) and VDRC (Vienna) for providing Drosophila stocks and reagents. U.B is supported by National Institutes of Health Grants RO1EY008152 and R01HL067395.
Appendix
MATERIALS AND METHODS
Fly Stocks
The following stocks were used in this paper: FRT40A and FRT80B (Bloomington), w; msn172 P{neoFRT}80B/TM6B (J. Treisman), yw; msnj1E2/TM3, Sb (Y.N. Jan), sac1L2F (H. Wei), sac1BG02228(Bloomington), bsk1 /Cyo (M. Seeger), UAS-DIAP1 (B. Hay), Pk61CEP837 (Szeged), hhts2 (K. Moses), dpp-lacZ (Bloomington), UAS-sac1RNAi (VDRC 37217), UAS-stt4RNAi (VDRC 15993), UAS-fwdRNAi (VDRC 27785), fwdEY05397 (Bloomington), puc-lacZ (Bloomington), UAS-ptcRNAi (NIG-FLY 2411R-1), UAS-smoRNAi (NIG-FLY 1156R-1), UAS-ptc-yfp, UAS-ci (K.Basler), UAS-hh (P. Ingham), Ay-Gal4 (Bloomington).
Immunohistochemistry and Embryonic ds-RNA Injections
We used the following antibodies: mouse anti anti-β-galactosidase (1:100; Promega); rat anti-Ci (1:50; R. Holmgren); rabbit anti–cleaved caspase-3 (1:200 ; Cell Signaling); rabbit anti-Hh (1:1,000 ; Ingham, P.); mouse anti-Ptc and mouse anti-Smo, anti-Crumbs, anti-Dl and anti-Notch (1:50; 1:20, 1:100, 1:20 and 1:20 Hybridoma Bank, Iowa); mouse anti-PI4P (1:100 ; Echelon), mouse anti-Wg (1:100; Hybridoma); rabbit anti-phospho JNK (1:50; Cell Signaling). Antibody staining were performed as described previously (Rogge et al., 1995), except anti-PI4P (Blagoveshchenskaya et al., 2002) was done using .3% Saponin in 1X TBS (Sigma S7900) during washes and antibody incubation. ds-RNA and denticle preparation experiments were performed as described previously (Kennerdell and Carthew, 2000).
Ay-Gal4 Flip out clones
Flip out clones were generated in the eye and in the salivary gland. In the eye discs, ey-flp was used to flip-out the stuffer cassette from act-FRTyFRT-Gal4 to generate act FRTGal4 which expresses Gal4 in all cells of the eye imaginal disc (Ito et al., 1997). In the salivary gland, hs-flp was used as a source for flp. The respective cross were maintained at 18°C and when the larvae reached mid second instar, a brief heat shock (10 minutes) was given at 37°C and when the larvae reached third instar, salivary glands were dissected out fixed and stained with appropriate antibodies.
Imaging
Samples were imaged using a BioRad Radiance 2000 confocal with LaserSharp 2000 acquisition software. Fluorescent intensity quantifications were analyzed by ImageJ software.
Shh reporter assay
Diced siRNA pools were generated using previously described methods (Myers et al., 2003). Gene-specific PCR primers were used to amplify ~550bp segments from the coding region of each of the gene indicated in Table S1. A second round of PCR was used to attach forward and reverse T7 polymerase binding sites to the gene-specific PCR products. This DNA was used as template for the production of long dsRNA using in vitro transcription (Ambion). The dsRNA products were processed into ~21bp fragments using recombinant RNAseIII enzyme, resulting in diced siRNA pools directed against each target gene (NEB).
To conduct mammalian Hh pathway reporter assays, diced siRNA pools were introduced into Shhh-LIGHT2 cells using a reverse transfection procedure. For each well of a 96-well plate, Lipofectamine 2000 (Invitrogen) was complexed with 10 pmol of siRNAs in 50 μL of Opti-MEM (Gibco) in individual wells. Early-passage Shh-LIGHT2 cells in log-phase growth were trypsinized, counted, and adjusted to a concentration of 200 cells/ μL. 100 μL of culture medium containing 2×104 cells was gently added to each well, atop the transfection mixture. One day after transfection, the culture medium was changed to DMEM containing 0.5% FBS with or without the addition of Shh-conditioned medium. After 24 h incubation with Shh-conditioned medium, cells were lysed and luciferase signals read using the Dual Luciferase Reporter Assay System (Promega). Data are reported as ratios of Hh-dependent firefly luciferase signal to constitutive Renilla luciferase signal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Mending and malignancy. Nature. 2004;431:402. doi: 10.1038/431402a. [DOI] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Hannah MJ, Allen S, Cutler DF. Selective and signal-dependent recruitment of membrane proteins to secretory granules formed by heterologously expressed von Willebrand factor. Mol Biol Cell. 2002;13:1582–1593. doi: 10.1091/mbc.01-09-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blero D, Payrastre B, Schurmans S, Erneux C. Phosphoinositide phosphatases in a network of signalling reactions. Pflugers Arch. 2007;455:31–44. doi: 10.1007/s00424-007-0304-5. [DOI] [PubMed] [Google Scholar]
- Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Schnieders EA, Emerick AW, Kunisawa R, Admon A, Thorner J. Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science. 1993;262:1444–1448. doi: 10.1126/science.8248783. [DOI] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. Embo J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Rogge R, Green PJ, Urano J, Horn-Saban S, Mlodzik M, Shilo BZ, Hartenstein V, Banerjee U. The role of yan in mediating the choice between cell division and differentiation. Development. 1995;121:3947–3958. doi: 10.1242/dev.121.12.3947. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Skwarek LC, Boulianne GL. Great expectations for PIP: phosphoinositides as regulators of signaling during development and disease. Dev Cell. 2009;16:12–20. doi: 10.1016/j.devcel.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wei HC, Sanny J, Shu H, Baillie DL, Brill JA, Price JV, Harden N. The Sac1 lipid phosphatase regulates cell shape change and the JNK cascade during dorsal closure in Drosophila. Curr Biol. 2003;13:1882–1887. doi: 10.1016/j.cub.2003.09.056. [DOI] [PubMed] [Google Scholar]
- Wong K, Meyers dd R, Cantley LC. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J Biol Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu Y, Ridgway ND, McMaster CR. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J Biol Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohya Y, Goebl M, Nakano A, Anraku Y. A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J Biol Chem. 1994;269:1166–1172. [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.