Summary
We have investigated determinants of polyhead formation in bacteriophage P22 in order to understand the molecular mechanism by which coat protein assembly goes astray. Polyhead assembly is caused by amino acid substitutions in coat protein at position 170, which is located in the β– hinge. In vivo scaffolding protein does not correct polyhead assembly by F170A or F170K coat proteins, but does for F170L. All F170 variants bind scaffolding protein more weakly than WT as observed by affinity chromatography with scaffolding protein-agarose and scaffolding protein shell re-entry experiments. Electron cryo-microscopy and 3D image reconstructions of F170A and F170K empty procapsid shells showed that there is a decreased flexibility of the coat subunits relative to WT. This was confirmed by limited proteolysis and protein sequencing, which showed increased protection of the A-domain. Our data support the conclusion that the decrease in flexibility of the A-domain leads to crowding of the subunits at the center of the pentons, thereby favoring the hexon configuration during assembly. Thus, correct coat protein interactions with scaffolding protein and maintenance of sufficient coat protein flexibility are crucial for proper P22 assembly. The coat protein β– hinge region is the major determinant for both features.
Keywords: virus assembly, cryo-reconstruction, scaffolding protein, suppressor substitutions, conformational switching
Introduction
Most viruses have evolved with capsids just large enough to enclose a complete viral genome, and given that the coding potential of each genome is quite limited, the capsids are often built from multiple copies of one or a few unique, virally-encoded proteins. Many dsDNA viruses, such as bacteriophage P22, have capsids composed of hundreds of chemically identical coat proteins organized as hexameric and pentameric morphological units (hexons and pentons, respectively). P22, which infects Salmonella enterica serovar Typhimurium, is an excellent model system for studying the assembly of tailed bacteriophage and Herpesviridae (Teschke and Parent, 2010). In vivo assembly of P22 virions involves co-polymerization of 415 coat proteins (gp5, the product of gene 5), 60–300 scaffolding proteins (gp8), the dodecameric portal complex (gp1), and a small number of ejection proteins (gp7, gp16 and gp20) into a precursor (procapsid) structure (Fuller & King, 1980, King et al., 1976). Procapsids mature into virions as DNA is packaged through the portal complex (Bazinet & King, 1988) and scaffolding protein concomitantly exits, most likely through holes in the isometric capsid (head) (Greene & King, 1994, Prasad et al., 1993). Maturation of P22 is accompanied by an increase in volume of the head by ~10% and a change in overall capsid morphology from one that is nearly spherical to one that is faceted (Earnshaw et al., 1976, Prasad et al., 1993). Subsequent addition of plug and needle proteins (gp4, gp10, and gp26) seal off the portal channel, and addition of tailspike proteins (gp9) complete the formation of mature, infectious phage (King et al., 1973, Strauss & King, 1984).
Scaffolding protein directs proper assembly of monomeric coat proteins to form a procapsid in which the coat proteins are organized with T=7 quasi-symmetry. In the absence of scaffolding protein in vivo, coat protein often assembles into aberrant spiral structures as well as T=4, icosahedral particles (Earnshaw & King, 1978, Thuman-Commike et al., 1998, Lenk et al., 1975). Three-dimensional (3D) image reconstructions computed from images of vitrified samples of scaffolding-containing and scaffolding-lacking procapsids (“shells”) show that the scaffolding protein contacts the inner surface of the coat protein at all three-fold-symmetric sites in the procapsid (Thuman-Commike et al., 1999, Thuman-Commike et al., 2000). Extraction of scaffolding protein from procapsids with GuHCl or bisANS, and also re-entry of scaffolding protein into empty shells, exhibits biphasic kinetics indicating there are two distinct populations of bound scaffolding proteins (Teschke & Fong, 1996, Greene & King, 1994, Teschke et al., 1993, Parker et al., 2001). Approximately 60 tightly-bound scaffolding proteins are necessary for assembly and bind with a Kd= ~ 0.1 – 0.3 μM (Parker et al., 2001). The remaining scaffolding proteins are loosely bound and bind with a Kd= ~ 28 μM (Parent et al., 2006). The loosely bound scaffolding proteins increase the overall stability of the procapsids and may also inhibit incorporation of extraneous host proteins (Parent et al., 2006, Earnshaw & Casjens, 1980).
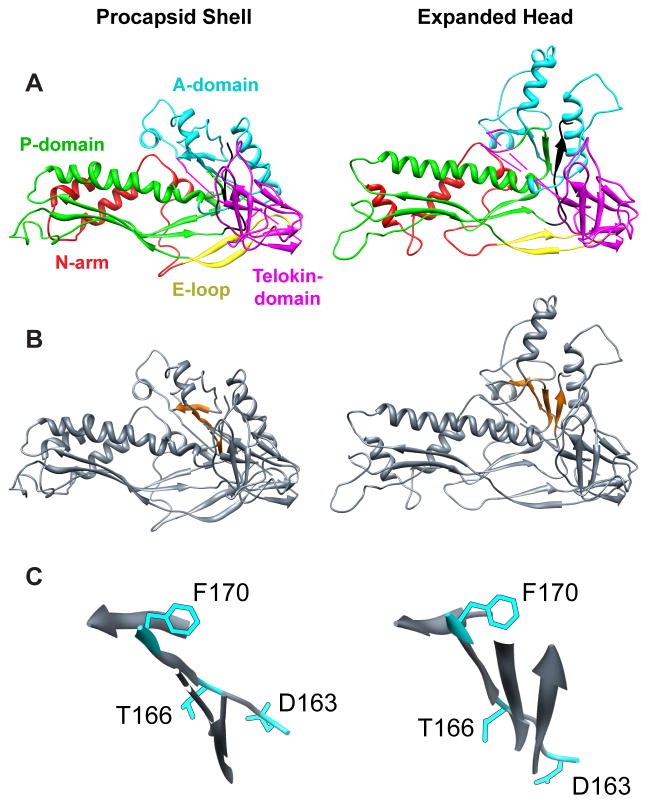
A nearly complete, pseudo-atomic model of the 47 kDa P22 coat protein based on 3D image reconstruction and homology modeling (Parent et al., 2010) revealed that the protein adopts a fold quite similar to the HK97 capsid protein (Wikoff et al., 2000), but unlike HK97 it also contains a telokin-like domain. The 430-residue P22 protein has a large, flexible region between residues 157 and 207 that is encompassed within the A-domain (residues 127 – 222) (Kang & Prevelige, 2005, Lanman et al., 1999). We have proposed that this region may control the conformational switching crucial for procapsid assembly and virion maturation (Parent et al., 2007b). Substitutions at three sites within this region (D163G, T166I, and F170L) were independently isolated as second site suppressors for multiple temperature-sensitive folding (tsf) substitutions, and termed global suppressors (su) of coat protein folding defects (Aramli & Teschke, 1999). In the absence of their tsf parents in vivo, the su substitutions result in the assembly of coat protein into aberrant forms. In vitro, D163G and T166I variants result in the production of spiral products and giant capsids, whereas the F170L variant also forms coat protein tubes (“polyheads”) (Parent et al., 2007b).
All three global su substitutions occur within a region of the coat protein that we call the β–hinge (Figure 1) (Teschke & Parent, 2010). The β– hinge is a four-strand, anti-parallel β-sheet whose strands comprise residues 167 – 170, 218 – 221, 355 – 357, and 421 – 423 in the procapsid (Figure 1) (Parent et al., 2010, Teschke & Parent, 2010). This hinge links the A-domain of the HK97-like core to the telokin-like domain and is believed to be a critical determinant of the conformational switching that occurs in the coat protein during assembly into procapsids, and in facilitating domain movements during maturation transmitted by changes in its conformation (Parent et al., 2007b, Teschke & Parent, 2010). The three global su substitutions highlight the importance of this region for allowing the coat protein monomer to fold correctly and assemble into a functional product. Given that F170L coat protein is the only known variant that leads to polyhead formation, we decided to explore the role of this residue and its location in dictating proper assembly. The leucine substitution for phenylalanine in the F170L variant is a conservative substitution since both are hydrophobic residues. In this study we employed site-directed mutagenesis to investigate how more radical substitutions (alanine and lysine) at this position affect conformational switching during assembly, and also how scaffolding protein regulates proper formation of procapsids.
Figure 1. β–hinge controls conformational switching.
(A) One pseudo-atomic model of the coat protein subunit color-coded by domain (N-arm, red; E-loop, yellow; P-domain, green; A-domain, cyan; telokin-domain, magenta) from the WT procapsid shell (left, PDB accession number 3IYI) and expanded head (right, PDB accession number 3IHI), aligned with respect to their telokin domains using Chimera (Pettersen et al., 2004). (B) Same as (A) but with the β– hinge (Teschke and Parent, 2010) highlighted in orange. (C) Enlarged views of the β –hinge from the procapsid shell and expanded head, in the same orientation as depicted in (B), to show the movement of the hinge upon the maturation reaction. The WT amino acids of the 3 global suppressors are shown as stick models in cyan.
Results
Scaffolding protein decreases F170L polyhead formation
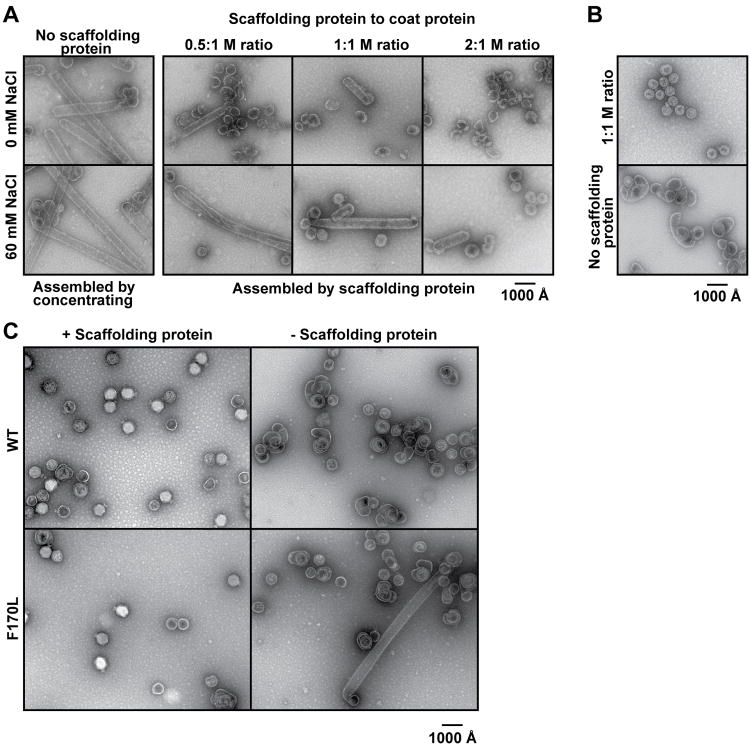
The affinity between coat protein and scaffolding protein can be reduced in vitro by increasing the anion concentration (Parker & Prevelige, 1998, Parent et al., 2005). We performed a series of in vitro assembly reactions to determine what, if any, effects scaffolding protein and NaCl concentrations have on F170L polyhead formation (Figure 2A). Polyheads, ranging in length between ~ 130 and 800 nm, formed when F170L coat protein was concentrated without scaffolding protein, whether or not NaCl was present (Figure 2A). The number and length of F170L polyheads decreased with increasing concentrations of scaffolding protein in the absence of NaCl. However, about twice as much scaffolding protein was required to produce a comparable reduction in polyhead formation when NaCl was present. This shows that scaffolding protein is not as effective in alleviating the assembly defect in the presence of NaCl. Hence, F170L polyhead formation depends on the absence of scaffolding protein or a decreased binding affinity between coat and scaffolding proteins. As a control experiment, in vitro assembly reactions were also performed with WT coat protein. Consistent with earlier studies, WT coat protein, when assembled in the presence of scaffolding protein and 60 mM NaCl, produced particles similar in size and morphology to procapsids that form in vivo (Figure 2B) (Fuller & King, 1980). Aberrant products, but not polyheads, are produced in vitro when WT coat protein is assembled without scaffolding protein. These results highlight both the requirement for scaffolding protein and the role of coat protein position 170 in directing assembly of native, functional procapsids (Prevelige et al., 1990, Teschke et al., 1993).
Figure 2. Scaffolding protein affects F170L polyhead formation.
All panels show representative micrographs of negatively stained samples. (A) In vitro assembly products obtained with F170L coat protein. The reactions were performed with varying concentrations of scaffolding protein in the presence or absence of 60 mM NaCl. (B) In vitro assembly products obtained with WT protein in the presence of 60 mM NaCl, with and without scaffolding protein. (C) Bottom fractions from sucrose gradients of lysates of phage infected cells expressing either WT or F170L coat protein, either with or without scaffolding protein expression, as described in the text.
F170L coat protein does not form polyhead structures in vivo in normal infections where scaffolding protein is present (Parent et al., 2007b), which suggests that the assembly defect caused by F170L only occurs in vitro. We analyzed cell lysates from complementation experiments to determine if polyheads form in vivo in the absence of scaffolding protein. In these complementation experiments, the phage carried amber mutations in genes 5 and 8 (5− am, 8− am) or in gene 5 alone (5− am), so that these proteins were not expressed in infected cells. The cells carried a plasmid that encoded for either WT or mutated gene 5. The soluble proteins from a clarified lysate of such phage-infected cells were applied to sucrose gradients where phage and large, aberrant particles sediment to the bottom upon centrifugation (Parent et al., 2007b). Material from the bottom of each gradient was examined by negative stain electron microscopy (Figure 2C). As expected, 5− am phage complemented with WT coat protein expressed from a plasmid produced phage and a small number of aberrant structures (Parent et al., 2007b). The 5− am 8− am double mutant phage complemented with WT coat protein generated empty shells and various aberrant structures, but not polyheads. Consistent with previous reports, complementation of 5− am phage with F170L coat protein resulted in production of some phage as well as aberrant structures (Parent et al., 2007b). In addition, F170L coat protein made polyheads in vivo when scaffolding protein was absent. These results recapitulate the in vitro assembly reactions shown in Figure 2A, and clearly demonstrate the importance of scaffolding protein for proper assembly. We have recently examined F170L polyheads using cryo-TEM and 3D image reconstruction methods and results show that these polyheads are helical assemblies of hexons (Parent et al., unpublished data). We propose that the F170L variation affects coat protein in one of two ways. Either it modifies the interactions between monomers within capsomers, causing the hexamer conformation to be favored, or it alters inter-capsomer interactions, increasing the probability that monomers added to the growing edge will assemble into a hexamer configuration.
Scaffolding protein does not affect formation of F170A or F170K polyheads
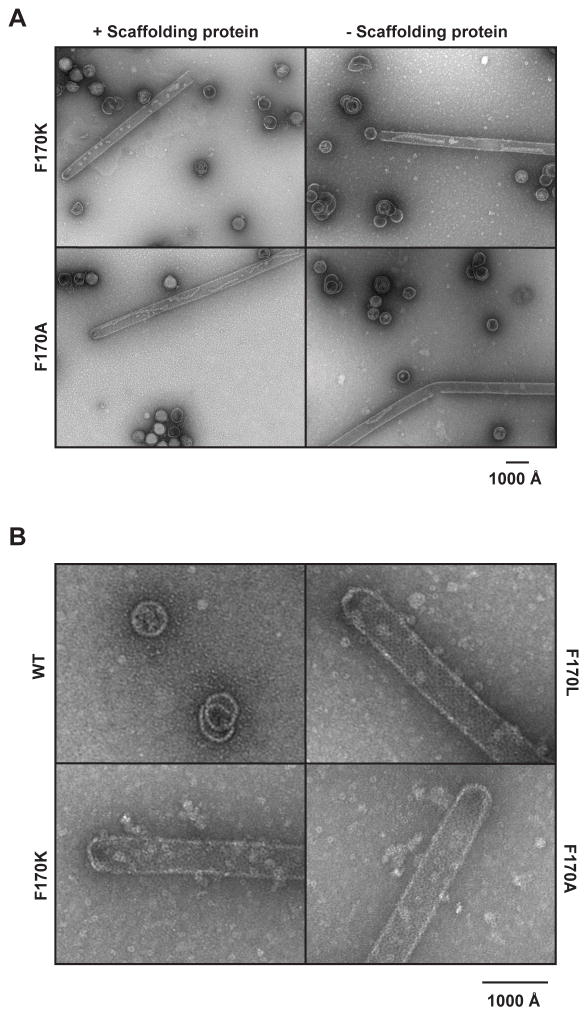
Alanine and lysine were introduced at position 170 to determine the effect of more dramatic amino acid substitutions on coat protein assembly. We investigated the products of infections from complementation experiments with F170A and F170K coat proteins, as described above. Though all three F170 variants made polyheads in the absence of scaffolding protein, F170A and F170K also made polyheads (up to 2 μm long, or more than double that seen with F170L) with scaffolding protein present (Figure 3, Table 1). F170A and F170K also produced mature, infectious phage in the presence of scaffolding protein, indicating these proteins sometimes assemble correctly. The variability in the length and diameter of the polyheads hindered attempts to quantify the amount of coat protein in polyheads versus correct assembly products. Qualitatively, F170A appears to have the most polyheads and they constitute a significant fraction of the coat protein. Preliminary examination of F170A polyheads via cryo-reconstruction methods shows that these are helical assemblies of hexons similar to those observed in F170L polyheads (Parent et al. unpublished data). All attempts thus far to examine the fine structure of F170K polyheads have been thwarted because they are very unstable. Unlike its effect on F170L, scaffolding protein does not prevent F170A and F170K coat proteins from forming polyheads, which suggests these variants may be inhibited in their ability to interact with scaffolding protein.
Figure 3. Effect of phage proteins on polyhead assembly in vivo.
Representative electron micrographs of negatively stained samples of the bottom fractions from sucrose gradients of lysates of (A) phage-infected cells that express coat protein F170K or F170A, either with or without scaffolding protein expression and (B) cells that express WT or the F170 variant coat proteins without phage infection.
Table 1.
Polyhead formation
| Coat type | In vivo | In vitro | ||
|---|---|---|---|---|
| With scaffolding protein | No scaffolding protein | With scaffolding protein | No scaffolding protein | |
| WT | - | - | - | - |
| F170L | - | + | - | + |
| F170A | + | + | x | x |
| F170K | + | + | x | x |
The “+” is conditions where polyheads form
x = do not assemble in vitro
The majority of polyheads formed from each of the coat protein variants had closed ends. We investigated the products of assembly of WT and the variant coat proteins in the absence of all other phage proteins to determine if capping required portal protein. Products were examined by negative stain electron microscopy (Figure 3B). Polyheads comprised of F170 variants were capable of capping with only coat protein present, indicating that the caps are solely coat protein.
Interactions between the F170 variant coat protein monomers and scaffolding protein
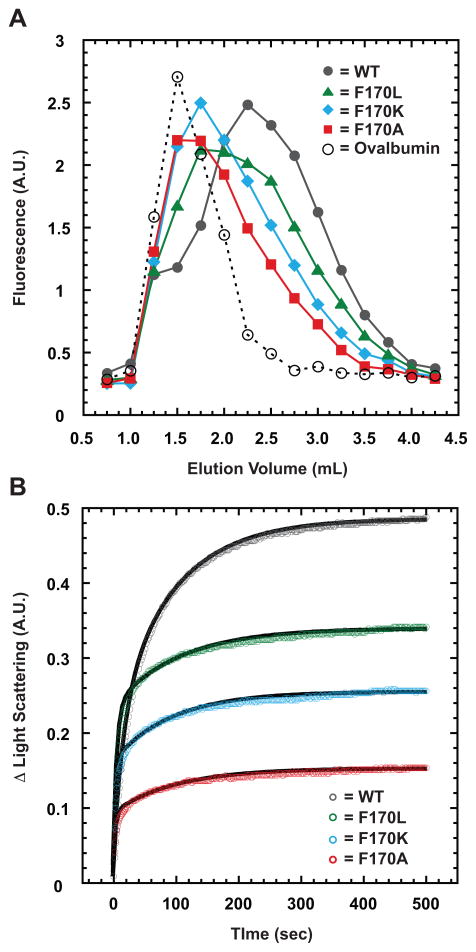
Since F170A and F170K coat proteins produce polyheads in vivo even with scaffolding protein present, we used weak-affinity chromatography to determine if monomers of these coat protein variants displayed any change in affinity for scaffolding protein (Zopf, 1990, Teschke & Fong, 1996). Refolded coat protein monomers (Materials and Methods) were applied to a scaffolding protein-affinity column in which retention of each type of monomer is directly related to the strength of its binding to scaffolding protein. The intrinsic tryptophan fluorescence of the coat protein monomers was used to monitor their elution from the column (Figure 4A), with ovalbumin serving as a negative control (elutes at 1.5 mL). WT monomers produced a broad elution peak centered at 2.25 mL, consistent with their having weak affinity for scaffolding protein. A small fraction (8.8%) of the WT protein eluted at 1.5 mL, indicating the presence of assembly incompetent monomers, which is consistent with previous studies that showed ~ 5% of monomers to be inactive (Parent et al., 2007a). F170L monomers eluted at 2.0 mL, which argues that they have slightly weaker affinity for scaffolding protein compared to WT. F170A and F170K monomers both eluted from the column near 1.75 mL, suggesting that they interact less strongly than either WT or F170L monomers. The elution profiles of all three F170 variants were broad and asymmetric, tailing to larger elution volumes, most likely the result of the weak binding affinity (Scopes, 1994). The ovalbumin control sample exhibited no tailing in its elution profile since it does not bind to scaffolding protein. These data indicate that all three coat protein variants interact with scaffolding protein, but not as strongly as WT coat protein.
Figure 4. F170 coat variants have less affinity for scaffolding protein compared to WT coat protein.
(A) Elution profile of WT and the F170 variant coat proteins from a metal affinity matrix bound with histidine-tagged scaffolding protein and monitored by tryptophan fluorescence. (B) Kinetics of scaffolding protein re-entry and binding to coat protein shells monitored by light scattering in a stopped-flow fluorometer. The experiment was repeated five times for all samples, with representative data shown after subtraction of initial light scattering caused by coat protein shells alone. Data points are plotted at 1 s intervals for the initial 20 s and at 2 s intervals thereafter. The black lines are the fit of the representative data to a first-order reaction with two exponentials.
Interaction between free scaffolding protein and assembled forms of F170 variant coat proteins
Scaffolding protein is known to re-enter empty procapsid shells (procapsids stripped of portal and pilot proteins as well as scaffolding protein), likely through the axial capsomer holes, and bind to the inner wall of the shell (Greene & King, 1994, Prasad et al., 1993, Thuman-Commike et al., 1999, Thuman-Commike et al., 2000). We used this scaffolding protein re-entry assay to examine the extent to which assembled F170 variants are impaired in their ability to interact with scaffolding protein. Empty procapsid shells were produced by taking procapsids assembled in vivo from WT or one of the three variant coat proteins and treating them with 0.5 M GuHCl and were further purified. These empty shells were then mixed with WT scaffolding protein in a stopped-flow fluorometer to determine the amount of scaffolding protein that interacts with properly folded, assembled coat variants. Re-entry of scaffolding protein into empty shells was monitored by light scattering (Figure 4B), which is known to display a direct, linear correlation to the amount of scaffolding protein inside procapsids (Greene & King, 1994, Parker et al., 1998).
The kinetics of scaffolding protein re-entry into each of the empty shells were fit to a first order decay with two exponentials. For WT shells, the fast phase of re-entry had a relaxation time of ~ 20 s, whereas all three of the F170 variants had relaxation times of ~ 5 s (Table 2). The relaxation times for the fast phase in all three variants were faster than WT; however, the differences were less than five-fold and are not likely significant. Based on measurements of the signal amplitude associated with the fast phase, only the F170L shells incorporated as much scaffolding protein as WT shells. The relaxation times for the slow phase in WT and all three variants were similar (ranging from 101.6 to 125.7 s), but the amplitudes associated with this phase significantly differ among the four coat proteins. Differences in amplitude likely reflect different numbers of scaffolding proteins that have re-entered the shells (Table 2). Because the initial light scattering of the empty capsids fell within ~ 7% for WT, F170L, and F170K, the observed changes in the amplitudes for the slow phase were not caused by differences in starting concentrations of the shells used in the experiments. The ~ 17 % higher initial light scattering of F170A compared to WT was attributed to the presence of a small percentage of polyheads in that sample, whereas no polyheads were present in the F170L or F170K samples (F170K polyheads are unstable and therefore none were present in the sample used for this experiment). Our results suggest that less than 30% of scaffolding protein re-entered F170A shells compared to WT.
Table 2.
Kinetics of scaffolding protein re-entry
| Phase 1 | Phase 2 | Relative scaffolding protein incorporation | |||
|---|---|---|---|---|---|
| Coat Protein | Relaxation Time (sec) | Amplitude (A.U.) | Relaxation Time (sec) | Amplitude (A.U.) | |
| WT | 20.2 +/− 1.9 | 0.212 +/− 0.017 | 101.6 +/− 8.7 | 0.248 +/− 0.014 | 1.0 |
| F170L | 7.0 +/− 0.3 | 0.204 +/− 0.007 | 119.1 +/− 10.5 | 0.099 +/− 0.003 | 0.66 |
| F170K | 4.5 +/− 0.5 | 0.150 +/− 0.005 | 107.7 +/− 8.0 | 0.081 +/− 0.005 | 0.50 |
| F170A | 5.4 +/− 1.6 | 0.084 +/− 0.004 | 125.7 +/− 27.1 | 0.053 +/− 0.005 | 0.30 |
Values are means +/− standard deviations in the measurements.
A.U. = arbitrary units
The kinetics of re-entry in WT and variant shells were also monitored for longer time periods in a steady state fluorometer. Relaxation times of ~ 20 and ~ 100 s were obtained for WT protein, which correlated well with the results from the stopped-flow experiment. All three F170 variants exhibited an additional, very slow phase (~ 2,000 – 3,000 s; data not shown). The amplitude associated with this additional phase in each variant comprised less than 20% of the total amplitude, so this phase alone could not account for the differences in the overall amplitudes of the different variants. In summary, the re-entry experiments indicate that, even when assembled as shells, the F170 variants bind less scaffolding protein inside the capsid as compared to WT.
Difference maps reveal that capsomers have altered conformations in the F170A and F170K variants
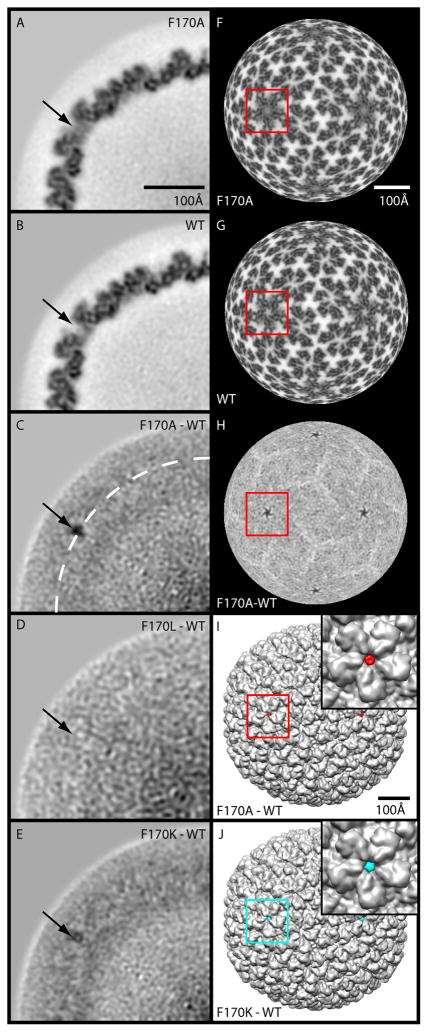
We used difference map analysis (Materials and Methods) to compare cryo-reconstructions of purified shells derived from all three coat protein variants to that of WT shells to determine if there are conformational changes that could account for the increased propensity of the F170 variants to produce polyheads (Table 3, Figure 5A–J) (Parent et al., 2010). For each difference map, an appropriately scaled (pixel size and relative density values) WT reconstruction was subtracted from one of the F170 variant maps. One-pixel thick (~ 1.62 Å), central (equatorial) sections through each of the difference maps reveal that, in F170A and F170K, the most significant changes occur near the 5-fold symmetry axes (Figure 5C, E). The F170L and WT maps show no significant differences indicating the two structures are essentially identical (Figure 5D). The F170A map differed most from WT, and F170K was intermediate between these extremes. A single residue change produces a mass difference likely too small to detect in cryo-reconstructions at moderate resolutions (i.e. > 5 Å). Hence, we attribute the differences seen in F170A and F170K compared to WT to effects that the substituted amino acids have on coat protein conformation and flexibility. Positive density features in the F170A and F170K minus WT difference maps reflect decreased coat protein flexibility in the variants since corresponding regions of the WT coat protein that are flexible would appear weak or absent in the WT reconstruction as a consequence of the averaging procedures used. Several minor, yet detectable changes were observed in other regions of the capsid. These appear in the difference maps as apposed pairs of positive and negative densities, which occur as a result of small, local movements of secondary structural elements, domains, or whole subunits (Supplementary movie). Overall, two primary types of structural change were observed in the variants: 1) a gain in density at the center of the pentons, which we propose to be due to a decrease in flexibility of part of the polypeptide chain, most likely in the highly flexible stretch of residues from 157 to 207 (Lanman et al., 1999, Kang et al., 2006), and 2) subtle, rigid-body domain movements in some, but not all of the individual hexon subunits, likely caused by substitutions in the hinge. The domain movements, most of which are 6 Å or less, are primarily confined between radii ~237 and 282 Å, and changes in the β-hinge affect the angle between the A- and telokin (Supplementary movie).
Table 3.
P22 image reconstruction statistics
| CP type | Micrographs | Particle Images† | Defocus Range(μm) †† | Resolution (Å) *† |
|---|---|---|---|---|
| WT* | 164 | 11,997 | 0.41 – 3.65 | 9.1 |
| F170L | 33 | 1,757 | 0.73 – 2.79 | 13.5 |
| F170K | 38 | 4,123 | 0.77 – 2.96 | 10.4 |
| F170A | 72 | 4,008 | 0.83 – 3.26 | 10.3 |
The data for the WT reconstruction is from a previous study (Parent et al., 2010)
Number of boxed particle images used to compute each 3D reconstruction
Range of objective lens underfocus settings used to record micrographs
Estimation of resolution based on FSC0.5 criterion (van Heel & Schatz, 2005)
Figure 5. Difference maps reveal conformational changes at 5-fold axes for F170A and F170K coat proteins.
(A) Projected density distribution of one quadrant in a planar, one-pixel thick (1.6 Å) equatorial section from the cryo-reconstruction of F170A shells. (B) Same as (A) for the WT shells. (C–E) Same as (A) for the F170A-WT, F170L-WT, and F170K-WT difference maps, respectively. High and low density features are rendered in black and white, respectively (A–H). Arrows point to the same position in each section and highlight the largest difference densities seen in (C) and (E). In (C) the white dashed circle corresponds to a radius of 271 Å. (F, G, H) Radial density projections (r=271 Å) for the maps corresponding to (A), (B), and (C). (I–J) Surface-shaded views of WT shell reconstruction (grey) with F170A-WT (red) and F170K-WT (cyan) difference densities superimposed. Insets show magnified views of the regions identified by the red or cyan boxes shown in colors consistent with Figure 4. The boxes highlight the same region in F–J. Magnification bar in (A) same in (B–E). Bar in (F) same for (G–H), and bar for (I) same as for (J).
The F170 variants show decreased flexibility of residues 157–207
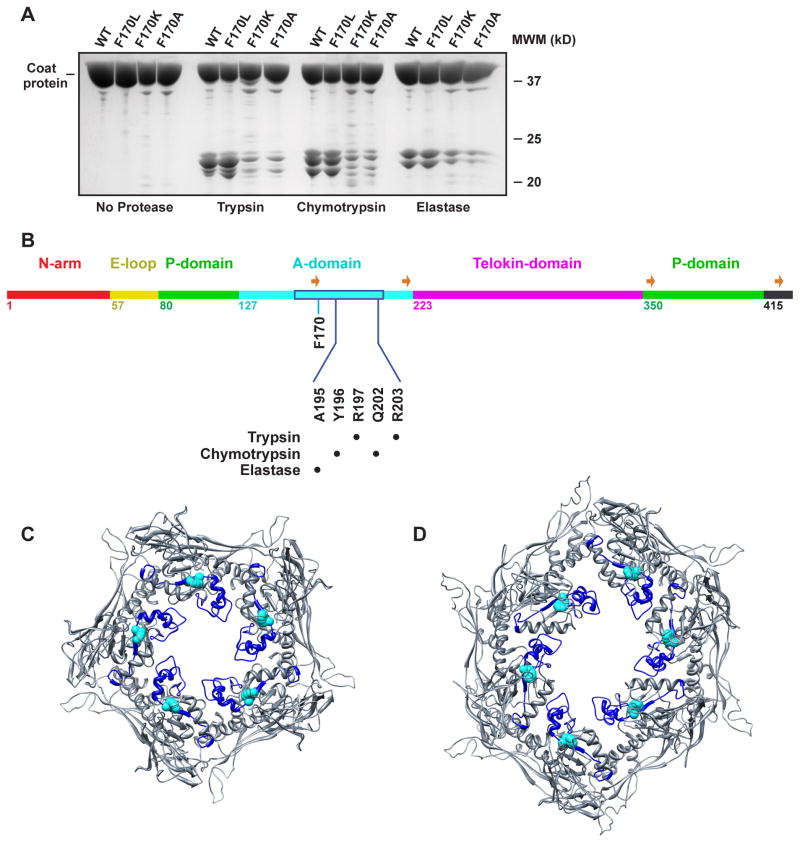
The large flexible region (residues 157–207) within the A-domain of coat protein is protease accessible in assembled procapsids (Lanman et al., 1999, Kang et al., 2006). We used limited proteolysis to probe for changes in the flexibility of coat protein suggested by the 3D reconstructions. WT and the F170 variant empty procapsid shells were digested using trypsin, chymotyrpsin and elastase (Materials and Methods). The cleavage patterns of WT and the F170 variants were similar for each of the proteases, with the major peptides having masses of 20–25 kDa. However, there were dramatic differences in the amount of these peptides (Figure 6A). F170K and F170A had significantly less cleavage products as compared to WT and F170L. Though the amounts of WT and F170L cleavage products were similar at 4 hr, a time course of proteolytic digestion showed that F170L is slightly less protease accessible compared to WT (data not shown). N-terminal sequencing was used to identify the cleavage sites for WT coat protein, which all mapped to the previously identified flexible loop (Figure 6B). Trypsin cleaves preferentially after R197 and R203. Chymotrypsin cleaves after Y196, and Q202. Elastase cleaves after A195, consistent with previous results (Lanman et al., 1999). Protection of the cleavage sites in F170K and F170A procapsid shells suggests that these substitutions cause an increase in rigidity of the loop region so that it is less accessible to these proteases. Quantification of the major peptides on SDS gels indicated F170K and F170A have ~ 20% less cleavage products compared to WT and F170L, consistent with observations seen in the density maps (Figure 5) and as shown in the supplementary movie. This may be due to preferential changes in conformation of pentameric subunits, but we cannot rule out that all coat protein subunits lose flexibility.
Figure 6. Limited proteolysis indicates that the A-domain in F170 variant shells have decreased flexibility.
(A) Tricine-SDS-PAGE of WT and F170 variant procapsid shells digested with trypsin, chymotrypsin, and elastase. (B) Primary sequence of the 430 amino acid P22 coat protein with the backbone color-coded by domain, listing the first amino acid in each domain. The location of the protease digestion sites are listed. The flexible region (Lanman et al., 1999, Kang et al., 2006) within the A-domain is boxed in blue. The four strands comprising the β–hinge are identified with orange arrows. (C,D) Pseudo-atomic models of WT procapsid shell penton and hexon, respectively, with the protease sensitive region shown in blue and a van der Waals representation of the side chain of F170 highlighted in cyan.
Discussion
Scaffolding protein affects assembly of phage coat proteins
Coat protein variants at position F170 result in assembly defects that lead to polyhead formation. To date, no other mutations or deletions in the bacteriophage P22 genome result in polyhead production. As demonstrated here, scaffolding protein can interact with F170L coat protein in a manner that permits formation of native procapsids. However, it interacts more weakly with F170A and F170K and cannot prevent them from assembling into aberrant structures. Scaffolding protein is proposed to bind to trimeric sites on the interior of the capsid at the strict and quasi three-fold axes (Thuman-Commike et al., 1999, Thuman-Commike et al., 2000). Close inspection of the four 3D reconstructions reveal no obvious differences at the trimer clusters (data not shown), yet scaffolding protein binding is clearly altered. Substitutions at the F170 position may cause small conformational changes in the protein that alter the binding site for scaffolding protein on the inside of the capsid shell. Given that scaffolding protein binding to coat protein likely involves interactions between charged residues (Parent et al., 2005, Parker & Prevelige, 1998, Sun et al., 2000), even a rather subtle change in conformation could affect the electrostatic potential of the inner capsid surface and go undetected in moderate resolution cryo-reconstructions.
Scaffolding proteins clearly have important roles in form determination for P22 and other dsDNA phage and herpesvirus (Dokland, 1999). The composition of assembly nuclei for P22 is not known for certain, but has been suggested to include five coat proteins and two or three scaffolding proteins (Prevelige et al., 1993). If scaffolding protein does dictate formation of the nuclei that lead to assembly of native procapsids and mature virions, the variant coat proteins may act in part by inducing the formation of defective nuclei. We propose that, since the variants F170A and F170K decrease the scaffolding protein binding affinity, the number of properly formed nuclei is reduced. F170L nucleates properly in the presence of scaffolding protein, likely because it has a slightly higher affinity for scaffolding protein compared to F170A and F170K, and off-pathway assemblies such as polyheads do not form under these conditions.
Coat protein conformation determines polyhead formation
F170 coat protein variants exhibit altered scaffolding protein affinity, but this property alone is insufficient to explain why these proteins assemble into polyheads. P22 polyhead formation has thus far only been observed to occur as a result of variations in the coat protein, and only at position 170. WT coat protein has never been observed to form polyheads, regardless of the presence or absence of scaffolding protein. Without scaffolding protein, WT coat protein will form empty, aberrant assemblies, consisting of both hexons and pentons, such as T=4 and T=7 icosahedral capsids or spiral structures (Thuman-Commike et al., 1998, Earnshaw & King, 1978, Lenk et al., 1975). Coat protein variants with a cs phenotype (T10I and R101C) interact weakly with scaffolding protein (Teschke & Fong, 1996) but do not form polyheads (Gordon, 1993). Thus, we propose that scaffolding protein plays an important role in regulating polyhead formation in the F170 variants, but it is not the sole determinant since the conformation of the β–hinge region is critical.
Additional density at the center of the pentons in F170A, and to a lesser extent in F170K, indicates that these substitutions reduce the inherent flexibility of a region of the protein. Careful inspection of the difference maps suggests that a region within the A-domain loses some flexibility. Indeed, a decrease in flexibility of the A-domain, specifically within the previously identified ‘flexible loop’ (amino acids 157–207) (Lanman et al., 1999, Kang et al., 2006), was confirmed by limited proteolysis and protein sequencing. It is notable that density occurs at the centers of pentons in the F170A and F170K procapsid shell reconstructions. The axial hole in the WT hexon is larger than in the penton, so a decrease in flexibility would not cause crowding or hinder assembly of hexamers subunits (Figure 6C,D). The hexons retain their axial holes and presumably allow scaffolding protein to enter and exit shells during re-entry experiments. A-domain flexibility may be needed to allow monomers to assemble into pentons and not just hexons as occurs in polyheads (Parent et al., unpublished data). Though no prominent differences were observed in the subunit conformations within hexons in WT and F170 variants, subtle changes were seen throughout the procapsid shell indicative of altered quaternary interactions (supplementary movie).
Polyhead formation in other phage
Changes in assembly conditions or mutations of various genes are known to promote polyhead formation in other tailed bacteriophage. For example, the major capsid proteins of bacteriophages T4 (Steven et al., 1976b), T7 (Steven et al., 1983), SPO1 (Parker et al., 1983), and λ (Georgopoulos et al., 1973) all assemble into polyheads composed of hexamers. Polyheads often result when scaffolding protein is altered or nucleation goes awry (Moody, 1999). In T4 phage, mutants defective in several proteins including gp20 (portal protein), gp22 (main scaffold protein), IPIII (internal protein of the core), and gp23 (the major capsid protein) all assemble as hexons into polyheads (Steven et al., 1976a). In vivo, T4 phage assembles via a scaffolding core composed of its main scaffold protein and five other proteins, including IPIII, which plays a major role in determining the shape of the capsid shell (Laemmli et al., 1970). In vitro, monomeric gp23 can be reversibly assembled into hexons and polyheads in the absence of gp20 or any of the core proteins (van Driel, 1977). This indicates that gp23 alone fully encodes hexon and polyhead formation (van Driel, 1977), but portal and scaffolding proteins may provide constraints needed to incorporate gp24 pentamers and therefore regulate assembly of closed capsids of the proper size and quasi-symmetry (Kellenberger, 1968).
The capsid proteins of T4 and P22 bacteriophages have common, HK97-like folds (Fokine et al., 2005, Jiang et al., 2003, Parent et al., 2010), but the assembly of these phage exhibit distinct differences. In T4 hexons and pentons are comprised of different proteins (gp23 and gp24, respectively), but in P22, both types of capsomer are made of gp5 (Casjens & King, 1974). Procapsids and polyheads of T4 phage are assembled from preformed capsomers (Stortelder et al., 2006), whereas procapsids of P22 are assembled by addition of coat protein monomers (Prevelige et al., 1993). Dynamic light scattering and sucrose gradient sedimentation data show that the F170 variants, at concentrations up to ~ 1 mg/mL, exist as monomers in solution and no hexons or pentons are detected (data not shown). These data suggest that the amino acid substitutions at F170 do not change the assembly mechanism to one in which hexons instead of monomers are added. In addition, T4 polyheads are open-ended structures (Laemmli et al., 1970), whereas P22 polyheads can cap in vitro and in vivo when only coat protein is present (Figure 2A, 3B). Given that P22 polyheads cap in the absence of portal protein, caps must contain pentons as well as hexons.
Bacteriophage T7 is a second example where scaffolding protein affects polyhead formation. Purified T7 head protein assembles into polyheads in vitro in the presence of polyethylene glycol or dextran and scaffolding protein (Cerritelli & Studier, 1996). Scaffolding protein is necessary to initiate assembly of T7 coat protein in vitro, yet decreasing the scaffolding:coat protein ratio increases the formation of polyheads. Initiation of procapsid-like particles was proposed to be more efficient at higher scaffolding protein ratios, thereby decreasing off-pathway polyhead production.
The β–hinge in P22 is crucial for proper folding and assembly of coat protein
The F170 residue lies in the coat protein β– hinge and single-site substitutions at this residue cause assembly defects. Previously, we proposed that the β-hinge functions in two ways (Teschke & Parent, 2010). First, it may serve as a ‘lock’ to stabilize the properly folded monomer since all domains of the protein, except the telokin domain, interconnect through the β– hinge (see Figure 4B, (Teschke & Parent, 2010). Second, the hinge may allow the A- and P-domains to reorient relative to each other during maturation (Teschke & Parent, 2010). In this study, we show that substitutions that disrupt the lock function of the β-hinge create monomers that are less able to assemble correctly. Impaired assembly of modified coat protein into the native capsid structure presumably occurs because conformational changes in the β– hinge are transmitted to distant portions of the polypeptide in ways that disrupt proper subunit-subunit interactions. One way may induce steric constraints that prevent formation of pentons, and another may decrease the binding affinity for scaffolding protein. Current cryo-reconstruction studies of several different P22 polyheads are aimed at further characterizing how scaffolding protein promotes proper nucleation and procapsid assembly.
Experimental Procedures
Bacteria and phage
Salmonella enterica serovar Typhimurium strain DB7136 (leuA414am, hisC525am, su0) has been previously described (Winston et al., 1979). Bacteriophage P22 strains used in these studies contained an amber mutation in gene 5 (5− am N114) or amber mutations in genes 5 (5− am N114) and 8 (8− am N123). Both strains carried the c1-7 allele, which prevents lysogeny, and an amber mutation in gene 13 (13− am H101) to block cell lysis. The bacteria were transformed with a plasmid (pHBW1) that encodes for WT coat protein or a coat protein variant, which confers ampR (Parent et al., 2007b). Phage carrying amber mutations in gene 5 were complemented by expression of coat protein from the various plasmids described below.
Generation of coat variants
A QuikChange site-directed mutagenesis kit (Stratagene) was used to alter pHBW1 to generate mutations in the coat gene that code for residue 170. The codon changes used for the QuikChange reactions are as follows; the WT TTC codon was changed to AAG for F170K and to GCC for F170A. The mutated plasmids were sequenced at the University of Connecticut Biotechnology Center. Generation of the F170L expression plasmid was described previously (Parent et al., 2007b).
Assembly reactions
Empty procapsid shells containing the coat protein substitutions were prepared as previously described (Parent et al., 2007b, Anderson & Teschke, 2003). Unfolded coat protein monomers were obtained by urea denaturation and were refolded by extensive dialysis against 20 mM phosphate buffer, pH 7.6 at 4 °C. After dialysis, the samples were centrifuged at 175,000 × g at 4 °C for 20 min in a Sorvall RP80AT3 rotor to remove aggregates and procapsids. For procapsid assembly experiments, coat protein monomers at a final concentration of 14.3 μM were mixed with scaffolding protein at concentrations corresponding to molar ratios of 0.5, 1, and 2 scaffolding:coat (7.0, 14.3 and 28.6 μM). Assembly reactions were performed in 20 mM sodium phosphate (pH 7.6) with and without 60 mM NaCl, at 20 °C for 150 min. Coat protein monomers were also assembled in the absence of scaffolding protein by centrifugation in a 10K MWCO Microcon (Millipore) at 14,000 × g, 4 °C for 15 minutes until the coat protein concentration exceeded 42.8 μM.
Analysis of in vivo phage production
Overnight cultures of DB7136 transformed with the plasmids encoding for WT or each of the variant coat proteins were grown at 30 °C in LB supplemented with 100 μg/mL ampicillin and were used to inoculate a culture into fresh media. The cells were grown at 30 °C to a density of 2 × 108 cells/mL, and induced with 1 mM IPTG. The cultures were divided and infected with phage carrying an amber mutation in gene 5 or phage with amber mutations in both genes 5 and 8. An additional culture was induced, but not infected with phage. The cultures were incubated at 30 °C for 4 hours.
Cells were concentrated by centrifugation at 13.2 K rpm, 4 °C for 5 minutes in a microfuge. Pelleted cells were resuspended in lysis buffer (0.1% Triton, 100 μg/mL lysozyme, 50 mM Tris HCl, 25 mM NaCl, 7 mM EDTA, pH 7.6), and frozen at −20 °C. Cells were thawed on ice and treated with 20 mM MgSO4, 100 μg/mL RNase and 100 μg/mL DNase. Samples were frozen and thawed two additional times to ensure complete lysis. Cell debris and insoluble aggregates were removed by centrifugation at 13.2 K rpm, 4 °C for 5 minutes. Assembled phage proteins were concentrated by centrifugation for 20 minutes at 175,000 × g in a Sorvall RP80AT3 rotor. The pellets were resuspended in a small volume of 20 mM sodium phosphate buffer, pH 7.6.
A 100 μL aliquot of the resuspended pellet was applied to a 2.2 mL, linear 5 –20% (w/w) sucrose gradient, prepared using a Gradient Master Model 106 (Biocomp Instruments). The gradients were centrifuged in a Sorvall RC M120EX centrifuge with a RP55S rotor for 35 minutes at 35,000 rpm at 20 °C and then fractionated from the top into 100 μL aliquots. Samples from each gradient fraction were analyzed by both SDS-PAGE and negative stain electron microscopy.
Negative stain electron microscopy
Aliquots (3 μL) of the in vitro assembly reactions and the lysates generated from phage-infected cells were applied to carbon-coated, 300-mesh copper grids, allowed to absorb for 1 minute, and the grids were then washed with 2–3 drops of water followed by staining with 1% aqueous uranyl acetate for 30 seconds. Excess stain was blotted off with filter paper and the grids were air-dried and viewed in a Phillips Model 300 TEM (nominal magnification of 71,300) or an FEI Technai Biotwin TEM (nominal magnification of 68,000), both operated at 80 kV.
Scaffolding protein column
Scaffolding protein with an N-terminal hexa-histidine tag at 0.17 μmol was applied to a 1 mL TALON superflow, metal-affinity column (Clontech) in 20 mM sodium phosphate buffer (pH 7.6) with 50 mM NaCl. The histidine tag was placed at the N-terminus because the C-terminus is required for coat protein binding (Parker et al., 1998). WT and the F170 variant coat proteins (100 μL each) were applied to the column and 250 μL fractions were collected. The concentration of the coat protein monomers (4.3 μM) applied to the column was kept low to avoid self-assembly as well as scaffold-mediated assembly. Ovalbumin was applied to the column at 4.5 μM. The elution profile of each protein was determined by fluorescence of each fraction using an SLM Aminco-Bowman 2 spectrofluorometer with the excitation wavelength at 280 nm, the emission wavelength set to 340 nm, and the bandpasses set to 1 and 8 nm.
Scaffolding protein re-entry into empty coat shells
Scaffolding protein was extracted from procapsids generated in vivo for each of the three F170 variants and treated with 0.5 M GuHCl (Greene & King, 1994). The kinetics of scaffolding protein reentry into the empty coat protein shells were measured by light scattering at 350 nm using an Applied Photophysics SX.18MV stopped-flow fluorometer and an SLM Aminco-Bowman 2 spectrofluorometer, both maintained at 20 °C. The excitation wavelength was set to 350 nm and a bandpass of 4 nm in the stopped-flow fluorometer. Light scattering was measured with the PMT at a 90° angle to the incident light, with no filter. In the steady state spectrofluorometer, the excitation and emission wavelengths were set to 350 nm with the bandpasses for each at 4 nm. Empty shells and scaffolding protein were mixed at a 1:1 molar ratio of monomers (final coat and scaffolding protein concentrations of 3.0 μM). Data were recorded at 0.5 s intervals for 500 s in the stopped-flow fluorometer and at 1 s intervals for one (WT) or two (F170 variants) hours in the spectrofluorometer. The light scattering of the coat protein shells alone was subtracted from the data. Relaxation times were determined with Kaleidagraph software (Abelbeck) using a first order rate equation with two exponentials:
where LSf is the final light scattering intensity, τ is 1/rate constant, t is time, and LS(t) is the change in light scattering intensity with time.
Cryo-TEM
Small (3.5 μL) aliquots of purified P22 particles (~8 mg/mL) were vitrified and examined as described previously (Baker et al., 1999). Briefly, samples were applied to Quantifoil holey grids that had been glow-discharged for ~15 s in an Emitech K350 evaporation unit. Grids were then blotted with Whatman filter paper for ~5 s, plunged into liquid ethane, and transferred into a precooled, FEI Polara, multi-specimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron-image film at 200 keV in an FEI Polara microscope under minimal-dose conditions (~15 e/Å2) at a nominal magnification of 39,000. The range of objective lens defocus settings, number of particles, and number of micrographs used to record each data set are listed in Table 2.
3D Image Reconstructions of Procapsid Shells
Micrographs exhibiting minimal astigmatism and specimen drift were selected for further processing and digitized at 6.35 μm intervals (representing 1.62 Å pixels at the specimen) on a Nikon Supercoolscan 8000 microdensitometer. The RobEM visualization and analysis software (http://cryoEM.ucsd.edu/programs.shtm) was used to estimate micrograph defocus and astigmatism, extract individual particle images, and preprocess the images. For each specimen, 150 particle images were used as input to the random-model computation procedure to generate an initial 3D density map at ~25 Å resolution (Yan et al., 2007a). Each map was then used to initiate determination and refinement of particle orientations and origins for the complete set of images using AUTO3DEM version 3.14 (Yan et al., 2007b). Phases and amplitudes of the particle structure factor data were corrected to compensate for the effects caused by the microscope contrast-transfer function (Bowman et al., 2002). The Fourier Shell Correlation (FSC0.5) criterion was used to estimate the resolutions limits of each 3D reconstruction (Table 3) (van Heel & Schatz, 2005).
Difference map calculation, visualization, and interpretation
Difference maps were computed to help identify changes that accompany amino acid substitutions at position 170. RobEM was used to subtract an appropriately scaled WT procapsid shell map (Parent et al., 2010) (density range and magnification) from each F170 map as previously described (Baker et al., 1999). Since the maps of the three F170 variants lead to cryo-reconstructions at different resolutions (Table 3), the WT map was computed to a resolution limit that corresponded to the map from which it was to be subtracted. The resultant difference density maps were visualized and analyzed using RobEM and Chimera (Pettersen et al., 2004). Flexible regions of a structure are inherently more difficult to detect than those that are stable, even in cryo-reconstructions at moderate resolution (Zhou, 2008). Hence, we interpreted any isolated, positive density features in the F170X-WT difference maps as identifying regions of the coat protein that are less flexible in the variant compared to WT. In several places, the difference maps showed weak yet clear pairs of closely juxtaposed positive and negative densities, which we interpreted as arising from small, rigid-body movements of subunits, domains, or sub-domains.
Protease Digestion
WT and F170 variant procapsid shells were digested using trypsin, chymotrypsin and elastase at 20 °C for 4 hr with an enzyme:substrate ratio of 1:20. Samples were quenched with reducing sample buffer, heated for 5 min at 95 °C, and analyzed by 16% Tricine-SDS-PAGE (Schagger & von Jagow, 1987). The peptides were transferred to a polyvinylidene difluoride (PVDF) blotting membrane in 20 mM Tris buffer, pH 8.3. Bands were N-terminally sequenced by automated Edman degradation at Tufts core facility.
Supplementary Material
Acknowledgments
We thank Dr. Marie Cantino for use of the University of Connecticut Electron Microscopy Center and her expert guidance in electron microscopy; members of the Teschke lab for helpful discussions; Mr. Norm H. Olson for guidance in cryo-TEM; Dr. Robert S. Sinkovits, Ms. Melissa L. Sinkovits, and Mr. Nicholos Brunn for preliminary analysis of micrographs; and the Tufts core facility for protein sequencing. This work was supported in part by NIH grants to CMT (R01 GM76661) and TSB (R37 GM-033050 and 1S10 RR-020016), NIH fellowship F32A1078624 to KNP, and support from UCSD and the Agouron Foundation to TSB to establish and equip cryoTEM facilities at UCSD.
Abbreviations
- gp
gene product
- WT
wild-type
- 3D
three-dimensional
- dsDNA
double-stranded DNA
- GuHCl
guanidine hydrochloride
- bisANS
1,1′-Bi(4-anilinonaphthalene-5-sulfonic acid)
- su
suppressor
- tsf
temperature-sensitive-folding
References
- Anderson E, Teschke CM. Folding of Phage P22 Coat Protein Monomers: Kinetic and Thermodynamic Properties. Virology. 2003;313:184–197. doi: 10.1016/s0042-6822(03)00240-x. [DOI] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem. 1999;274:22217–22224. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. [erratum appears in Microbiol Mol Biol Rev 2000 Mar;64(1):237] Microbiology & Molecular Biology Reviews. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J Mol Biol. 1988;202:77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An Antibody to the Putative Aphid Recognition Site on Cucumber Mosaic Virus Recognizes Pentons but Not Hexons. Journal of Virology. 2002;76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996;258:286–298. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- Dokland T. Scaffolding proteins and their role in viral assembly. Cellular & Molecular Life Sciences. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison SC. Assembly of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J Mol Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, King J. Regulation of coat protein polymerization by the scaffolding protein of bacteriophage P22. Biophys J. 1980;32:381–401. doi: 10.1016/S0006-3495(80)84963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos CP, Hendrix RW, Casjens S, Kaiser AD. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973;76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Gordon CL. Biology. Cambridge, MA: Massachusettes Institute of Technology; 1993. Analysis of Conditional-Lethal Mutations of the Phage P22 Coat Protein. [Google Scholar]
- Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Kang S, Hawkridge AM, Johnson KL, Muddiman DC, Prevelige PJ. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass-spectrometry. J Proteome Res. 2006;5:370–377. doi: 10.1021/pr050356f. [DOI] [PubMed] [Google Scholar]
- Kang S, Prevelige PJ. Domain Study of Bacteriophage P22 Coat Protein and Characterization of the Capsid Lattice Transformation by Hydrogen/Deuterium Exchange. J Mol Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. Studies on the morphopoiesis of the head of phage T-even: V. The components of the T4 capsid and of other, capsid-related structures. Virology. 1968;34:549–561. doi: 10.1016/0042-6822(68)90074-3. [DOI] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc London B. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22 II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK, Molbert E, Showe MK, Kellenberger E. Form-determining Function of the Genes Required for the Assembly of the Head of Bacteriophage T4. J Mol Biol. 1970;49:99–113. doi: 10.1016/0022-2836(70)90379-7. [DOI] [PubMed] [Google Scholar]
- Lanman J, Tuma R, Prevelige PE., Jr Identification and characterization of the domain structure of bacteriophage P22 coat protein. Biochemistry. 1999;38:14614–14623. doi: 10.1021/bi9915420. [DOI] [PubMed] [Google Scholar]
- Lenk E, Casjens S, Weeks J, King J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology. 1975;68:182–199. doi: 10.1016/0042-6822(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Moody MF. Geometry of Phage Head Construction. J Mol Biol. 1999;293:401–433. doi: 10.1006/jmbi.1999.3011. [DOI] [PubMed] [Google Scholar]
- Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Phage P22 procapsids equilibrate with free coat protein subunits. J Mol Biol. 2007a;365:513–522. doi: 10.1016/j.jmb.2006.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Polyhead formation in phage P22 pinpoints a region in coat protein required for conformational switching. Mol Microbiol. 2007b;65:1300–1310. doi: 10.1111/j.1365-2958.2007.05868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Zlotnick A, Teschke CM. Quantitative analysis of multi-component spherical virus assembly: scaffolding protein contributes to the global stability of phage P22 procapsids. J Mol Biol. 2006;359:1097–1106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- Parker MH, Brouillette CG, Prevelige PJ. Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry. 2001;40:8962–8970. doi: 10.1021/bi0026167. [DOI] [PubMed] [Google Scholar]
- Parker MH, Casjens S, Prevelige PE., Jr Functional Domains of Bacteriophage P22 Scaffolding Protein. J Mol Biol. 1998;281:69–71. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- Parker MH, Prevelige PE., Jr Electrostatic interactions drive scaffolding/coat protein binding and procapsid maturation in bacteriophage P22. Virology. 1998;250:337–349. doi: 10.1006/viro.1998.9386. [DOI] [PubMed] [Google Scholar]
- Parker ML, Ralston EJ, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. II. Head structure and DNA size. J Virol. 1983;46:250–259. doi: 10.1128/jvi.46.1.250-259.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Jr, Marieta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J, Towse SA, Thomas GJ. Conformational states of the bacteriophage P22 capsid subunit in relation to self-assembly. Biochemistry. 1990;29:5626–5633. doi: 10.1021/bi00475a030. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis for the Separation of Proteins in the Range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Scopes RK. Protein Purification: Principles and Practice. Springer Sciences and Business Media LLC; New York, NY: 1994. [Google Scholar]
- Steven AC, Aebi U, Showe MK. Folding and capsomere morphology of the P23 surface shell of bacteriophage T4 polyheads from mutants in five different head genes. J Mol Biol. 1976a;102:373–400. [PubMed] [Google Scholar]
- Steven AC, Couture E, Aebi U, Showe MK. Structure of T4 polyheads. II. A pathway of polyhead transformation as a model for T4 capsid maturation. J Mol Biol. 1976b;106:187–221. doi: 10.1016/0022-2836(76)90307-7. [DOI] [PubMed] [Google Scholar]
- Steven AC, Serwer P, Bisher ME, Trus BL. Molecular architecture of bacteriophage T7 capsid. Virology. 1983;124:109–120. doi: 10.1016/0042-6822(83)90294-5. [DOI] [PubMed] [Google Scholar]
- Stortelder A, Hendriks J, Joost BB, Bulthuis J, Gooijer C, van der Vies SM, van der Zwan G. Hexamerization of the Bacteriophage T4 Capsid Protein gp23 and Its W13V Mutant Studied by Time-Resolved Tryptophan Fluorescence. J Phys Chem B. 2006;110:25050–25058. doi: 10.1021/jp064881t. [DOI] [PubMed] [Google Scholar]
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PEJ, Krishna NR. Structure of the Coat Protein-binding Domain of the Scaffolding Protein from a Double-stranded DNA Virus. J Mol Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- Teschke CM, Fong DG. Interactions between coat and scaffolding proteins of phage P22 are altered in vitro by amino acid substitutions in coat protein that cause a cold-sensitive phenotype. Biochemistry. 1996;35:14831–14840. doi: 10.1021/bi960860l. [DOI] [PubMed] [Google Scholar]
- Teschke CM, King J, Prevelige PE., Jr Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene-5-sulfonic acid) Biochemistry. 1993;32:10658–10665. doi: 10.1021/bi00091a016. [DOI] [PubMed] [Google Scholar]
- Teschke CM, Parent KN. Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010 doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jakana J, McGough A, Prevelige PE, Chiu W. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J Virol. 2000;74:3871–3873. doi: 10.1128/jvi.74.8.3871-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PEJ. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel R. Assembly of bacteriophage T4 head-related structures. Assembly of polyheads in vitro. J Mol Biol. 1977;114:61–72. doi: 10.1016/0022-2836(77)90283-2. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Winston R, Botstein D, Miller JH. Characterization of amber and ochre suppressors in Salmonella typhimurium. Journal of Bacteriology. 1979;137:433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D, Ohlson S. Weak-affinity chromatography. Nature. 1990;346:87–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.