Abstract
The recently developed Multiple Displacement Amplification technique (MDA) allows for the production of a large quantity of high quality genomic DNA from low amounts of the original DNA. The goal of this study was to evaluate the performance of the MDA technique to amplify genomic DNA of siphonapterids that have been stored for long periods in 70% ethanol at room temperature. We subjected each DNA sample to two different methodologies: (1) amplification of mitochondrial 16S sequences without MDA; (2) amplification of 16S after MDA. All the samples obtained from these procedures were then sequenced. Only 4 samples (15.4%) subjected to method 1 showed amplification. In contrast, the application of MDA (method 2) improved the performance substantially, with 24 samples (92.3%) showing amplification, with significant difference. Interestingly, one of the samples successfully amplified with this method was originally collected in 1909. All of the sequenced samples displayed satisfactory results in quality evaluations (Phred ≥ 20) and good similarities, as identified with the BLASTn tool. Our results demonstrate that the use of MDA may be an effective tool in molecular studies involving specimens of fleas that have traditionally been considered inadequately preserved for such purposes.
Findings
The Neotropical region (excluding Mexico) contains 52 genera and about 280 species of fleas, and one of the most important families is Tungidae [1-3]. Females of the genus Tunga Jarocki, 1838 (Tungidae) penetrate into the skin of their hosts, including armadillos, anteaters, rodents, pigs, humans, dogs and other domestic animals [4]. After mating, gravid females undergoes hypertrophy, becoming a neosome 5-10 mm in size [5-7]. Taxonomic knowledge regarding siphonapterids is based primarily on morphological analysis of specimens permanently mounted on slides or preserved in 70% ethanol [3]. With the advent of the polymerase chain reaction (PCR) and other molecular approaches, DNA has become an excellent tool to study phylogenetic relationships [8]. However, in fleas few studies have analyzed genetic diversity in preserved specimens [9-11].
Many species of fleas have been identified from a small number of adult specimens mounted on permanent slides or adults and neosomes preserved in 70% ethanol at room temperature in scientific collections [12]. The DNA of samples preserved under these conditions for long periods is generally of poor quality and is often degraded, preventing its use in molecular studies [13]. Recently, some newly-developed methods have demonstrated high efficiency in the amplification of whole genomes from small amounts of template DNA. One of theses is the multiple displacement amplification method (MDA), a process that uses the bacteriophage φ29 DNA polymerase and exonuclease-resistant, thiophosphate-modified, degenerate hexamers to amplify genomic DNA from crude or pure sources and that has been used in some studies [14-19]. The enzyme φ29 DNA polymerase replicates DNA at a constant temperature of 30°C in a few hours without a thermal cycler and includes more complete genome coverage and unbiased amplification compared with PCR-based methods [14]. The goal of this study was to evaluate the use of MDA for the amplification of genomic DNA from siphonapterids preserved in scientific collections for long periods in 70% ethanol at room temperature.
Twenty-six of the samples analyzed were obtained from specimens deposited in the ectoparasites collection of the Department of Parasitology at the Universidade Federal de Minas Gerais and the Museum of Zoology at the Universidade de São Paulo. All of the samples were preserved in 70% ethanol at room temperature (Table 1). Samples used as controls [two Ctenocephalides felis felis (Bouché, 1835) and two Tunga penetrans (L., 1758)], were collected and preserved in absolute ethyl-alcohol and stored at - 20°C.
Table 1.
Sampling data for the species analyzed.
| Samples | Siphonapterids | Stages | Location | Hosts | Collection Date | Scientific collection1 |
|---|---|---|---|---|---|---|
| 1 | C. felis felis | 1 female | Belo Horizonte/MG | Canis familiaris | 2008 | UFMG |
| 2 | C. felis felis | 1 female | Serra do Cipó/MG | Leopardus pardalis | 1998 | UFMG |
| 3, 4 | C. felis felis | 2 females | Belo Horizonte/MG | C. familiaris | 1988 | UFMG |
| 5, 6 and 7 | C. felis felis | 3 females | Ouro Preto/MG | C. familiaris | 1979 | UFMG |
| 8 and 9 | T. penetrans | 1 female, 1 male | Vale do Mucuri/MG | ___ | 2007 | UFMG |
| 10, 11 and 12 | T. penetrans | 3 neosomes | Brasília/DF | Tamandua tetradactyla | 2002 | UFMG |
| 13 | T. caecata | 1 neosome | Santa Bárbara/MG | Akodon cursor | 2007 | UFMG |
| 14 | T. caecata | 1 neosome | Nova Lima/MG | Rhipidomys mastacalis | 2006 | UFMG |
| 15 | T. caecata | 1 neosome | Nova Lima/MG | R. mastacalis | 2005 | UFMG |
| 16 and 17 | T. caecata | 2 neosomes | Curitiba/PR | Rattus norvegicus | 1990 | UFMG |
| 18 | T. caecata | 1 neosome | Ouro Preto/MG | R. rattus | 1989 | UFMG |
| 19 | T. caecata | 1 neosome | Caratinga/MG | Nectomys squamipes | 1980 | UFMG |
| 20 | T. terasma | 1 female | Alegre/ES | Dasypus novemcinctus | 2003 | UFMG |
| 21 | T. terasma | 1 neosome | Unaí/MG | Priodontes maximus | 1996 | UFMG |
| 22 | T. travassosi | 1 neosome | Belo Horizonte/MG | Dasypus novemcinctus | 1966 | UFMG |
| 23 | T. travassosi | 1 neosome | São Paulo/SP | Dasypus novemcinctus | 1916 | USP |
| 24 | T. bondari | 1 neosome | Franca/SP | Cariama cristata | 1909 | USP |
| 25 and 26 | Tunga sp. | 2 neosomes | Itatiaia/RJ | Delomys dorsalis | 2000 | UFMG |
1 Department of Parasitology of the Universidade Federal de Minas Gerais (UFMG) and Museum of Zoology of the Universidade de São Paulo (USP). All samples were preserved in 70% ethanol and stored at room temperature.
Genomic DNA was isolated using the DNeasy Tissue Kit (QIAGEN). Samples of adult fleas or neosomes (Figure 1) were used as the sources for DNA extraction. The specimens (a flea or a neosome) were dissected, mechanically shredded, and incubated with lysis buffer containing Proteinase K for 4 h. Afterwards, we followed the recommended protocols supplied by the manufacturer [10]. Following extraction, we subjected each sample to two different methodologies: (1) amplification of mitochondrial 16S sequences without MDA; (2) amplification of 16S after MDA. The exoskeletons obtained from the dissection were permanently mounted on slides [3]. The Repli-g® Fast Mini kit (QIAGEN) was used for MDA of siphonapterid DNA, according to the manufacturer's instructions. The MDA-DNA preparations were stored at -20°C.
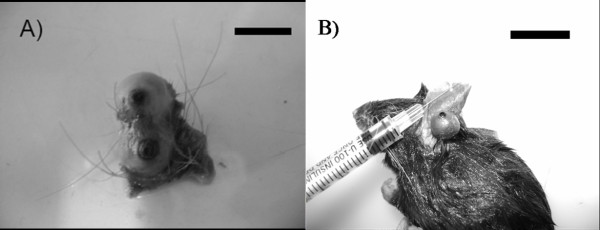
Figure 1.
Neosomes of some tungids. A) Tunga travassosi, female. Bar: 8 mm B) Tunga caecata, female. Bar: 12 mm.
PCR amplification of DNA from single flea or neosome samples was performed using the primers Sen-mt16S TAC ATA ACA CGA GAA GAC C and Rev-mt16S GTG ATT GCG CTG TTA TCC, as previously described [20]. The HotStar MasterMix Kit was used for PCR reactions, according to the manufacturer's instructions (QIAGEN). The samples were scored as positive for amplification if a PCR product of approximately 200 bp was detected. No-template control reactions were routinely performed.
PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN). The BigDye® Terminator v3.1 Cycle Sequencing Kit (APPLIED BIOSYSTEMS) was used for sequence determination by following the manufacturer's instructions. Sequencing was performed in an automatic sequencer (3130 × l APPLIED BIOSYSTEMS).
The sequences produced were analyzed using the CodonCode Aligner software (Phred algorithm) for the analysis of sequencing quality. Additionally, the samples obtained were submitted to the BLASTn tool from the National Center of Biotechnology Information (NCBI) to evaluate sequence similarity. The chi-square test with the Yates correction was used to compare the two methodologies: without or with MDA.
Morphological analysis identified the following species [3,21]: Tunga penetrans, T. bondari Wagner, 1932, T. caecata (Enderlein, 1901), T. terasma Jordan, 1937, T. travassosi Pinto & Dreyfus, 1927, Tunga sp. and C. felis felis (Table 1).
Without the use of MDA, only four samples (15.4%) displayed successful amplification of the 16s sequences (one of C. felis felis, collected in 2008; one of T. penetrans, collected in 2007 and two of T. caecata, collected in 2005 and 2007) (Figure 2). In contrast, application of the MDA technique permitted amplification from 24 samples (92.3%), presenting significant differences between the methods employed (χ2 = 27.92; p < 0.01). These samples varied greatly in their collection times: T. penetrans (N = 5, in 2002 and 2007); T. travassosi (N = 2, in 1916 and 1966); T. caecata (N = 6, in 1970, 1980, 1990, 2005, 2006, 2007); T. terasma (N = 2, in 1996, 2003); T. bondari (N = 1, in 1909); Tunga sp. (N = 2, in 2000); C. felis felis (N = 6, in 1979, 1988, 1998, 2008) (Figure 3). Both amplification protocols (with and without the use of MDA) generated fragments of approximately 200 bp when performed on positive control samples.
Figure 2.
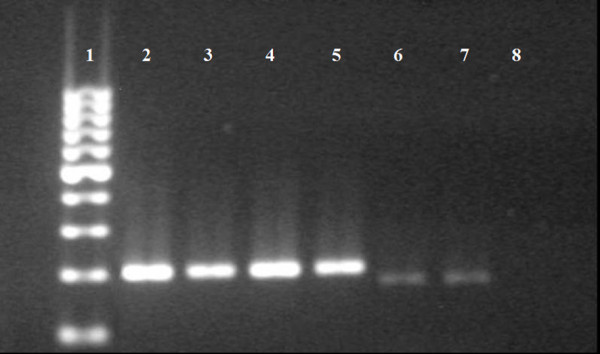
Amplification, without the use of the MDA technique, of the 200 bp fragment of the 16S mtDNA of samples from scientific collections preserved in 70% ethanol and stored at room temperature. 1) 100 bp DNA Marker; 2) Tunga penetrans (control); 3) Ctenocephalides felis felis (control); 4) C. felis felis (2008); 5) T. penetrans (2007); 6 and 7) T. caecata (2005 and 2007); 8) Negative control.
Figure 3.
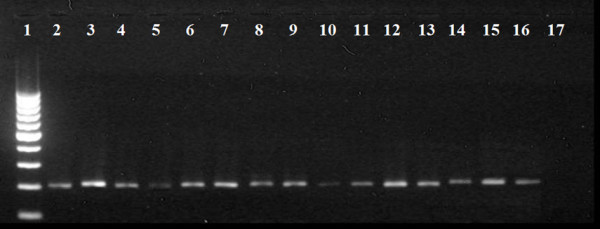
Amplification, with the use of the MDA technique, of the 200 bp fragment of the 16S mtDNA gene of samples from scientific collections preserved in 70% ethanol and stored at room temperature. 1) 100 bp DNA Marker; 2) Tunga penetrans (control); 3) Ctenocephalides felis felis (Control); 4 and 5) T. penetrans (2007 and 2002); 6 and 7) T. travassosi (1966 and 1916); 8, 9 and 10) T. caecata (2007, 1980 and 1989); 11 and 12) T. terasma (1996 and 2003); 13) T. bondari (1909); 14 and 15) Tunga sp. (2000); 16) C. felis felis (1988); 17) Negative control.
All the sequences obtained showed satisfactory quality (Phred ≥ 20) (Figure 4). The samples of T. penetrans evaluated using the BLASTn tool showed a 100% degree of similarity with the reference sequence. The other samples did not have specific sequence references, but the primary matches identified were always to samples belonging to the same genus (Figure 5).
Figure 4.
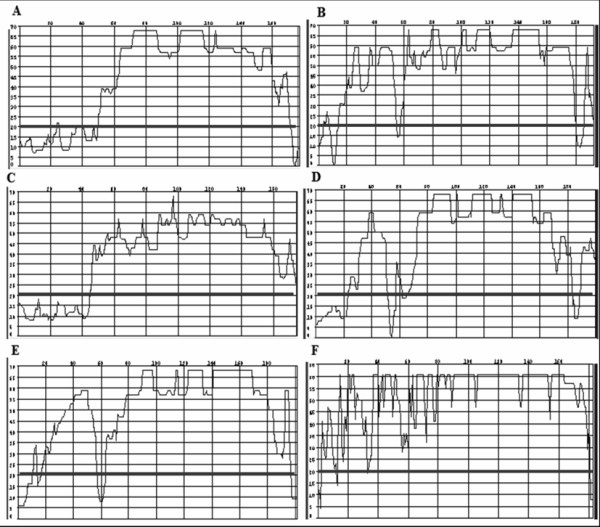
Analysis of sequence quality of the 16S mtDNA gene fragment using the CodonCode Aligner software. The black line denotes Phred = 20. A) Tunga penetrans; B) Ctenocephalides felis felis; C) T. caecata; D) T. travassosi; E) Tunga sp.; F) T. terasma.
Figure 5.

Analysis of similarity between some of the sequences (177 pb) of the 16S mtDNA gene fragment from fleas preserved in scientific collections and sequences deposited in GenBank, as analyzed using the BLASTn tool. A) Tunga penetrans collected in 2002; B) T. bondari collected in 1909.
Collections are vital for taxonomic research and the study of biodiversity [22]. Otherwise, insects preserved in absolute or 70% ethanol and maintained at room temperature show a loss in the quality and quantity of DNA, compromising their use in genetic analysis [13]. Our results support this observation, as we were only able to amplify the 200 bp fragment corresponding to the 16S region of the mitochondrial rDNA from recently preserved samples.
In this study, we demonstrate the highly effective use of a commercially available MDA-based method for whole genome amplification of siphonapterid DNA from samples of adults and neosomes preserved in 70% ethanol at room temperature. The use of the MDA technique prior to the amplification of the target DNA (mtDNA 16S) allowed us to detect the 200 bp fragment in samples preserved under non-ideal conditions for long periods, including from one specimen collected in 1909. MDA-based commercial kits have been successfully used in the amplification of total genomic DNA from a wide range of organisms, including humans [23], dogs [17], pygmy elephantids [19], nematodes [18], mites [16] and insects [15]. A question raised by some authors is the possibility of amplification of contaminating DNA, instead of DNA from the target organism [18]. The similarity analysis performed using the BLASTn tool excluded the possibility of contamination, since the sequences obtained showed a 100% similarity to known samples (T. penetrans). For samples lacking reference sequences in the NCBI database, the primary matches identified were all from specimens belonging to the same genus, reinforcing the absence of contamination.
One of the possible applications of the MDA technique is its use in the amplification of DNA from samples with historical value that have been maintained in scientific collections [15,16,18]. The results obtained here support this idea, as this is the first reported use of the MDA technique in the amplification of DNA from fleas stored in museums. It is important to point out that, concerning tungid fleas, a large part of samples preserved in scientific collections is constituted by neosomes removed from several sites of their hosts (Figures 1, 2). Although some museums do not allow dissection of type specimens due to the risks of damage [24], sometimes it is necessary to dissect neosomes for identification, since that a dissected specimen is not a damaged specimen. Among siphonapterids, many species are morphologically indistinguishable (eg, some female fleas Rhopalopsyllidae) or have their identification difficult due to loss of structures (eg, loss of legs and bristles in Tungidae, during the formation of the neosome) [3,21]. Moreover, the morphological characteristics used in the identification of species of fleas are often autapomorphic, having limited use for phylogenetic reconstruction [11]. On that account, sometimes fleas need to be genotyped. Otherwise, fleas act as vectors of several diseases, including bacteria, Protozoa and helminths [25-27], in which the quantity of endosymbionts available for molecular analysis is often limited.
The combined application of MDA and conventional protocols for siphonapterid DNA amplification may be a valuable new tool for molecular studies concerning taxonomy, phylogeny and epidemiology, involving samples of fleas that have, until now, been considered inadequately preserved for such purposes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PML and DMA contributed equally to this work by performing all experiments and writing the manuscript.
Contributor Information
Daniel M Avelar, Email: aveldan@gmail.com.
Pedro M Linardi, Email: linardi@icb.ufmg.br.
Acknowledgements
This research was part of the PhD thesis of D.M.A. in Parasitology/Post-graduate program in Parasitology/Instituto de Ciências Biológicas/Universidade Federal de Minas Gerais/UFMG. The authors wish to thank Dr. Carlos José Einecker Lamas from the Museum of Zoology of USP for the T. bondari and T. travassosi samples used in this study and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (National Council of Scientific and Technological Development) for the research fellowship for P.M.L.
References
- Hopkins GHE, Rothschild M. An Illustrated Catalogue of the Rothschild Collection of Fleas (Siphonaptera) in the British Museum (Natural History), Vol. I, Tungidae and Pulicidae. British Museum (Natural History), London; 1953. p. 361. [Google Scholar]
- Lewis RE. Résumé of the Siphonaptera (Insecta) of the world. J. Med.l Entomol. 1998;35:377–389. doi: 10.1093/jmedent/35.4.377. [DOI] [PubMed] [Google Scholar]
- Linardi PM, Guimarães LR. Sifonápteros do Brasil. Museu de Zoologia- USP/FAPESP, São Paulo, Brazil; 2000. p. 291. [Google Scholar]
- Pampiglione S, Fioravanti ML, Gustinelli A, Onore G, Mantovani B, Luchetti A, Trentini M. Sand flea (Tunga spp.) infections in humans and domestic animals: state of the art. Med. Vet. Entomol. 2009;23:172–186. doi: 10.1111/j.1365-2915.2009.00807.x. [DOI] [PubMed] [Google Scholar]
- Audy JR, Radovsky FJ, Vercammen-Grandfean PH. Neosomy: radical intrastadial metamorphosis associated with arthropod symbioses. J. Med. Entomol. 1972;9:487–494. doi: 10.1093/jmedent/9.6.487. [DOI] [PubMed] [Google Scholar]
- Rothschild M. Neosomy in fleas, and the sessile life-style. J. Zool. 1992;226:613–629. doi: 10.1111/j.1469-7998.1992.tb07504.x. [DOI] [Google Scholar]
- Witt LH, Linardi PM, Meckes O, Schwalfenberg S, Ribeiro RA, Feldmeier H, Heukelbach J. Blood-feeding of Tunga penetrans males. Med. Vet. Entomol. 2004;18:439–441. doi: 10.1111/j.0269-283X.2004.00533.x. [DOI] [PubMed] [Google Scholar]
- Bybee SM, Zaspel JM, Beucke KA, Scott CH, Smith BW, Branham MA. Are molecular data supplanting morphological data in modern phylogenetic studies? Syst. Entomol. 2010;35:2–5. doi: 10.1111/j.1365-3113.2009.00496.x. [DOI] [Google Scholar]
- Vobis M, D'Haese J, Mehlhorn H, Heukelbach J, Mencke N, Feldmeier H. Molecular biological investigations of brazilian Tunga isolates from man, dogs, pigs and rats. Parasit. Res. 2005;96:107–112. doi: 10.1007/s00436-005-1320-z. [DOI] [PubMed] [Google Scholar]
- Vobis M, D'Haese J, Mehlhorn H, Mencke N, Blagburne BL, Bond R, Denholm I, Dryden MW, Payne P, Rust MK, Schroeder I, Vaughn MB, Bledsoe B. Molecular phylogeny of isolates of Ctenocephalides felis and related species based on analysis of ITS1, ITS2 and mitochondrial 16S rDNA sequences and random binding primers. Parasit. Res. 2004;4:219–226. doi: 10.1007/s00436-004-1201-x. [DOI] [PubMed] [Google Scholar]
- Whiting MF, Whiting AS, Hastriter MW, Dittmar K. A molecular phylogeny of fleas (Insecta: Siphonaptera): origins and host associations. Cladistics. 2008;24:677–707. doi: 10.1111/j.1096-0031.2008.00211.x. [DOI] [Google Scholar]
- Levi HW. The care of alcoholic collections of small invertebrates. Syst. Zool. 1966;15:183–188. doi: 10.2307/2411389. [DOI] [PubMed] [Google Scholar]
- Post RJ, Flook PK, Millest AL. Methods for the preservation of insects for DNA studies. Biochem. Syst. Ecol. 1993;21:85–92. doi: 10.1016/0305-1978(93)90012-G. [DOI] [Google Scholar]
- Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochotegui-Escalante N, Black WC IV. Amplifying whole insect genomes with multiple displacement amplification. Insect Mol. Biol. 2003;12:195–200. doi: 10.1046/j.1365-2583.2003.00401.x. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. Multiple displacement amplification in combination with high-fidelity PCR improves detection of bacteria from single females or eggs of Metaseiulus occidentalis (Nesbitt) (Acari: Phytoseiidae) J. Invertebr. Pathol. 2004;86:111–116. doi: 10.1016/j.jip.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Short AD, Kennedy LJ, Forman O, Barnes A, Fretwell N, Wiggall R, Thomson W, Ollier WER. Canine DNA subjected o whole genome amplification is suitable for a wide range of molecular applications. J. Hered. 2005;96:829–835. doi: 10.1093/jhered/esi126. [DOI] [PubMed] [Google Scholar]
- Skantar AM, Catar LK. Multiple displacement amplification (MDA) of total genomic DNA from Meloidogyne spp. and comparison to crude DNA extracts in PCR of ITS1, 28S D2-D3 rDNA and Hsp90. Nematology. 2005;7:285–293. doi: 10.1163/1568541054879584. [DOI] [Google Scholar]
- Poulakakis N, Parmakelis A, Lymberakis P, Mylonas M, Zouros E, Reese DS, Glaberman S, Caccone A. Ancient DNA force reconsideration of evolutionary history of Mediterranean pygmy elephantids. Biol. Lett. 2006;2:451–454. doi: 10.1098/rsbl.2006.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A, Kornfield I. An improved amplification and sequencing strategy for phylogenetic studies using the mitochondrial large subunit rRNA gene. Genome. 1996;39:793–797. doi: 10.1139/g96-099. [DOI] [PubMed] [Google Scholar]
- Johnson PT. A classification of the Siphonaptera of South America with descriptions of new species. Mem. Entomol. Soc. Wash. 1957;5:1–298. [Google Scholar]
- Winston JE. Archives of a small planet: The significance of museum collections and museum-based research in invertebrate taxonomy. Zootaxa. 2007;1668:47–54. [Google Scholar]
- Maragh S, Jakupciak JP, Wagner PD, Rom WN, Sidransky D, Srivatava S, O'Connel CD. Multiple strand displacement amplification of mitochondrial DNA from clinical samples. BMC Med. Genet. 2008;9:7. doi: 10.1186/1471-2350-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Aguiar AP, Amorim DS. Knowledge of insect diversity in Brazil: chalenges and advances. Neotrop. Entomol. 2009;38:565–570. doi: 10.1590/S1519-566X2009000500001. [DOI] [PubMed] [Google Scholar]
- Jenkins DW. Pathogens, Parasites, and Predators of Medically Important Arthropods. Bull. WHO. 1964;30(Suppl):1–150. [PMC free article] [PubMed] [Google Scholar]
- Beard CB, Butler JF, Becnel JJ. Prevalence and biology of endosymbionts of fleas (Siphonaptera: Pulicidae) from dogs and cats in Alachua County, Florida. J. Med. Entomol. 1990;27:1050–1061. doi: 10.1093/jmedent/27.6.1050. [DOI] [PubMed] [Google Scholar]
- de Avelar DM, Bussolotti AS, Ramos MCA, Linardi PM. Endosymbionts of Ctenocephalides felis felis (Siphonaptera: Pulicidae) obtained from dogs captured in Belo Horizonte, Minas Gerais, Brazil. J. Invertebr. Pathol. 2007;94:149–152. doi: 10.1016/j.jip.2006.10.001. [DOI] [PubMed] [Google Scholar]