Abstract
RNA polymerase (Pol) III transcribes many noncoding RNAs (e.g. tRNAs) important for translational capacity and other functions. Here, we localized Pol III, alternative TFIIIB complexes (BRF1/2) and TFIIIC in HeLa cells, determining the Pol III transcriptome, defining gene classes, and revealing ‘TFIIIC-only’ sites. Pol III localization in other transformed and primary cell lines revealed novel and cell-type specific Pol III loci, and one miRNA. Surprisingly, only a fraction of the in silico-predicted Pol III loci are occupied. Many occupied Pol III genes reside within an annotated Pol II promoter. Outside of Pol II promoters, occupied Pol III genes overlap with enhancer-like chromatin and enhancer-binding proteins such as ETS1 and STAT1. Remarkably, Pol III occupancy scales with the levels of nearby Pol II, active chromatin and CpG content. Taken together, active chromatin appears to gate Pol III accessibility to the genome.
Introduction
RNA synthesis in mammals is conducted by three RNA Polymerases, termed Pols I, II, and III, with an additional two polymerases (IV and V) present in plants. Pol III transcribes small noncoding RNAs important for translational capacity1, such as the 5S rRNA, RNase P, RNase MRP, and all tRNAs. In addition, Pol III also transcribes a growing list of noncoding RNAs with alternative functions2, providing interesting connections to the biology of splicing (U6), viral RNAs (VA-I/II), microRNAs, DNA repeat-derived RNAs (SINEs, including mammalian-wide interspersed repeat [MIR] and Alu elements), neuronal disease and mRNA translation (BC200), Pol II transcriptional regulation (7SK, BC2), spermatogenesis (BC1), and multidrug resistance (Vault). However, the full repertoire of Pol III genes in the human genome is not known, and must be determined in multiple cell types to understand the full scope of Pol III biology. Also of high interest is Pol III regulation—whether the Pol III transcriptome is constitutive, or instead highly regulated—and if regulated, by what mechanism(s)? For example, there are 513 predicted tDNAs (genes encoding tRNAs) in the contiguous (hg18) genome, not including tRNA pseudogenes (172 total), but the fractional usage in different cell types is entirely unknown. Also, high levels of Pol III transcription and tRNA pools are correlated with the growth of cancer cells3,4. Clearly, a better understanding of Pol III dynamics and regulation is needed for understanding normal Pol III biology, and its misregulation in cancer and disease.
Extensive work on Pol III genes in yeasts, invertebrates, and vertebrates has revealed the factors required for directing Pol III to target genes5–7, and have defined the three ‘Types’ of Pol III genes in humans (Fig. 1a) based on 1) the presence and positions of cis regulatory elements, and 2) the requirement for particular basal or accessory transcription factors. Briefly, 5S rRNA is the sole Type 1 gene, uniquely requiring TFIIIA. Type 1 and Type 2 genes both require TFIIIC, a basal factor and targeting complex which recognizes gene-internal A-box and B-box elements at Type 2, but not Type 1 genes. The TFIIIB complex includes the TATA-binding protein (TBP), needed for TATA/promoter recognition and Pol III initiation. Type 2 and 3 genes utilize alternative assemblies of TFIIIB: BRF1 for Type 2 and BRF2 for Type 3 genes. Type 3 genes lack an internal A- or B-box, and lack reliance on TFIIIC—relying instead on upstream proximal and distal sequence elements (PSE and DSE) and specific factors (OCT1, SNAP, others) for targeting. Notably, Type 3 Pol III promoters resemble Pol II genes in their architecture, which utilizes upstream regulatory elements rather than gene-internal elements.
Figure 1.
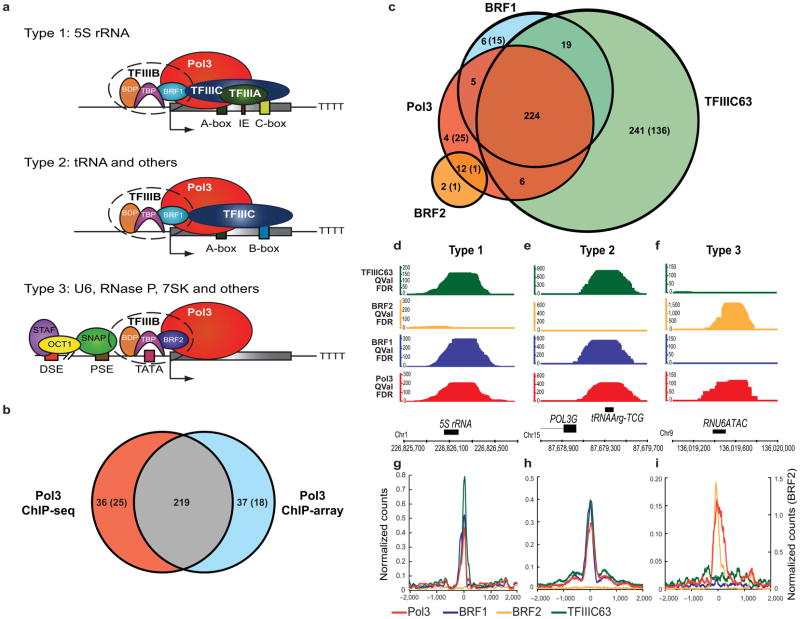
Occupancy analysis of Pol III and associated machinery in HeLa cells. (a) Pol III transcribed genes are classified into three Types based on sequence elements and transcription factors. IE, intermediate element. See Introduction for details. (b) Comparison of Pol III enriched regions obtained by ChIP-array (threshold 8.5-fold over background) and ChIP-seq (FDR<1%). Most sites appearing unique in the ChIP-array can be explained by data thresholding; they are scored as occupied after lowering the threshold for Pol III in the ChIP-seq format, and meet the high threshold (FDR<1%) in ChIP-seq experiments with other Pol III factors (TFIIIB or TFIIIC). (c) Intersections of Pol III-enriched regions (FDR<1%) with regions enriched by the basal transcription machinery (BRF1, BRF2 and TFIIIC; FDR<1%). In the Venn diagrams, the numbers outside parentheses denote known annotated Pol III genes, whereas the numbers within parentheses denote novel Pol III targets. (d–f) Genomic visualization of one random example of each of the three Types of Pol III genes. The y-axis depicts QValFDR significance values, which correspond to p-values (50=10−5, 100=10−10). Values are depicted for Pol III (red), BRF1 (blue), BRF2 (yellow) and TFIIIC63 (green). The x-axis is the physical map (hg18). (g–i) Class average maps of Pol III (red), BRF1 (blue), BRF2 (yellow) and TFIIIC63 (green) at Type 1, Type 2 and Type 3 genes. The y-axis is the number of read counts at each nucleotide normalized to the number of genes in the class. The x-axis is the distance from the transcription start site (TSS) of the Pol III gene.
Here, we applied genomics approaches toward the following goals: 1) to define human Pol III transcriptomes by occupancy of the Pol III machinery, 2) to discover new or alternative Pol III loci, 3) to classify all Pol III genes by the specialized Pol III machinery present, and 4) to provide new insights regarding the placement and regulation of Pol III genes in chromosomes/chromatin.
Results
Localization of Pol III in HeLa reveals gene classes
To define Pol III transcriptomes we applied chromatin immunoprecipitation (ChIP) of Pol III machinery to determine occupied loci, as RNA sequencing cannot determine all active tDNA loci due to the small fraction (21%) of uniquely mappable tDNAs (see Methods). Thus, Pol III occupancy of the unique flanking region (by ChIP-array or ChIP-seq) was a proxy measurement of gene activity. We chose HeLa cells for our initial Pol III transcriptome, and localized RNA Pol III itself (RPC32 subunit) by standard ChIP-array approaches, probing the unique portion of the human genome at ~150 bp resolution (Agilent Technologies). A threshold of 8.5-fold enrichment yielded 271 sites bound by Pol III and included the vast majority of formerly-verified unique Pol III genes, a few candidate unique loci, and approximately half of the predicted tDNAs (Supplementary Data 1). With an FDR of 1%, ChIP-seq revealed 257 loci bound by Pol III in HeLa cells, which overlap 255 annotated Pol III genes and 25 unannotated loci (Table 1, full datasets in Supplementary Data 1). Bound loci occasionally encompass closely-linked tDNAs (within 600 bp); for Pol III, 20 bound loci each contain 2–4 tDNAs. Loci occupied in ChIP-array largely overlapped with those identified by ChIP-seq (p-value<10−7, Fig. 1b). A small number of Pol III genes reside in non-unique regions (5S, small NF90-associated RNA [snaR] genes, certain tDNAs; Supplementary Data 1) but are not included in the analyses below. Taken together, two genomics formats yielded similar Pol III-occupied loci in HeLa cells.
Table 1.
Pol III Genes and Pol III-related Repeats in the Human Genome and Enrichment with Pol III Initiation Machinery in HeLa ChIP-seq
| Pol III Promoter Type | Class of RNA | Subclassa | Number in Human hg18 Reference Genomeb | Number and Percent with Attributec | Number and Percent Occupied with Pol III or BRF1/2 (FDR<1%) in HeLa | ||
|---|---|---|---|---|---|---|---|
| Type 1 | 5S | Consensus | 17 | 17d | 100.0% | 17e | 100.0% |
| 5S-related | 1,260 | 175d | 13.9% | 0e | 0% | ||
| Type 2 | tRNA | Consensus | 513 | 490f | 95.5% | 242 | 47.1% |
| tRNA-pseudogeneg | 172 | 120f | 69.8% | 2 | 1.2% | ||
| tRNA-relatedg | 1,224 | 227f | 18.5% | 0 | 0% | ||
| Alu | Dimerh | 1,099,242 | 638,006f | 58.0% | 13 | 0.001% | |
| Monomerh | 90,882 | 30,301f | 33.3% | 2 | 0.002% | ||
| MIR | MIR | 587,443 | 65,530f | 11.5% | 1 | 0.0002% | |
| snaR | Consensus | 21 | 21f | 100.0% | 21e | 100.0% | |
| snaR-related | 7 | 5f | 71.4% | 2e | 28.5% | ||
| HVG or Vault | Consensus | 3 | 3f | 100.0% | 3 | 100.0% | |
| HVG-related | 1 | 1f | 100.0% | 0 | 0% | ||
| 7SL or SRP | Consensus | 2 | 0f | 0% | 1 | 50% | |
| 7SL-related | 954 | 128f | 13.4% | 0 | 0% | ||
| BC200 | Consensus | 1 | 1f | 100.0% | 1i | 100.0% | |
| BC200-related | 253 | 141f | 55.9% | 0 | 0% | ||
| Type 3 | Y | Consensus | 4 | 4j | 100.0% | 4 | 100.0% |
| Y-related | 1,123 | 0j | 0% | 0 | 0% | ||
| U6 | Consensus | 9 | 5j | 55.5% | 5 | 55.5% | |
| U6-related | 1,670 | 0j | 0% | 0 | 0% | ||
| 7SK | Consensus | 1 | 1j | 100.0% | 1 | 100.0% | |
| 7SK-related | 713 | 1j | 0.1% | 0 | 0% | ||
| RNase P | Consensus | 1 | 1j | 100.0% | 1 | 100.0% | |
| RNase P-related | 1 | 0j | 0% | 0 | 0% | ||
| RNase MRP | Consensus | 1 | 1j | 100.0% | 1 | 100.0% | |
| RNase MRP-related | 6 | 0j | 0% | 0 | 0% | ||
| U6atac | Consensus | 1 | 1j | 100.0% | 1 | 100.0% | |
| U6atac-related | 39 | 0j | 0% | 0 | 0% | ||
| tRNASeC | Consensus | 3k | 1f,j | 33.3% | 1k | 33.3% | |
‘Consensus’ subclass is based on high sequence identity (using BLAT, genome.ucsc.edu) to the validated RNA, except tRNAs from the Genomic tRNA database (GtRNAdb, gtrnadb.ucsc.edu) and snaRs from others31.
Based on UCSC Genome Browser (genome.ucsc.edu) tracks ‘RNA Genes’ and ‘RepeatMasker’ (www.repeatmasker.org) (except for tRNAs and snaRs, see footnote a).
Attributes determined with ‘Consensus’ and ‘Patser’ programs (rsat.ulb.ac.be/rsat; see Methods). See Supplementary Data 1 for consensus matrices.
Contains an internal C-box
Mapped allowing multiple alignments with Bowtie (bowtie-bio.sourceforge.net; see Methods and Supplementary Data 1.)
Contains an internal B-box
‘tRNA-pseudogene’ from GtRNAdb and ‘RNA Genes’; ‘tRNA-related’ from ‘RepeatMasker’ track (genome.ucsc.edu), not overlapping ‘Consensus’ or ‘tRNA-pseudogene.’
‘Monomer’ refers to free left and right element (FLAM, FRAM); ‘Dimer’ means the Alu has a FLAM and a FRAM
Enriched in Pol III ChIP-array but not ChIP-seq
Contains a TATA (within 50 bp upstream) and PSE (between 100 bp and 25 bp upstream)
These are a subset of the tRNAs in Type 2 Promoter section.
To classify Pol III genes (Fig. 1a), we localized BRF1 (Types 1 and 2), BRF2 (Type 3) and TFIIIC (TFIIIC63 subunit, Types 1 and 2, not 3) by ChIP-seq in HeLa cells. With an FDR of 1%, we obtained 242, 16 and 549 occupied loci for BRF1, BRF2 and TFIIIC, respectively. Venn diagrams (Fig. 1c), examples (Fig. 1d–f) and class average maps (Fig. 1g–i) reveal two important features. First, BRF1 and BRF2 are mutually exclusive, supporting earlier work on individual genes; here we demonstrate this exclusion genome-wide and reveal all separate Type 2 and Type 3 genes in HeLa cells. Second, the majority of TFIIIC-bound loci lack Pol III (Fig. 1c), an observation of possible high interest given the known roles of TFIIIC-only sites in genome organization in lower eukaryotes8–11, addressed further later. A compilation of gene types and occupancy, including repetitive elements, is provided in Table 1 and Supplementary Data 1. Notably, we verify the single selenocysteine tDNA as the only Type 3 tRNA gene in the genome, with clear Brf2 occupancy (Supplementary Data 1).
Cell type variation in Pol III loci
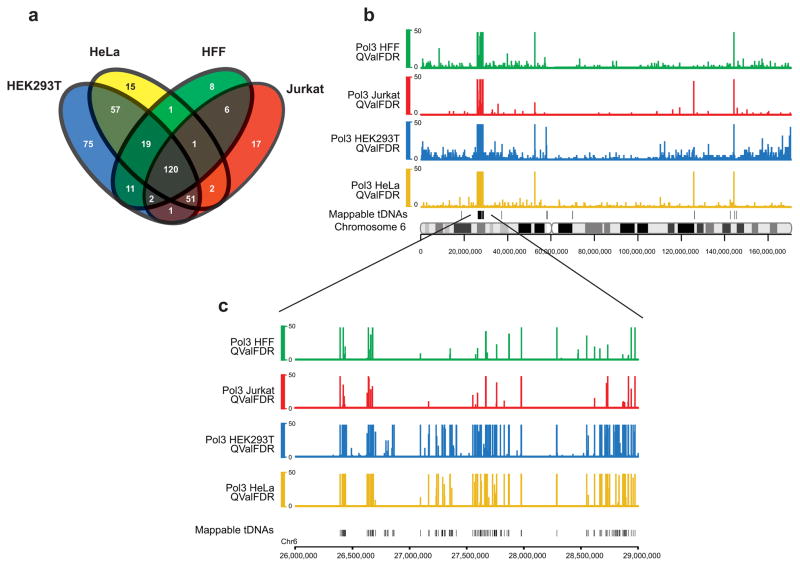
To explore the dynamic and cell type-specific Pol III transcriptome, we then performed ChIP-seq of Pol III in three other cell types: human embryonic kidney HEK293T cells (bearing adenovirus and T antigen), human foreskin fibroblasts (HFF; immortalized with hTERT, but non-transformed), and Jurkat T cells. For comparisons, we intersected the top 400 enriched loci from each cell type (Fig. 2a), which overlapped 336, 266, 200, or 168 predicted Pol III genes in HEK293T, HeLa, Jurkat, and HFF cells, respectively. Notably, 120 genes are clearly occupied in all four cell types. In addition, HEK293T cells display a large number (75, see Fig. 2a) of unique loci (primarily tDNAs). Region p22.1 of chromosome 6, which harbors the majority of genomic tDNAs, illustrates cell type variation (Fig. 2b,c). Of 24 genes displaying variance, 9 HEK293T genes and one HeLa gene (Supplementary Data 1) met a stringent threshold for differential occupancy (>38-fold enriched over background in HEK293T or >14-fold in HeLa, while <3-fold in other cell types) in a qPCR format. Thus, although the Pol III transcriptomes from these cell types show considerable overlap, cell-type specific Pol III-bound loci do exist. In addition, we observed that three transformed cell lines share a set of 51 genes not occupied in HFF (Fig. 2a).
Figure 2.
Differential Pol III occupancy in various cell types. (a) Intersection analysis of the known Pol III occupied genes between the four cell types. Pol III bound regions (top 400 enriched loci) were filtered for unannotated regions and only known genes were used for the analysis. Total numbers of genes for HEK293T, HeLa, Jurkat, and HFF were 336, 266, 200, and 168, respectively. (b) A snapshot of chromosome 6 showing Pol III occupancy in HeLa (yellow), HEK293T (blue), Jurkat (red) and Human Foreskin Fibroblasts (HFF, green) and mappable tDNAs on a separate track. (c) A zoomed-in view of a tDNA cluster in 6p22.1 showing Pol III occupancy in the four cell types. Note that many tDNAs are differentially enriched with Pol III between cell types.
Occupied tDNAs often reside in Pol II promoters
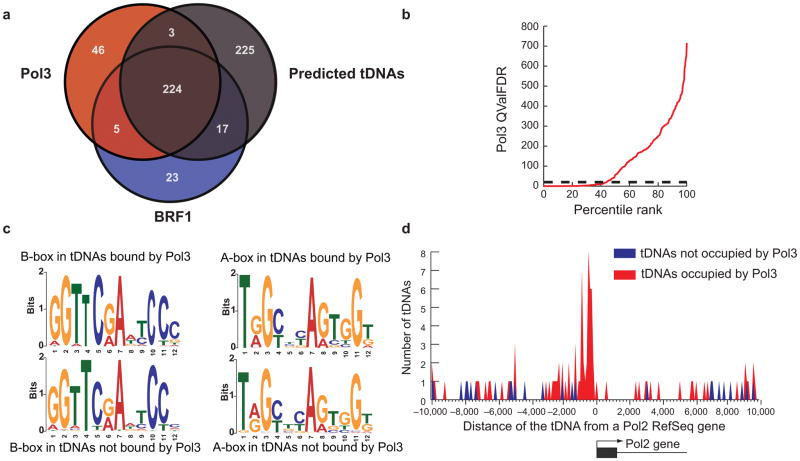
A particularly interesting observation was that only a portion of the in silico predicted tDNAs (ranging from ~30–60%) were occupied by Pol III in the different cell lines (52% in HeLa, Fig. 3a). This observation does not derive from data thresholding—rather, percentile rank analysis suggests two types of tDNAs: occupied or unoccupied, with variation in the occupied class (HeLa, Fig. 3b). This differential occupancy is not a mapping artifact, as most tDNAs lacking Pol III enrichment can be mapped at >85% efficiency (Supplementary Fig. 1). These occupancy differences (occupied vs. unoccupied) are also not explained by predicted TFIIIC affinity, as MEME12 analysis revealed nearly identical A- and B-box elements at occupied versus unoccupied tDNAs in HeLa cells (Fig. 3c).
Figure 3.
Genomic features of Pol III-occupied and unoccupied tDNAs in HeLa cells. (a) Venn Diagram illustrating that predicted tDNAs are bound by Pol III or Pol III transcription factors (BRF1). These 469 tDNAs represent 467 mappable, predicted tDNAs plus two Pol III-enriched tRNA-pseudogenes. (b) All predicted mappable tDNAs were ranked by their Pol III occupancy (x-axis) and plotted against Pol III QValFDR (y-axis), to show that ~50% of tDNAs are unoccupied by Pol III. The dotted line represents FDR 1% cutoff (QValFDR 20). (c) Sequence Discovery (MEME)12 analysis of the regulatory sequence elements (A-box and B-box) of tDNAs bound or not bound by Pol III. Note that the sequences are nearly identical between the two classes of tDNAs. (d) Pol III bound (red) or unbound tDNAs (blue) were clustered based on their distance from the nearest Pol II gene TSS in 100 bp bins. Note that Pol III bound tDNAs cluster within 1 kb of the Pol II gene TSS (p-value<10−7), whereas the unbound tDNAs show no such clustering. A tDNA was classified as Pol III-bound if it had RPC32 and/or BRF1 occupancy. A promoter was defined as being ±2 kb from a Pol II gene TSS.
Remarkably, in HeLa cells 53 occupied Pol III genes (19%) reside just upstream (within 2 kb) of an annotated Pol II gene, a highly statistically significant enrichment in location (p-value<10−7). In striking contrast, only 3 predicted tDNAs within 2 kb of a Pol II gene were unoccupied by Pol III, and two of those three tDNAs flank Pol II genes that are inactive in HeLa cells. Notably, histogram plots of all occupied tDNAs residing near Pol II genes reveals a pair of peaks at −300 and −900 (Fig. 3d), reflecting the relatively common presence of two tandem tDNAs (~600 bp apart) just upstream of a Pol II gene. In contrast, tDNAs lacking Pol III do not cluster near Pol II genes (Fig. 3d). Finally, adjacent Pol II and Pol III genes are often (71%) divergent (see Discussion).
Occupied tDNAs coincide with regions of active chromatin
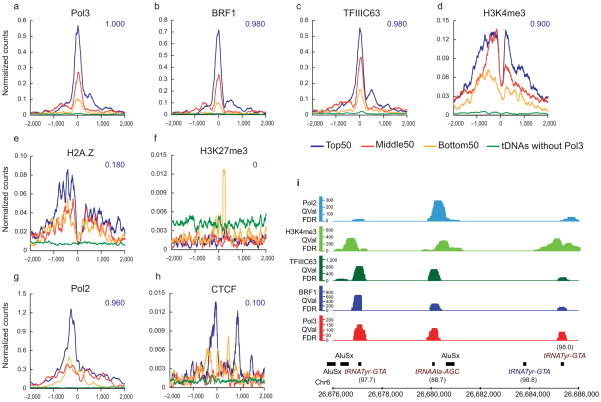
Interestingly, intersection analyses revealed Pol III co-incident with Pol II protein, H3K4me1, H3K4me3, H2A.Z, CTCF, and H3.3, at levels above our p-value (<10−3) and enrichment (10-fold above random) cutoffs (Supplementary Data 2, chromatin ChIP-seq datasets from others13–16). Of particular interest, the extent of Pol III occupancy scaled with the level of regional Pol II and active chromatin (Fig. 4a–h). To reveal this, we separated Pol III-occupied loci into four classes: the top 50 occupied loci, the middle 50 occupied loci, the bottom 50 occupied loci (remaining above the FDR 1% cutoff), and Pol III-unoccupied tDNAs. These four classes were compared to levels of Pol II, chromatin modifications, and chromatin factors (class average map, centered on the Pol III gene TSS). Remarkably, the levels of Pol II, positive histone modifications and H2A.Z all scaled with Pol III occupancy. Also, CTCF was observed at a small subset (10%) of the tDNAs with the highest Pol III occupancy. In contrast, repressive H3K27me3 was more prevalent at predicted tDNAs lacking occupancy (below our cutoff) and was not correlated with Pol III (Fig. 4f).
Figure 4.
Chromatin features at Pol III-bound tDNAs in HeLa cells. (a–h) Pol III-bound genes were binned into three categories: Top 50 (blue), Middle 50 (red) and Bottom 50 (yellow) based on their Pol III levels, and class average maps were plotted for various factors and chromatin marks13–16. Class average plots of factors at tDNAs without Pol III (green) serve as negative controls. The x-axis is the distance from the Pol III TSS and the y-axis is the number of read counts at each nucleotide normalized to the number of regions in the class. The intersection fraction for each of these factor-bound regions with the Top 50 class is indicated in blue. (i) Example of a genomic locus showing a tDNA cluster with differential occupancy of Pol III, BRF1, TFIIIC63, H3K4me3 and Pol II. Note that the tRNATyr gene lacking Pol III machinery also lacks Pol II and H3K4me3. The percent mapping efficiency of each of the tRNA genes is indicated in parentheses (see Methods).
The correlations described above would not be surprising if all tDNAs simply resided within active annotated Pol II promoters. However, most tDNAs occupied by Pol III actually reside outside annotated Pol II gene promoters (201, 82% in HeLa cells). Therefore, we separated occupied Pol III genes into two classes: those within annotated Pol II promoters, and those outside, and again examined how Pol III occupancy scaled with Pol II and chromatin attributes. Remarkably, active tDNAs outside annotated Pol II promoters still strongly correlated with adjacent Pol II and chromatin modifications typical of a Pol II promoter or enhancer (including H3K4me1) (Supplementary Fig. 2 and Supplementary Data 2). In contrast, unoccupied tDNAs lack adjacent Pol II or active chromatin (Fig. 4d–h), and instead bear higher levels of H3K27me3 (Fig. 4f). A clear example of this partitioning is observed in Figure 4i, where the two Pol III-occupied genes encoding tRNATyr-GTA genes also bear Pol II and active chromatin, whereas the single Pol III-unoccupied gene encoding tRNATyr-GTA lacks these factors or attributes. Thus, active tDNAs outside of annotated Pol II promoters are found in a chromatin region that resembles an active Pol II gene promoter or enhancer. We note that Type 3 genes (BRF2-containing) show similar active chromatin profiles to Type 2 genes.
We find a considerable fraction (~30%) of Pol III-occupied loci reside within CpG islands, a highly significant overlap (p-value<10−6), and a striking correlation exists between Pol III occupancy and CpG content at Pol II promoters. Others have separated promoters into three types based on their CpG density17: high CpG (HCP), intermediate CpG (ICP), and low CpG (LCP). Interestingly, Pol III-occupied promoters intersect well with HCPs (typically associated with constitutively active genes), moderately with ICPs, and are anti-correlated with LCPs (Supplementary Data 2). Taken together with the results above, Pol III occupancy is apparently enabled by active Pol II promoter-like chromatin.
Jurkat and HeLa show similar Pol III chromatin
Genome-wide chromatin maps in HeLa and primary CD4+ T cells are extensive, whereas those in Jurkat, HFF and HEK293T cells are lacking. Jurkat T cells are similar to CD4+ T cells (Jurkat is also a CD4+ T cell18), though proliferative and (in our hands) technically more amenable to Pol III occupancy analysis. ChIP-seq of Pol III in Jurkat cells yielded 211 occupied loci (FDR of 10%) which overlap 182 annotated genes. This list had high overlap (88%) with the list from HeLa cells and showed similar trends; Pol III-occupied loci were correlated with (and scaled with) Pol II and with active Pol II promoter-like chromatin (Fig. 5a, Supplementary Figs. 3, 4; chromatin ChIP-seq datasets from others19–22), and anti-correlated with additional repressing modifications (e.g. H3K36me3; Supplementary Data 2). However, with resting CD4+ T cells, the correlations of Pol III with adjacent Pol II were less strong, and correlations of Pol III with adjacent H2A.Z were stronger, possibly reflecting the ‘poised’ nature of many genes in resting CD4+ cells. Consistent with this notion, Pol II levels rise and H2A.Z levels fall adjacent to tDNAs when CD4+ T cells are activated (Supplementary Data 2; Pol II and H2AZ data from others23).
Figure 5.
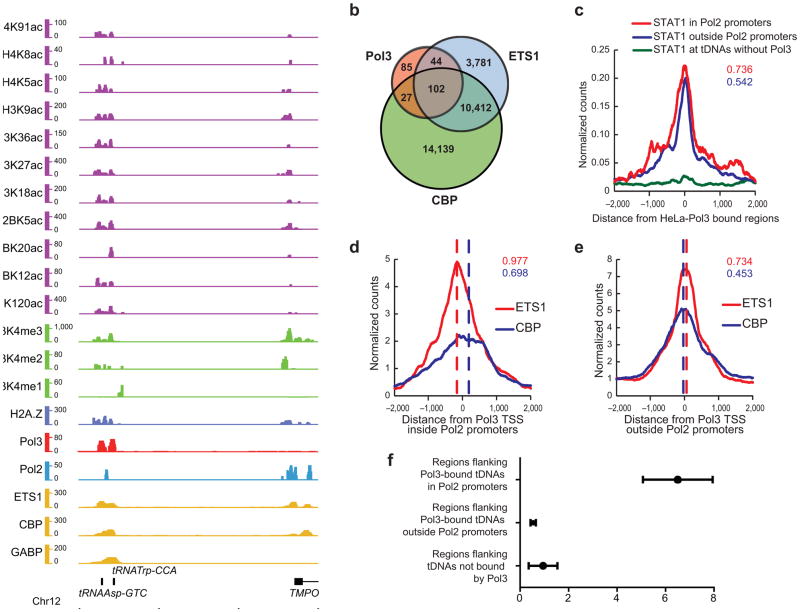
Chromatin marks and factors associated with Pol III-bound regions in Jurkat cells. (a) Example of a genomic locus showing a tDNA bound by Pol III (red) in Jurkat cells. Also shown are active acetylation (purple) and methylation (green) marks, H2A.Z (blue), Pol II (turquoise) and transcription factors (yellow). This putative enhancer is ~12 kb upstream of a Pol II gene TSS. Chromatin and factor ChIP-seq from others19–22. (b) Intersection analysis of Pol III bound regions (FDR<10%) with regions bound by ETS1 (FDR<1%; filtered for regions overlapping DNase I hypersensitivity in CD4+ cells41) and CBP (FDR<1%) in Jurkat cells. (c) Pol III-bound genes in HeLa (FDR<1%) were binned into those inside (red) or outside (blue) annotated Refseq promoters, and class average map of STAT1 was plotted; tDNAs without Pol III (green) serve as a negative control. The numbers denote the fraction of total Pol III genes occupied by STAT1 for loci either inside (red) or outside (blue) Refseq promoters. (d,e) Class average maps of ETS1 (red) and CBP (blue) at Pol III-bound genes inside (d) or outside (e) Refseq promoters in Jurkat cells. The red and blue dashed lines indicate approximate peaks of the corresponding factors. The intersection fractions of ETS1 (FDR<1%) and CBP (FDR<1%) regions with Pol III regions in Jurkat are indicated in red and blue respectively. (f) The graph represents the average HeLa RNA-seq read per kb score for each 1 kb region flanking tDNAs with error bars representing s.e.m. Note that transcripts are seen near tDNAs in Pol II promoters (from neighboring Pol II genes), but are not seen near tDNAs outside Pol II promoters.
Occupied Pol III loci correlate with STAT1 and ETS1
Our results prompt two questions regarding the establishment of Pol III-correlated chromatin: 1) are there specific DNA binding factors/activators that co-localize with Pol III-occupied regions, and 2) might the non-annotated loci bearing Pol III and Pol II represent enhancers or enhancer-like regions. Here, we compared our datasets to the extensive transcription factor binding profiles in T cells (Jurkat and CD4+)—except for STAT1, where binding profiles13,24 in HeLa were directly compared. Although certain transcription factors have been linked to Pol III regulation (p53, c-MYC, RB)25,26, the lack of genome-wide ChIP-seq datasets prevented comparison. However, we found a striking overlap between Pol III occupancy and particular general transcription factors: STAT1 (ref. 13) (in HeLa, FDR 1%, overlap 161/278 loci, p-value<10−5) and ETS1 (ref. 22) (in Jurkat, overlap 144/182 loci, p-value<10−5; Fig. 5b). However, using the list of STAT1-occupied sites generated by Robertson et al.24, (FDR 0.1%), we derive an overlap of 254/278. Notably, STAT1 binding sites reside very near the Pol III gene TSS, both for Pol III genes within and outside of annotated Pol II gene promoters (Fig. 5c and Supplementary Fig. 4f). ETS1 has different properties at promoters versus enhancers22. For example, sites within Pol II promoters are typically consensus, and a feature of promoter-localized ETS1 is that it lacks precise co-alignment with CBP occupancy. In contrast, ETS1 sites at enhancers display considerable variation from consensus, and often physically partner with other transcription factors (e.g., RUNX1). Also, ETS1 at enhancers is aligned more precisely with CBP occupancy22. In keeping with these promoter and enhancer differences, we find that Pol III-occupied annotated promoters bearing ETS1 are typically consensus sites (data not shown), and lack precise alignment with CBP (Fig. 5d). In contrast, the tDNAs not adjacent to annotated Pol II genes coincide with enhancer-like ETS1 sites (data not shown) that are well aligned with CBP (Fig. 5e). This raises the possibility that particular enhancer-binding proteins such as STAT1 and ETS1 help nucleate open chromatin at promoters or enhancers, which then promotes Pol III occupancy (see Discussion). We also see marked overlap with SRF (FDR 1%, 52/182 loci) and moderate overlap with GABP (FDR 1%, 29/182 loci). However, we observe little overlap with NRSF (FDR 1%, 10/182) in Jurkat cells (Supplementary Fig. 4g–i; data from others21).
Enhancers strongly correlate with three chromatin attributes27: H3K4me1, H3K27ac, and DNase I hypersensitivity, with maps available in CD4+ T cells or HeLa cells. We see highly significant overlap between Pol III occupancy at unannotated regions and the presence of H3K4me1 (p-value<10−5) and H3K27ac (p-value<10−5) (Supplementary Data 2; Supplementary Fig. 3b,i,r). Furthermore, of the 150 occupied tDNAs in Jurkat cells, 145 overlap DNase I hypersensitive sites (p-value<10−5), whereas only 96 of the 321 unoccupied tDNAs overlap. Notably, 59 of those hypersensitive sites become enriched with Pol III in HeLa cells.
Occupied tDNAs correlate with enhancer-like chromatin, which typically does not involve the production of RNA. However, the presence of Pol II at these loci prompted us to address whether transcription by Pol II occurs from these enhancer-like loci. Here we performed RNA-seq of total RNA in HeLa and quantified reads in the region flanking the tDNAs. This region contains the peak of Pol II (Supplementary Fig. 2d), but not the tDNA itself (or Pol III), so that only reads from Pol II were compiled. At these tDNAs in enhancer-like chromatin, we do not generally observe RNA transcripts in the aforementioned region (Fig. 5f). This suggests that ‘active’ chromatin modifications, including those involved in Pol II initiation (e.g., H3K4me3), but not Pol II transcripts per se, best correlate with Pol III occupancy.
Novel Pol III genes and miRNAs
ChIP-seq data yielded 42 candidate novel Pol III loci in HeLa cells. From these, 13 were tested by qPCR and 10 enriched at least 3.5-fold above background for Pol III occupancy. One interesting unannotated locus is about 2 kb upstream of the SLC7A2 TSS, which shows nearby STAT1, H3K4me3, Pol II, and a transcript by RNA-seq (Supplementary Fig. 5a–c). Another candidate novel locus resides on chromosome 5 among multiple repeats. Here, BRF2 and Pol III co-localized over an L1M5 LINE, with STAT1, Pol II and H3K4me3 nearby (Supplementary Fig. 5d–f). This could represent a novel Type 3 gene, of which there are only 14 currently known. We also identify one MIR highly enriched with Pol III, BRF1, and TFIIIC in the first intron of POLR3E (Supplementary Fig. 5g,h). These positives also include other loci in promoters of Pol II-transcribed genes, such as ADARB1, FZR1, and U1, or near other retroviral elements (e.g., MER41C LTR) (Supplementary Fig. 6a,b). Furthermore, we curiously observe six annotated tRNA-pseudogenes enriched at high levels in various cell types, and verify two by qPCR (Supplementary Fig. 6c, Supplementary Data 1). Finally, we also observe 17 Pol III genes within Pol II transcriptional units (mostly in introns; data not shown).
For repetitive regions, we applied mapping algorithms that allow multiple alignments (Bowtie; bowtie-bio.sourceforge.net) which revealed Pol III enrichment (and often other Pol III factors, see Supplementary Data 1) at snaR-class genes, at all of the consensus 5S rDNA genes on chromosome 1 (but no other 5S-related genes), at multiple Alus, and at 35 tDNAs that map inefficiently. However, the HPV-18 and 45S rDNA loci were not occupied.
Pol III has been reported to transcribe multiple miRNAs on the chr19 miRNA cluster (C19MC)28, driven by Alu promoters in HEK293T cells, which would represent the first example of Pol III-driven miRNAs. However, after remapping our reads for HeLa, HEK293T, and HFF, allowing multiple alignments, we see no enrichment of Pol III at any region (or miRNA) in this cluster (data not shown); a negative result supported by recent evidence showing transcription of C19MC by Pol II instead29. Another study30 supported two additional Pol III transcribed loci, SNAR-A31 (also known as CBL-1) and MIR886 [ref. 32] (also known as CBL-3), but did not test for occupancy by Pol III. Here, we show clear occupancy of both loci (Supplementary Fig. 7), and also observed transcripts lacking a 5′ cap derived from the MIR886 locus by RNA-seq (see Methods and Supplementary Fig. 7d), thus providing the first direct evidence for occupancy of and transcription by the Pol III machinery of a miRNA in mammals.
TFIIIC-only and TFIIIB–TFIIIC sites
Given their role in chromatin organization in yeast cells8–11, we identified 307 loci in HeLa cells that are bound by TFIIIC, but not Pol III or BRF1/2. TFIIIC-only sites partition into two classes: loci adjacent to Alu and/or MIR repeats (181) and those that lack repeats (126). Certain loci were adjacent to both an MIR and an Alu element, which accounts for the higher number of total TFIIIC-only loci (377) depicted in Figure 1c. Notably, 101 TFIIIC-only loci reside within 2 kb of the TSS of an annotated Pol II gene, 60% of which have HCP Pol II promoters, a highly significant enrichment (p-value<10−5), while 206 reside in unannotated regions (Supplementary Fig. 8a). Those within annotated Pol II genes show correlations with active promoter chromatin (Supplementary Fig. 8b–i and Supplementary Data 2). Those in unannotated regions also show strong correlations with active chromatin, though weaker than with annotated Pol II genes (Supplementary Data 2). Interestingly, TFIIIC-only sites near Alu and MIR elements typically have a B-box (164/181), and bear high levels of H3K4me1, H3K4me3 and Pol II, whereas those distal from repeats are generally void of positive marks, and typically lack a consensus B-box element (27/126 have a B-box) (Supplementary Fig. 8b–i), raising the possibility that TFIIIC cooperates with other factors for binding at these loci. Here, MEME12 and TOMTOM33 analysis revealed the consistent presence of a G/A-rich site with significant (p-value 0.00028) similarity to the binding site of KLF4 (Supplementary Fig. 8j). Nineteen sites in our HeLa datasets contain both TFIIIB (BRF1) and TFIIIC, but lack Pol III enrichment (at our threshold of FDR 1%). However, the majority (14/19) of these sites become enriched with Pol III in HEK293T cells, or in HeLa cells if compared with a lower threshold (FDR 5% Pol III ChIP-seq) or our Pol III ChIP-chip dataset (data not shown). At present, it is not clear whether these sites are truly different in their mode of Pol III recruitment, or simply represent sites near our occupancy cutoff thresholds.
Discussion
Our datasets and analyses address the scope and regulation of human RNA Polymerase III transcriptomes, providing multiple insights. First, we observe the close proximity of Pol III genes to Pol II genes genome-wide. This result is in keeping with previous work at the Pol III-transcribed U6 gene (a Type 3 gene), which has proximal Pol II that assists in Pol III expression34. Second, we show genome-wide a highly significant overlap of Pol III with active chromatin. Here, previous work at U6 supports the notion that nearby chromatin remodeling promotes U6 expression35, and our work extends this concept genome-wide to all Type 2 and 3 genes, and also to many active histone modifications and composition. However, most occupied Pol III genes reside at regions outside of annotated Pol II genes—yet those regions still bear high levels of H3K4me3 and Pol II protein, properties typical of promoters. Interestingly, these unannotated regions also have properties of enhancers27, as they contain H3K4me1, H3K27ac, and overlap with enhancer-binding proteins and DNase I hypersensitive sites. In addition, we find few, if any, transcripts adjacent to most of these unannotated Pol II peaks, and they generally lack a long downstream open reading frame. Thus, it is not entirely clear whether one should consider these regions new unannotated promoters, or instead a sub-class of enhancers that contain Pol II and H3K4me3, resembling Pol II ‘poised’ promoters. Here, we speculate that the promoter-like chromatin formed nearby either ‘poised’ or active Pol II might be sufficient for enabling Pol III occupancy. In addition, it will be of interest to determine whether these unannotated promoters/enhancers produce a functional transcript in particular cell types, or whether they are typical enhancers, activating another Pol II gene in the larger region. Regardless, the overlap of Pol III-occupied tDNAs with active chromatin is striking. Notably, Pol III occupancy scaled with active chromatin marks and proximal Pol II, but not with RNA transcript levels for the Pol II gene, emphasizing the connection of Pol III occupancy with active chromatin. Finally, DNA hypomethylation may also contribute to the active chromatin state, as occupied Pol III genes correlated with high CpG content regions (which are typically unmethylated) and as STAT1 consensus sites are strongly correlated with DNA hypomethylation36.
Occupied Pol III genes often reside 300–900 bp upstream of the Pol II TSS—close enough to overlap with promoter proximal chromatin, but generally not within the promoter proximal region where the Pol II basal machinery assembles. Furthermore, tRNAs residing in Pol II promoters are typically transcribed away from the Pol II gene (divergent orientation), with a significant bias (p-value 0.006). We suggest that these properties allow the Pol III gene to benefit from promoter chromatin dynamics while avoiding interference with the transcription of the Pol II gene itself. We note that previous Pol III transcriptomes in S. cerevisiae10,37,38 showed that virtually all predicted Pol III genes (including tDNAs) were occupied by Pol III, arguing against appreciable Pol III regulation by chromatin or position relative to Pol II genes. Moreover, S. cerevisiae lacks key modifications of vertebrate heterochromatin (H3K9me3, H3K27me3, DNA methylation), suggesting that Pol III in human cells may encounter chromatin obstacles not present in lower yeasts.
Our work also reveals moderate variation in Pol III occupancy among cell types, and cell-type ‘specific’ occupancy of a small number of loci. Here, we note that specificity is defined using a stringent criterion, but very low occupancy of these ‘specific’ loci may exist in other cell types. A key issue is the basis for cell-type variation and specificity. One possibility is that as each cell type varies its repertoire of Pol II gene transcription and active enhancers, the Pol III genes overlapping that permissive chromatin gain access to their machinery. By this model, Pol III relies on both general Pol II transcription factors and cell-type specific factors to create open/active chromatin. Although active chromatin may gate Pol III access, it does not constitute all of Pol III regulation—the activity of occupied Pol III genes is likely still regulated by other factors such as the general Pol III repressor Maf139. Furthermore, we emphasize that these models are based on extensive sets of strong correlations, but genetic experiments are required to determine their dependency relationships.
Interestingly, tDNAs are thought to have expanded in the genome via retrotransposition40, and retrotransposons often insert in regions of open chromatin. One interpretation of our data is that the juxtaposition of active Pol III genes (~300) with active Pol II chromatin is largely a consequence of this initial accessibility during transposition, which would imply that these regions were accessible in the germline at some point during evolution. However, there are an additional ~1396 tRNA-derived elements in the genome, and these are generally not occupied by Pol III machinery (~0.1% occupied). Furthermore, as a class these elements are not coincident with active chromatin (data not shown), raising the possibility that transposition may have occurred in the germline into inactive chromatin, with inactive chromatin preventing their subsequent expression and contribution to fitness, allowing sequence drift. Alternatively, the transposition may have occurred into active chromatin, but these regions were later converted into heterochromatin, with similar consequences. Regardless, we observe active chromatin coincident with active Pol III genes, and not with tRNA-derived elements.
Our work also reveals many new Pol III-occupied loci in multiple cell types, which also require functional work in vivo. For the three new loci clearly enriched with Pol III machinery (Supplementary Fig. 5), we propose new names with a ‘P3’ (Pol III) designation: the MIR in the POLR3E intron, MIRP3; the chr8 locus conserved in primates, CPP3; LINE L1M5 locus, L1M5P3. Furthermore, we show the transcription of a miRNA, clarifying and extending earlier work30,32. Here, it will be of interest to determine the Pol III transcriptomes of pluripotent cell lines and/or early embryos to determine if additional noncoding RNAs are produced by Pol III.
Methods
Cell culture and cross-linking
We used cross-linked HeLa S3 cells (Biovest International) for ChIP-array and most HeLa ChIP-seq. For a second replicate of RPC32 ChIP, we obtained HeLa cells from ATCC (Cat. CCL-2.2) and cultured them in DMEM, 10% (v/v) FBS, 10 mM glutamine. We harvested cells at ~80% confluence and cross-linked them in 1% (v/v) formaldehyde for 30 min. We cultured and cross-linked Jurkat, HEK293T, and human foreskin fibroblast (with hTERT) cells as for HeLa (except we grew Jurkat in RPMI). We obtained Jurkat E6-1 cells from ATCC (Cat. TIB-152).
Chromatin immunoprecipitation
We used Standard Agilent Technologies Mammalian ChIP-on-chip protocol version 10.0 (www.Agilent.com) for ChIP-array. For ChIP-seq, we made the following modifications. We lysed nuclei in 50 mM Tris-HCl pH 8, 100 mM NaCl, 10 mM EDTA, 1% (w/v) SDS. We sheared chromatin by sonicating (Misonix) 10–20 times on setting 4–5 to an average shear length of 200–400 bp. For each IP, we bound 100ul Dynabeads (Invitrogen) to 5–10 ug antibody in dilution buffer (15 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% (w/v) Triton X-100, 2 mg ml−1 BSA) for 5 h to overnight. We pre-cleared chromatin sonicate from 20–40 × 106 cells in 1.4 ml dilution buffer with 50 ul Dynabeads for 1 h, and transferred it to bead-antibody complexes for overnight immunoprecipitation. We washed and eluted the immunoprecipitate and reversed cross-links in 200 mM NaCl as described42. We purified DNA from eluate with phenol-chloroform-isoamyl (25:24:1, pH 8; Invitrogen), and Qiagen PCR Purification. We used these antibodies for immunoprecipitation: RPC32 (Santa Cruz Biotechnologies, sc-21754), anti-RPB90 (i.e., BRF1), BRF2 (abcam, ab17011) and TFIIIC63 (Bethyl Laboratories, A301-242A).
ChIP-array
We designed ten ~1 million feature custom microarrays tiling the non-repetitive human genome (average resolution of 150 bp) from the Agilent ChIP database. We carried out RPC32 ChIP eluate amplification, labeling, array hybridization and wash according to Agilent Mammalian ChIP-on-chip protocol 10.0. We scanned arrays with Agilent Technologies’ Scanner C (Cat. G2505C) and performed Feature Extraction with Agilent Technologies FE version 10.1.1.22 using default ChIP settings. We analyzed arrays using DNA Analytics’ ChIP-chip analysis module (Agilent) and determined bound regions using default settings.
RNA-seq
We seeded 107 HeLa S3 cells per plate on two 15 cm dishes overnight. We washed cells with warm PBS, added 5 ml Trizol (Invitrogen) per plate, and purified total RNA. We subjected RNA to RiboMinus (Invitrogen) or double 7meG-cap purification44 plus RiboMinus before preparing RNA-seq libraries.
Sequencing
We used the Illumina GA2 with standard protocols for preparing and sequencing libraries. Read numbers are unique satellite-filtered reads (26–36 bp): Input HeLa 20,691,965; RPC32 HeLa 13,082,194; BRF1 HeLa 10,986,064; BRF2 HeLa 11,053,174; TFIIIC HeLa 16,076,219; RPC32 Jurkat 18,173,688; RPC32 human foreskin fibroblast 8,917,992; RPC32 HEK293T 7,762,672; Total RNA HeLa 4,392,375; Capped RNA HeLa 5,934,673. For instances where multiple alignments were desired, we remapped reads with Bowtie (bowtie-bio.sourceforge.net), retaining either the top 5 or 15 alignments.
Analysis of ChIP-seq data
We analyzed ChIP-seq data with the USeq package (useq.sourceforge.net). We used Jurkat input reads21,22 to analyze RPC32 ChIP data from Jurkat, HFF, and HEK293T as well as ChIP-seq tags for CD4+ and Jurkat from others19–23,43, unless provided. We used HeLa input reads from this study to analyze all HeLa datasets produced in this study as well as HeLa ChIP-seq tags from others13–16, unless provided. We created the ‘Random regions’ file for IntersectRegions by pooling data from this study and others13,14,19,20,22 and merging all 110 bp windows with at least one read; this is an estimate of the uniquely mappable genome. We removed duplicate alignments of RPC32 HeLa replicate 2, HEK293T, and HFF datasets. We performed Venn intersections using a Perl script (available upon request). We determined mapping efficiency of a tDNA by tiling every possible 36 bp sequence within the tDNA (plus 100 bp upstream and downstream sequence for ChIP-seq). The expected number of mappable reads for 100% efficiency is the number of tiles. We aligned the tiles uniquely with Bowtie (bowtie-bio.sourceforge.net) and calculated a percent of observed mapped tiles over expected. We only used tDNAs overlapping the ‘Random regions’ file for analysis. See Supplementary Data 1 for lists and further descriptions. We used the Integrated Genome Browser (IGB, http://igb.bioviz.org/) for screen shots.
Consensus binding sites
We used ‘Consensus’ and ‘Patser’ (rsat.ulb.ac.be/rsat) to determine position-weight-matrices for consensuses in Table 1 from Pol III-enriched genes, and to search for the consensus in repeats. We downloaded sequences for repeats in Table 1 from UCSC (genome.ucsc.edu) tracks ‘RepeatMasker’ (www.repeatmasker.org) and ‘RNA Genes,’ except tDNAs were from the Genomic tRNA database (gtrnadb.ucsc.edu). We analyzed A- and B-box of tDNAs as well as TFIIIC-only and KLF4 consensuses with MEME and TOMTOM (meme.sdsc.edu).
Real-Time PCR
For qPCR reactions, we used 1/50 to 1/100 of ChIP eluate, 500 nM primers, and iQ SYBR Green Supermix (Bio-Rad) in a total volume of 20 ul. We serially diluted ChIP input DNA for a standard curve. We designed primers of annealing temperature 62–65°C with a single melt curve peak (Supplementary Table 1). We analyzed PCR results with iCycler (Bio-Rad).
Statistical analysis
Others have detailed a full description of statistical methods used in USeq programs45. To generate p-values and fold enrichment over random in intersections of datasets, as well as determining statistical significance of position of Pol III genes with respect to Pol II Refseq TSS, we used IntersectRegions (USeq package), which uses multiple permutation tests of random regions, as described46. We generated the p-value for the divergent orientation of Pol II and nearest Pol III gene with the binomial p-value function in ‘R’ (www.r-project.org).
Data availability
We deposited data in the Gene Expression Omnibus (GEO) under accessions GSE20309 and GSE20609. The processed data is available for programmatic access using the GenoPub DAS/2 data distribution server (Description: http://bioserver.hci.utah.edu/BioInfo/index.php/Software:DAS2, GenoPub web app: http://bioserver.hci.utah.edu:8080/DAS2DB/genopub, and the DAS/2 Data Access URL: http://bioserver.hci.utah.edu:8080/DAS2DB/genome). One can use DAS/2 compliant genome browsers such as IGB (http://igb.bioviz.org/) to view the datasets, found under Homo sapiens→H_sapiens_Mar_2006→Cairns Lab→Oler_2010.
Supplementary Material
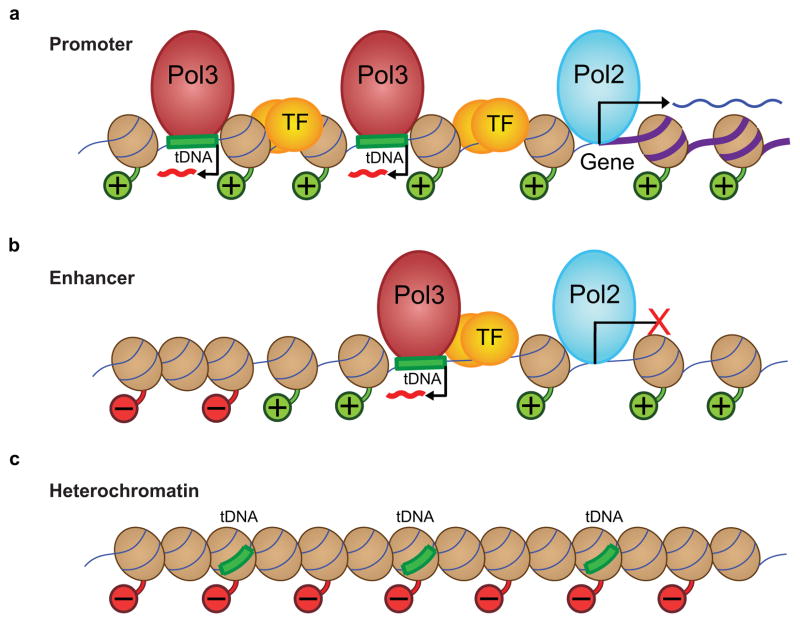
Figure 6.
Model depicting how chromatin features affect Pol III recruitment, at three different regions: promoters, enhancer-like promoters, and heterochromatin. (a) Chromatin features at an annotated Pol II gene promoter, where ~20% of active tDNAs with Pol III (red spheroid) reside, typically in a divergent orientation and about 300–400 bp from the peak of Pol II. This region contains transcription factors (TF, transcription factors and enhancer binding proteins, yellow spheres) and a large region of active chromatin, where nucleosomes (brown spheroids) have histone tails with positive/active modifications (green histone tails with ‘+’ balls). Active tDNAs are frequently found in pairs (shown) or clusters. At promoters, Pol II produces an RNA transcript from the annotated gene (purple thick DNA). (b) About 80% of active tDNAs are located distal to annotated Pol II promoters, and these regions have features both of Pol II promoters and enhancers. They contain active local chromatin, nearby Pol II, and TFs close to the Pol III gene. Although Pol II is present and there is evidence of Pol II initiation (H3K4me3), stable transcripts by Pol II are not detected (although it cannot be ruled out that unstable transcripts are produced). Outside of the local active region, we see facultative heterochromatin, represented by red histone tails and ‘−’ balls. (c) Inactive tDNAs lack active chromatin marks, TFs, or Pol II, and instead are associated with repressive chromatin marks (red histone tails and ‘−’ balls), suggesting a more tightly packed chromatin environment that is inaccessible to Pol III machinery.
Acknowledgments
We thank D. Ayer and S. Lessnick at the Huntsman Cancer Institute (HCI) for cells, B. Dalley (HCI) for his expertise in Illumina sequencing, and D. Nix (HCI) for suggestions for ChIP-seq analysis. We thank R. Roeder (Rockefeller University) for the gift of anti-RPB90 antibody. Financial support was from the Howard Hughes Medical Institute (HHMI) (supplies, genomics resources), the National Institutes of Health (GM38663 to B.J.G., CA42014 to the Huntsman Cancer Institute for support of core facilities, and CA63640 to C.H.H.), Huntsman Cancer Institute/Huntsman Cancer Foundation, and Agilent Technologies Foundation (supplies). B.R.C. is an investigator with the HHMI.
Footnotes
Author Contributions
B.R.C and A.J.O., overall scope and design. A.J.O., overall experimental execution. D.N.R. and A.W., ChIP-array experiments. C.A.N. and C.H.H., cap-purified RNA. P.A.C., qPCR determinations. P.C.H., K.J.C. and B.J.G, ETS1 and CBP datasets and analysis. A.J.O., R.K.A., and B.R.C. data analysis and interpretation. A.J.O. and R.K.A., figures, tables and data organization. B.R.C. and A.J.O. wrote the manuscript, with comments from all authors.
References
- 1.White RJ. RNA Polymerase III Transcription. Springer-Verlag; New York, NY: 1998. [Google Scholar]
- 2.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 3.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–9. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–4. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- 5.Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 6.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 7.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 8.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Simms TA, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:2078–86. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–27. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–48. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 13.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson AG, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–17. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 18.Koka P, et al. Increased expression of CD4 molecules on Jurkat cells mediated by human immunodeficiency virus tat protein. J Virol. 1988;62:4353–7. doi: 10.1128/jvi.62.11.4353-4357.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–34. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenhorst PC, et al. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barski A, et al. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–51. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 25.Felton-Edkins ZA, et al. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–4. [PubMed] [Google Scholar]
- 26.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 27.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 29.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–73. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35:6249–58. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Listerman I, Bledau AS, Grishina I, Neugebauer KM. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan CC, et al. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol. 2007;27:8729–38. doi: 10.1128/MCB.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–71. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 37.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci U S A. 2003;100:14695–700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harismendy O, et al. Genome-wide location of yeast RNA polymerase III transcription machinery. Embo J. 2003;22:4738–47. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–3. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–61. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 41.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon M, et al. Genome-Wide Dynamics of SAPHIRE, an Essential Complex for Gene Activation and Chromatin Boundaries. Mol Cell Biol. 2007;27:4058–69. doi: 10.1128/MCB.02044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc Natl Acad Sci U S A. 2003;100:7033–8. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We deposited data in the Gene Expression Omnibus (GEO) under accessions GSE20309 and GSE20609. The processed data is available for programmatic access using the GenoPub DAS/2 data distribution server (Description: http://bioserver.hci.utah.edu/BioInfo/index.php/Software:DAS2, GenoPub web app: http://bioserver.hci.utah.edu:8080/DAS2DB/genopub, and the DAS/2 Data Access URL: http://bioserver.hci.utah.edu:8080/DAS2DB/genome). One can use DAS/2 compliant genome browsers such as IGB (http://igb.bioviz.org/) to view the datasets, found under Homo sapiens→H_sapiens_Mar_2006→Cairns Lab→Oler_2010.