Abstract
Advances in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) have allowed for the direct analysis of biological molecules from tissue. Although most of the early studies of direct tissue profiling by MALDI-TOFMS have focused on proteins and peptides, analysis of lipids has increased dramatically in recent years. This review gives an overview of the factors to consider when analyzing lipids directly from tissue and some recent examples of the use of MALDI-TOFMS for the direct profiling of lipids in tissue.
Keywords: Lipids, Direct profiling, Tissue, MALDI, MS/MS
1. Introduction
After water, lipids are the most common biomolecules found in the brain (12%) and make up 50% of its dry weight. Furthermore, lipids are the major building blocks of biomembranes, play a key role in signal transduction, and are an important reservoir of energy in biological systems. Due to these important functions of lipids in organisms, the field of lipidomics has increased significantly in importance in the biological sciences [1–6]. Altered levels of lipids are found in many pathological conditions such as Alzheimer's disease [7,8], Down syndrome [9], diabetes [10], Stargardt disease-3 macular dystrophy [11], and Niemann-Pick disease [12]. Additionally, changes in the levels of lipids, in particular ceramides and glycerophospholipids, have been observed in apoptosis or cell death [13–15].
Lipids represent a large and very diverse group of biomolecules that have one or both of the following properties: soluble in organic solvents and presence of long hydrocarbon chains [2]. Lipids can be organized into eight categories: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides [16]. Most of the experimental work discussed in this review will focus on glycerophospholipids (GP) and sphingolipids (SP). Glycerophospholipids are widely abundant in nature, especially in biological membranes, and are divided into classes based upon their head group as follows: glycerophosphocholines (PCs), glycerophosphoethanolamines (PEs), glycerophosposerines (PSs), glycerophosphoglycerols (PGs), glycerolphosphoinositols (PIs), and glycerophosphates (PAs). Sphingolipids contain a sphingoid base backbone and include sphingomyelins (SMs), sulfatides (STs), ceramides, cerebrosides, and gangliosides. Until recently the study of lipid composition and distribution in tissue was a complex and time-consuming undertaking, as intricate techniques had to be used to extract and separate lipid species. However, recent advances in mass spectrometry, mainly matrix-assisted laser desorption/ionization (MALDI), have made it possible to directly probe biomolecules, including lipids, from tissue. This technique and examples of its use in tissue analysis are discussed below.
2. Method
2.1. MALDI-TOFMS
MALDI is a modification of laser desorption/ionization mass spectrometry (LDI-MS) that permits the analysis and detection of intact high-mass biomolecules, such as proteins, peptides, oligonucleotides, and lipids [17]. MALDI was developed simultaneously in the mid 1980's by Tanaka [18] and Hillenkamp [19]. In the Tanaka study, proteins and polymers up to 100,000 Da were analyzed using a matrix of glycerol containing a metal powder. In the same year, the use of a solid organic acid matrix, nicotinic acid, was reported by Hillenkamp for the analysis of proteins up to 67,000 Da. In MALDI, the analyte molecule is mixed with an excess of chemical matrix prior to analysis (molar matrix to analyte ratio: 103–105:1). The chemical matrix is typically a low-molecular weight solid organic acid with strong absorption at the wavelength of the laser being used. In MALDI, the matrix absorbs the laser energy and vaporizes, carrying off some of the analyte molecules. Although the ionization mechanism of MALDI is not clearly understood, the matrix is believed to aid in the ionization of analyte molecules in the gas phase. Traditionally, MALDI has been coupled to time-of-flight (TOF) mass analyzers, and thus this review will mainly focus on direct profiling of lipids using MALDI-TOFMS [20,21]. However, MALDI has been successfully coupled to several additional types of mass analyzers including, ion traps and Fourier transform ion cyclotron resonance (FT-ICR) [17]. Although MALDI-TOFMS has been used extensively for the analysis of peptides and proteins, its use for lipid analysis has recently increased dramatically [22,23].
2.2. Direct profiling by MALDI-TOFMS
MALDI-TOFMS has become a valuable technique for the direct analysis of biomolecules from tissue [24–27]. Caprioli was the first to demonstrate that biomolecules, mainly proteins, could be directly detected in tissue using MALDI [28], followed by Sweedler who probed peptides in neurons and ganglia [29]. MALDI is well-suited for in situ tissue analysis due to its high sensitivity, large tolerance for salts and other contaminants, and a wide mass range with little fragmentation. There are two general types of experiments conducted using MALDI for in situ tissue analysis: profiling and imaging. In profiling experiments, the matrix is deposited directly onto specific regions of interest in the tissue section. The matrix is typically dissolved in a water/organic solvent mixture and deposited onto the tissue section as droplets. Due to the solubility of some biomolecules in the matrix solution droplet, the spatial resolution of this method is usually limited to the size of the matrix droplet. Next, the tissue section is mass analyzed and mass spectral profiles are generated that allow for the comparison of different regions in tissue, i.e. white vs. gray matter in normal brain tissue or cancerous vs. non-cancerous cells when studying pathological changes. In imaging experiments, the matrix is applied over the entire tissue section and individual mass spectra are automatically acquired across the entire tissue section. The data can then be use to generate 2D ion intensity maps (images), in which the individual mass spectra represent pixels. The spatial resolution of this technique is limited to the laser spot size, but spatial resolution below 100 μm is easily obtained. As in profiling experiments, the addition of the matrix also plays a key role in spatial resolution and in the quality of the data. Typically, the matrix is applied by either a spraying technique [30–33] (capillary electrospray emitters, TLC sprayer, airbrush sprayer, oscillating capillary nebulizer), in which the tissue is coated with small aerosolized matrix particles or by using automated matrix spotters [34–36] that deposit an ordered array of small matrix spots (less than 200 μm) across the tissue section. Recently, a method employing the sublimation of matrix [37] and a solvent-free technique using dry matrix particles [31] have been developed for coating tissue and have shown great promise for lipid analysis. In this review, we will focus primarily on profiling experiments, but will briefly mention recent imaging experiments for lipid analysis.
Despite the success of MALDI for the direct profiling of peptides and proteins in tissue, it is only recently that lipid species have begun to be profiled in situ. This is probably because most of the lipid species in tissue have a molecular weight below 1000 Da. In this mass range, identification of analytes by MALDI can be difficult due to matrix ions or background interference ions from the preparation of tissue sections, i.e. stains, optimal cutting temperature compound, etc. Some of the specific factors that need to be considered for direct profiling of lipids in tissue are discussed in the section below.
3. Factors for direct profiling of tissue lipids by MALDI-TOFMS
3.1. Tissue preparation
Appropriate tissue preparation is important to maintaining the spatial resolution of the biomolecules of interest [24]. First, tissue is removed and immediately frozen in liquid nitrogen, dry-iced chilled isopentane, etc., and then stored at −80 °C until sectioning. Next, frozen tissue sections are cut into thin sections, usually 10–20 μm, in a cryostat. Typically, tissue samples are attached to the cryostat sample stages using optimal cutting temperature compound (OCT). However, care must be taken not to contaminate the tissue with OCT, because of previous studies [24] showing that OCT interference can reduce the quality of the mass spectra. This is especially important for lipids in which the m/z range of interest is under 1000. One alternative to OCT is to attach the tissue samples to the cryostat sample stages using ice slush made from distilled water [38]. In this method, the ice slush only comes in contact with the tissue blocks at the surface opposing the sample stages, and is frozen into a thin layer of ice within 5 s. After cutting, tissue sections are collected directly onto MALDI sample targets. The final step is to add the matrix directly to the tissue sections prior to insertion into the mass spectrometer.
3.2. Ionization mode
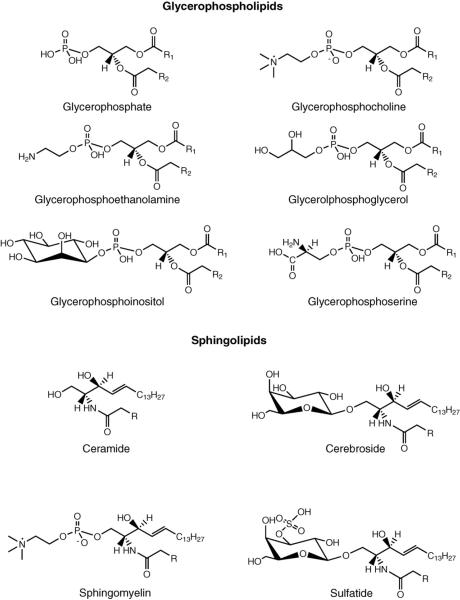
In direct tissue analysis, the ionization mode (positive vs. negative) selected greatly determines which classes of lipids are detected. Since tissue contains a wide variety of lipid classes and this technique is in situ by nature, the ionization efficiency of the different lipid classes will play as important role as to the amount of each lipid classes in terms of what species are detected by mass spectral analysis. Fig. 1 shows the structures of some major classes of glycerophospholipis and sphingolipids that are encountered by direct tissue analysis. The presence of a positively charged quaternary amine group in PCs and SMs makes their ionization in positive ion mode a fait accompli, while the acidic head groups of PIs, PSs, STs aid their ionization in negative ion mode. Several studies [38–44] focusing on lipid species have been conducted analyzing tissue directly by MALDI-TOFMS in positive ion mode. As expected, mass spectra produced from these studies have been dominated by protonated, sodiated, and potassiated adducts of PCs and SMs. Additionally, MALDI-TOFMS studies of standard lipid mixtures have shown that in positive ion mode phosphatidylcholines and sphingomyelins suppress the detection of other lipid classes at similar concentrations [45,46]. One example of the suppression of other lipid classes by PCs is a study profiling glycerophospholipids in rat brain tissue [39]. Although PCs and PEs have similar concentrations in brain tissue, the relative abundance of PE species are weak compared to PC species in the mass spectral profiles recorded in positive ion mode. One way to cancel the innate advantage that the lipid species containing quaternary amines groups have in positive ion mode, is to conduct analysis in negative ion mode. The use of negative ion mode has allowed for the profiling of PEs, PSs, PIs, STs, ganglioside, and cardiolipin species directly from tissue [40,41,47,48].
Fig. 1.
Major classes of glycerophospholipids and sphingolipids.
3.3. Matrix selection
Proper matrix selection is one of the keys to a successful MALDI experiment and is especially important in direct tissue analysis due to the complex nature of the sample. Earlier MALDI studies of lipid standards demonstrated 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA), and 6,7-dihydroxycoumarin (esculetin) as suitable matrices for several classes of glycerophospholipid and sphingolipid species [49,50]. DHB has emerged as one of the most commonly used matrices for lipid analysis due to relatively low matrix interference and the ability to be used in both positive and negative ion mode [22]. The matrix, ρ-nitroaniline (PNA), has been shown to improve the sensitivity for phospholipids species detection in negative ion mode [46]. 2,6-Dihydroxyacetophenone (DHA) has been employed successfully as a matrix for the direct analysis of several classes of lipids (PCs, PEs, PSs, PIs, SMs, cardiolipin, gangliosides) from tissue sections in both positive and negative ion mode [38–42,47,48]. However, one drawback of DHA is that it sublimates in high vacuum (e.g., 1 × 10−6 torr or lower) and thus the sample should be analyzed as soon as possible after it is inserted in the mass spectrometer. Recently, metal/particle matrices have been used for the direct analysis of lipids in tissue, including implanted gold clusters [51], gold nanoparticles [52], and graphite [53]. In all of these studies, mass spectra contained strong mass peaks associated with cerebrosides species in positive ion mode. This is in contrast to what is observed when using traditional organic acid matrices (e.g. DHB) for direct tissue analysis in which PC and SM species are the dominant signals in that lipid mass range in positive ion mode and the cerebrosides species are not usually detected. Ionic liquid matrices have been successfully used to analyze standards from several different phospholipid classes in both positive and negative ion mode and compared to DHB offers improved reproducibility and similar or increased sensitivity [54].
3.4. Laser selection
Commercial MALDI-TOF mass spectrometers typically employ ultraviolet lasers (nitrogen λ = 337 nm, Nd:YAG λ = 355 nm, 266 nm) for ionization. All of the examples of direct profiling of lipids by MALDI-TOFMS discussed in the next section are conducted on instruments using ultraviolet lasers. However, several type of infrared (IR) lasers have also been used for MALDI ionization [55–59]. Additionally, IR lasers have been used for direct analysis of lipids in animal and plant tissue [60–63]. One of the main advantages of using IR lasers is the ability to analyze lipids in tissue directly without the need for a matrix addition step. As mentioned above, the addition of the matrix to the tissue section is one of the most crucial steps in sample preparation and great care has to be taken to ensure that analyte delocalization does not occur. In two recent studies [60,61], glycerophospholipids and sphingolipids species were profiled in situ by MALDI-TOFMS using IR lasers without a matrix addition step. In one study [60], an Nd:YAG laser pumped optical parametric oscillator (λ = 2.8–3.2 μm) was coupled with an ion mobility time-of-flight mass spectrometer for the direct analysis of PCs and SMs species in rat brain tissue sections in positive ion mode. In the other study [61], an Er:YAG laser (λ = 2.94 μm) was used with a MALDI-TOFMS for the analysis of rat brains in both positive and negative ion mode. Overall, around 100 lipid species were assigned including PCs, SMs, PEs, PIs, PSs, STs, PAs, and gangliosides.
4. Examples of direct profiling of tissue lipids by MALDI-TOFMS
4.1. Profiling the distribution of glycerophospholipids and sphingolipids in tissue
Several studies [39–41,43,44,47] have been conducted using MALDI-TOFMS to directly profile glycerophospholipid and sphingolipid distribution in a variety of tissue samples. In one study [43], MALDI-TOFMS was employed to analyze the PC species in leg muscle for a mouse model of Duchenne muscular dystrophy. Mass spectral profiles were generated in positive ion mode for control and diseased areas in the leg muscle. Several PC species were identified, but two in particular were observed to be different between the two areas. PC 16:0/18:2 was more intense in the control area, while PC 16:0/18:1 was stronger in the diseased area. Based upon this ratio, the researchers were able to map areas of damage and regeneration in muscle tissue that agreed with optical images and thus could be a useful method in the future to track the progress of regeneration. Another example of direct profiling of lipids by MALDI is an extensive study [44] to record the distribution and ratio of PC and SM species in lens tissue. In this study, both fresh and formaldehyde fixed tissue was analyzed in positive ion mode with PNA matrix. Fig. 2 illustrates mass spectral profiles from two different areas in bovine lens (fresh and fixed) and the corresponding ratios of PCs to SMs for several different regions. The resulting ratios for PCs and SMs for the different regions agreed with previous extraction experiments and there was no major difference between the calculated ratios for PCs and SMs with fresh vs. fixed tissue, thus showing the robustness of MALDI profiling for different tissue preparations.
Fig. 2.
Comparison of mass spectral profiles from slices of fresh and fixed halves of the same bovine lens. The two mass spectra at top A and B correspond to the outermost region of the cortex. (A) Fresh tissue; (B) tissue fixed in 2.5% formaldehyde buffered solution (pH 7.4). The two mass spectra at the bottom were obtained from the nuclear region. (C) Fresh tissue; (D) fixed as in (B). The ratios of PCs to SMs at different locations in the lens are also labeled. Reprinted with permission from [44]. Copyright 2004 American Chemical Society.
Several studies [39,41,47] have been conducted that profile the regional distribution of glycerophospholipids and sphingolipids in rat brain using both positive and negative ion mode, mainly focusing on the difference between white and gray matter regions. In positive ion mode, mass spectra were dominated by PCs and SMs with PCs species showing the greatest regional differences. The three major PC species detected were PC 32:0, PC34:1, PC 36:1 and each had a different distribution pattern. PC 32:0 was heavily concentrated in gray matter; PC 34:1 was observed in similar amounts in both white and gray matter, and PC 36:1 was overwhelmingly present in white matter. In negative ion mode, PEs, PIs, PSs, STs, and gangliosides were recorded. PIs were more abundant in gray matter regions, while STs were heavily concentrated in white matter. Diacyl PE species were observed to be at higher levels in gray matter, while PE plasmalogen species were recorded at higher levels in white matter. Gangliosides also illustrated a clear distinction between white and gray matter with mainly GM1 being detected in white matter and GM1, GD1, and GT1 all being observed in gray matter.
Although not as widely used as MALDI-TOFMS, MALDI FTMS has recently been used to profile glycerophospholipids in situ in different types of tissue [64]. In this study, mouse brain, heart, and liver tissue were profiled and produced complex mass spectra that were able to be sorted by a computational approach. The major advantage of MALDI FTMS is the increase in reproducibility and mass resolution that improves mass accuracy. A resolving power of over 12,000 was used in this work and allowed for the assignment of several PC, PE, PA, PS, PG, and PI species.
4.2. In situ structural characterization of lipids by tandem MS studies
Profiling of lipids in tissue by MALDI-TOFMS allows for the assignment of lipids; however, to confirm the fatty acid groups of the lipids tandem MS analysis is necessary. Tandem MS studies have been conducted on glycerophospholipids and sphingolipids in rat brain using a MALDI-TOF/TOF mass spectrometer [40,42,47]. These studies were conducted in positive and negative ion mode and allowed for the rapid structural characterization of the lipid species directly from tissue. In the studies [40,42] conducted using positive ion mode, mainly PC and SM species were characterized. Fig. 3 illustrates product-ion spectra for the protonated, potassiated, and lithiated adducts of PC 32:0 from rat brain sections in positive ion mode. The product-ion spectra of PC 32:0+H in Fig. 3a and PC 32:0+K in Fig. 3b contain few fragment peaks and only allow for the assignment of the phosphocholine head group. In order to gain more structural information, lithium adducts can be generated by adding lithium salt to the matrix prior to deposition onto the tissue. Fig. 3c shows a MALDI-TOF/TOF mass spectrum of [PC 32:0 + Li]+ (m/z = 740.514), which contains several fragment peaks that provide structural information. The mass peak at m/z = 681.536 corresponds to the loss of trimethylamine, while the mass peaks at 557.560 and 551.516 Da corresponds to the loss of the phosphocholine head group. Structural information enabling the identification of the acyl groups of the PC specie is provided by the mass peaks at 484.349, 478.320, and 425.311 Da. These mass peaks are assigned as follows: 484.349 [M+Li—C16H32O2]+, 478.320 [M+H—C16H32O2]+, 425.311 [M+Li—N(CH3)3—C16H32O2]+. Based upon these fragment peaks observed [PC 32:0 + Li]+ can be more completely assigned as PC 16:0/16:0.
Fig. 3.
MALDI/TOF-TOF product-ion spectra of (a) PC 32:0+H mass peak, (b) PC 32:0+K mass peak, and (c) PC 32:0+Li mass peak. (c) Reprinted with permission from [42]. Copyright 2005 Elsevier.
Additional tandem MS studies [40,47] were conducted in negative ion mode to confirm the assignment of several PE, PI, PS, and ST species in tissue sections of rat brain. Fig. 4a illustrates a product-ion spectrum of PE 40:6a–H that contains several structurally pertinent fragment peaks. These fragment peaks are attributed to the loss of the acyl group at sn-1 as a ketene, the loss of the acyl group at sn-1, the loss of the acyl group at sn-2 as a ketene, the loss of the acyl group at sn-2, docosahexaenate (22:6) anion, and stearate (18:0) anion and are assigned as follows: 524 [M – H – R′1 CH=C=O]−, 506 [M–H–R1CO2H]−, 480 [M – H – R′2 CH=C=O]−, 462 [M–H–R2CO2H]−, 327 [C22H31O2]−, 283 [C18H35O2]−. Based upon the fragmentation pattern in Fig. 4a, the PE specie was assigned as PE 18:0a/22:6. A product-ion spectrum of PI 38:4–H in Fig. 4b has several diagnostic ions. The mass peaks at m/z 241 (inositolphosphate-H2O), m/z 259 (inositolphosphate) and 223 (inositolphosphate-2H2O) confirm the presence of the inositol polar head group, while the carboxylate anions of the acyl groups are recorded at the following m/z 283 (18:0) and 303 (20:4). Additional mass peaks, attributed to the loss of the acyl group at sn-2 [M–H–R2CO2H]− and the loss of the inositol head group and the fatty acid at sn-2 [M–H–R2CO2H–C6H10O5]−, were observed at 581 and 419 Da. Based upon the product-ion spectrum in Fig. 4b, the PI species is assigned as 18:0–20:4 PI. Fig. 4c illustrates a product-ion spectrum for PS 40:6 H. The presence of a [M–H–87]− mass peak at 747.5 Da represents the loss of the serine head group and confirms the assignment of the PS species. Additionally the carboxylate anions of the acyl groups were recorded at the following m/z sn-1 283 (18:0) and sn-2 327 (22:6). Furthermore mass peaks associated with the loss of acyl groups were observed at 419 m/z [M—H—C3H5NO2—R2CO2H]− corresponding to the loss of the serine head group and the acyl group at sn-2, at m/z 463 attributed to the loss of the serine head group and the acyl group at sn-1, and at m/z 437 corresponding to the loss of the serine head group and the acyl group at sn-2 as a ketene. Based upon the fragmentation pattern in Fig. 4c, the PS species was assigned as 18:0–22:6 PS. In one experiment, the use of tandem MS studies in negative ion was able to structurally assign 32 species of PE, PG, PI, PS, and ST directly from rat brain tissue [47].
Fig. 4.
MALDI/TOF-TOF product-ion spectra of (a) PE 40:6a–H mass peak, (b) PI 38:4–H mass peak, and (c) PS 40:6–H mass peak. Reprinted with permission from [47]. Copyright 2007 Elsevier.
Recently, a MALDI-ion trap mass spectrometer has been used for tandem MS studies of lipids directly from tissue [65,66]. In these studies, SM, PC, PS, and cerebroside species from spinal cord and sciatic nerve tissue were identified based upon MS2 and MS3 data.
4.3. Two-dimensional analysis of phospholipids by MALDI-IM-TOFMS
Due to the in situ nature of direct profiling of tissue by MALDITOFMS, purification and chromatographic separation steps are not possible since the spatial location of the bioanalyte is paramount. Therefore, without the power of chromatography, the mass spectra acquired can be extremely complex and difficult to interpret. This is particularly true at m/z less than 2 kDa where lipid, peptide, and matrix peaks can overlap. Ion mobility (IM) spectrometry is a robust method that allows for the rapid separation and analysis of a wide range of compounds [67]. In this technique, ions can be generated by several methods such as pyrolysis, electrospray, and laser desorption prior to entering a gas-filled mobility drift cell region. In the drift cell, ions obtain an average drift velocity from an electric field based upon their collision cross section (Ω) or shape, which allows for the separation of different shaped molecules [67,68]. The coupling of MALDI-MS with IM spectrometry [69,70] has allowed for a wide range of samples to be analyzed and offers the potential for real-time “chromatography” that operates within the several hundred microsecond time interval between the application of each focused laser desorption pulse to the sample.
MALDI-IM-TOFMS results in the rapid 2D analysis of biological families, in which compounds of similar chemical types (lipids, peptides, oligonucleotides, etc.) fall along trend lines plotted in two-dimensional graphs of ion mobility drift time as a function of m/z [71,72]. Direct tissue analysis by MALDI-IM-TOFMS has produced 2D graphs, in which the native lipids are separated from the native peptides and also from matrix mass peaks [38,52,73]. In one recent study [74], MALDI-IM TOFMS was used for the analysis of complex mixtures of phospholipids and allowed for the fast 2D separation of phopholipid species based upon drift time and m/z. The change in drift time (i.e. collision cross section of the ion) of phospholipids was due to the radyl chain length and degree of unsaturation, the head group, and the cationization of individual species. Additional experiments in this study [74] were conducted in which phospholipids were directly profiled from rat brain tissue sections. Fig. 5 illustrates an overlay of two MALDI-IM 2D spectra of the cerebral caudate-putamen region in rat brain tissue with DHB matrix without cesium (blue area) and with cesium (red area) in positive ion mode. The addition of cesium allows for the assignment of other phospholipid species which were not observed with only DHB matrix due to suppression by PC and SM species. Based upon the drift time observed for each lipid species due to radyl chain, head group, and cationization (mainly due to cesium), assignments were simplified and allowed for the identification of 22 phospholipids species including PCs, PEs, PSs, PIs, and SMs.
Fig. 5.

Overlay of two MALDI-IM 2D plots of the cerebral caudate-putamen region in rat brain tissue with DHB matrix without cesium (blue area) and with cesium (red area). Reprinted with permission from [74]. Copyright 2008 Elsevier (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article).
5. Future outlook
The use of MALDI-TOFMS for direct profiling of lipids in tissue should continue to grow in the future. It offers researchers a quick method to compare different regions of tissue and different disease states. Additionally, building upon earlier profiling studies of lipids in tissue, MALDI imaging experiments for lipids in tissue have increased dramatically in recent years [31–33,37,52,53,73,75–77]. Fig. 6 shows the type of high quality images that can be generated for individual lipid species in tissue using MALDI, in which clear anatomical distribution of these species are observed. This figure compares an optical image of a mouse brain sagittal section with three 2D ion intensities maps generated by MALDI imaging. The MALDI images were recorded in positive ion mode and sublimation of the matrix was employed to coat the tissue. Recent studies using MALDI have produced images tracking the distribution of lipid species in colon cancer tissue [77] and in a mouse model of Tay-Sachs/Sandhoff disease [33]. The further application of MALDI imaging of lipids in tissue will provide a valuable tool for researches in biological sciences.
Fig. 6.
Comparison of (a) an optical image of a mouse brain section with three MALDI images of (b) m/z 760.6, (c) m/z 826.6, (d) m/z 834.6 from a corresponding mouse brain section in positive ion mode. Reprinted with permission from [37]. Copyright 2007 Elsevier.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, NIH. The authors thank the Office of National Drug Control Policy (ONDCP) for instrumentation funding, without which this and other projects could not have been accomplished.
Footnotes
This paper is part of the special issue “Lipidomics: Developments and Applications”, X. Han (Guest Editor).
References
- [1].Holthuis JCM, Levine TP. Nat. Rev. Mol. Cell Biol. 2005;6:209. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- [2].Watson AD. J. Lipid Res. 2006;47:2101. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- [3].van Meer G. EMBO J. 2005;24:3159. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wenk MR. Nat. Rev. Drug Discov. 2005;7:594. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- [5].Piomelli D, Astarita G, Rapaka R. Nat. Rev. Neurosci. 2007;8:743. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- [6].Han X. Front. Biosci. 2007;12:2601. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Han X, Holtzman DM, McKeel DW., Jr. J. Neurochem. 2001;77:1168. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- [8].Han X, Holtzman DM, McKeel DW, Jr., Kelley J, Morris JC. J. Neurochem. 2002;82:809. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- [9].Murphy EJ, Schapiro MB, Rapoport SI, Shetty HU. Brain Res. 2000;867:9. doi: 10.1016/s0006-8993(00)02205-8. [DOI] [PubMed] [Google Scholar]
- [10].Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Biochemistry. 2007;46:6417. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McMahon A, Jackson SN, Woods AS, Kedzierski W. FEBS Lett. 2007;581:5459. doi: 10.1016/j.febslet.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He X, Chen F, McGovern MM, Schuchman EH. Anal. Biochem. 2002;306:115. doi: 10.1006/abio.2002.5686. [DOI] [PubMed] [Google Scholar]
- [13].Thomas RL, Jr., Matsko CM, Lotze MT, Amoscato AA. J. Biol. Chem. 1999;274:30580. doi: 10.1074/jbc.274.43.30580. [DOI] [PubMed] [Google Scholar]
- [14].Cui Z, Houweling M. Biochim. Biophys. Acta. 2002;87:1585. doi: 10.1016/s1388-1981(02)00328-1. [DOI] [PubMed] [Google Scholar]
- [15].Fuchs B, Schiller J, Cross MA. Chem. Phys. Lipids. 2007;150:229. doi: 10.1016/j.chemphyslip.2007.08.005. [DOI] [PubMed] [Google Scholar]
- [16].Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr., Murphy RC, Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. J. Lipid Res. 2005;46:839. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- [17].Dass C. Principles and Practice of Biological Mass Spectrometry. Chapter 2. John Wiley & Sons; New York, NY: 2001. [Google Scholar]
- [18].Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yohida T. Rapid Commun. Mass Spectrom. 1988;2:151. [Google Scholar]
- [19].Karas M, Hillenkamp F. Anal. Chem. 1988;60:2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- [20].Wiley WC, McLaren IH. Rev. Sci. Instrum. 1955;26:1150. [Google Scholar]
- [21].Cotter RJ. Anal. Chem. 1999;71:445a. doi: 10.1021/ac9904617. [DOI] [PubMed] [Google Scholar]
- [22].Schiller J, Sub R, Arnhold J, Fuchs B, LeBig J, Muller M, Petkovic M, Spalteholz H, Zschornig O, Arnold K. Prog. Lipid Res. 2004;43:449. doi: 10.1016/j.plipres.2004.08.001. [DOI] [PubMed] [Google Scholar]
- [23].Schiller J, Suss R, Fuchs B, Muller M, Zschornig O, Arnold K. Front. Biosci. 2007;12:2568. doi: 10.2741/2255. [DOI] [PubMed] [Google Scholar]
- [24].Schwartz SA, Reyzer ML, Caprioli RM. J. Mass Spectrom. 2003;38:699. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- [25].Chaurand P, Schwartz SA, Reyzer ML, Caprioli RM. Toxicol. Pathol. 2005;33:92. doi: 10.1080/01926230590881862. [DOI] [PubMed] [Google Scholar]
- [26].McDonnell LA, Heeren RMA. Mass Spec. Rev. 2007;26:606. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- [27].Fournier I, Wisztorski M, Salzet M. Expert Rev. Proteomics. 2008;5:413. doi: 10.1586/14789450.5.3.413. [DOI] [PubMed] [Google Scholar]
- [28].Caprioli RM, Farmer TB. J. Giles. Anal. Chem. 1997;69:4751. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- [29].Rubakhin SS, Li L, Moroz TP, Sweedler JV. J. Neurophysiol. 1998;81:1251. doi: 10.1152/jn.1999.81.3.1251. [DOI] [PubMed] [Google Scholar]
- [30].Jurchen JC, Rubakhin SS, Sweedler JV. J. Am. Soc. Mass Spectrom. 2005;16:1654. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- [31].Puolitaival SM, Burnum KE, Cornett DS. J. Am. Soc. Mass Spectrom. 2008;19:882. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Garrett TJ, Prieto-Conway MC, Kovtoun V, Bui H, Izgarian N, Stafford G, Yost RA. Int. J. Mass Spectrom. 2007;260:166. [Google Scholar]
- [33].Chen Y, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM, Merrill AH, Jr., Sullards MC. Anal. Chem. 2008;80:2780. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- [34].Aerni H-R, Cornett DS, Caprioli RM. Anal. Chem. 2006;78:827. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- [35].Baluya DL, Garrett TJ, Yost RA. Anal. Chem. 2007;79:6862. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- [36].Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. J. Proteome Res. 2008;7:969. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- [37].Hankin JA, Barkley RM, Murphy RC. J. Am. Soc. Mass Spectrom. 2007;18:1646. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jackson SN, Wang H-YJ, Woods AS, Ugarov M, Egan T, Schultz JA. J. Am. Soc. Mass Spectrom. 2005;16:133. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- [39].Jackson SN, Wang H-YJ, Woods AS. Anal. Chem. 2005;77:4523. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- [40].Woods AS, Wang H-YJ, Jackson SN. Curr. Pharm. Des. 2007;13:3344. doi: 10.2174/138161207782360636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Woods AS, Jackson SN. AAPS J. 2006;8:E391. doi: 10.1007/BF02854910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jackson SN, Wang H-YJ, Woods AS. J. Am. Soc. Mass Spectrom. 2005;16:2052. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- [43].Touboul D, Piednoel H, Voisin V, La Porte SD, Brunelle A, Halgand F, Laprevote O. Eur. J. Mass Spectrom. 2004;10:657. doi: 10.1255/ejms.671. [DOI] [PubMed] [Google Scholar]
- [44].Rujoi M, Estrada R, Yappert MC. Anal. Chem. 2004;76:1657. doi: 10.1021/ac0349680. [DOI] [PubMed] [Google Scholar]
- [45].Petkovic M, Schiller J, Muller M, Benard S, Reichl S, Arnold K, Arnhold J. Anal. BioChem. 2001;289:202. doi: 10.1006/abio.2000.4926. [DOI] [PubMed] [Google Scholar]
- [46].Estrada R, Yappert MC. J. Mass Spectrom. 2004;39:412. doi: 10.1002/jms.603. [DOI] [PubMed] [Google Scholar]
- [47].Jackson SN, Wang H-YJ, Woods AS. J. Am. Soc. Mass Spectrom. 2007;18:17. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
- [48].Wang H-YJ, Jackson SN, Woods AS. J. Am. Soc. Mass Spectrom. 2007;18:567. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Harvey DJ. J. Mass Spectrom. 1995;30:1311. [Google Scholar]
- [50].Harvey DJ. J. Mass Spectrom. 1995;30:1333. [Google Scholar]
- [51].Tempez A, Ugarov M, Egan T, Schultz JA, Novikov A, Della-Negra S, Lebeyec Y, Pautrat M, Caroff M, Smentkowski VS, Wang H-YJ, Jackson SN, Woods AS. J. Proteome Res. 2005;4:540. doi: 10.1021/pr0497879. [DOI] [PubMed] [Google Scholar]
- [52].Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. J. Mass Spectrom. 2007;42:1093. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cha S, Yeung ES. Anal. Chem. 2007;79:2373. doi: 10.1021/ac062251h. [DOI] [PubMed] [Google Scholar]
- [54].Li YL, Gross ML, Hsu F-F. J. Am. Soc. Mass Spectrom. 2005;16:679. doi: 10.1016/j.jasms.2005.01.017. [DOI] [PubMed] [Google Scholar]
- [55].Overberg A, Karas M, Bahr U, Kaufmann R, Hillenkamp F. Rapid Commun. Mass Spectrom. 1990;4:293. [Google Scholar]
- [56].Berkenkamp S, Karas M, Hillenkamp F. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7003. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Overberg A, Karas M, Hillenkamp F. Rapid Commun. Mass Spectrom. 1991;5:128. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- [58].Cramer R, Hillenkamp F, Haglund RF, Am RFJ. Soc. Mass Spectrom. 1996;7 doi: 10.1016/S1044-0305(96)00111-0. [DOI] [PubMed] [Google Scholar]
- [59].Caldwell KL, Murray KK. Appl. Surf. Sci. 1998;127–129:242. [Google Scholar]
- [60].Woods AS, Ugarov M, Jackson SN, Egan T, Wang H-YJ, Murray KK, Schultz JA. J. Proteome Res. 2006;5:1484. doi: 10.1021/pr060055l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dreisewerd K, Lemaire R, Pohlentz G, Salzet M, Wisztorski M, Berkenkamp S, Fournier I. Anal. Chem. 2007;79:2463. doi: 10.1021/ac061768q. [DOI] [PubMed] [Google Scholar]
- [62].Dreisewerd K, Draude F, Kruppe S, Rohlfing A, Berkenkamp S, Pohlentz G. Anal. Chem. 2007;79:4514. doi: 10.1021/ac070191p. [DOI] [PubMed] [Google Scholar]
- [63].Li Y, Shrestha B, Vertes A. Anal. Chem. 2008;80:407. doi: 10.1021/ac701703f. [DOI] [PubMed] [Google Scholar]
- [64].Jones JJ, Borgmann S, Wilkins CL, O'Brien RM. Anal. Chem. 2006;78:3062. doi: 10.1021/ac0600858. [DOI] [PubMed] [Google Scholar]
- [65].Garrett TJ, Yost RA. Anal. Chem. 2006;78:2465. doi: 10.1021/ac0522761. [DOI] [PubMed] [Google Scholar]
- [66].Landgraf RR, Garrett TJ, Calcutt NA, Stacpoole PW, Yost RA. Anal. Chem. 2007;79:8170. doi: 10.1021/ac0713555. [DOI] [PubMed] [Google Scholar]
- [67].Baumbach JI, Eiceman GA. Appl. Spectrosc. 1999;53:338A. doi: 10.1366/0003702991947847. [DOI] [PubMed] [Google Scholar]
- [68].McDaniel EW, Mason EA. The Mobility and Diffusion of Ions in Gases. Wiley; New York, NY: 1973. p. 68. [Google Scholar]
- [69].Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, Gonin M, Schultz JA. Anal. Chem. 2000;72:3965. doi: 10.1021/ac0005619. [DOI] [PubMed] [Google Scholar]
- [70].Von Helden G, Wyttenbach T, Bowers MT. Science. 1995;267:1483. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- [71].Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, Fuhrer K, Gonin M, Schultz JA. Anal. Chem. 2004;76:2187. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- [72].Fenn LS, McLean JA. Anal. Bioanal. Chem. 2008;391:905. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- [73].McLean JA, Ridenour WB, Caprioli RM. J. Mass Spectrom. 2007;42:1099. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- [74].Jackson SN, Ugarov M, Post JD, Egan T, Langlais D, Schultz JA, Woods AS. J. Am. Soc. Mass Spectrom. 2008;19:1655. doi: 10.1016/j.jasms.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang H-YJ, Jackson SN, Post J, Woods AS. Int. J. Mass Spectrom. 2008;278:143. doi: 10.1016/j.ijms.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shimma S, Sugiura Y, Hayasaka T, Zaima N, Matsumoto M, Setou M. Anal. Chem. 2008;80:878. doi: 10.1021/ac071301v. [DOI] [PubMed] [Google Scholar]
- [77].Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. J. Chromatogr. B. 2007;855:98. doi: 10.1016/j.jchromb.2007.02.037. [DOI] [PubMed] [Google Scholar]