Abstract
Hyperactivation of phosphatidylinositol-3 kinase (PI3K) can occur as a result of somatic mutations in PIK3CA, the gene encoding the p110α subunit of PI3K. The HER2 oncogene is amplified in 25% of all breast cancers and some of these tumors also harbor PIK3CA mutations. We examined mechanisms by which mutant PI3K can enhance transformation and confer resistance to HER2-directed therapies. We introduced the PI3K mutations E545K and H1047R in MCF10A human mammary epithelial cells that also overexpress HER2. Both mutants conferred a gain of function to MCF10A/HER2 cells. Expression of H1047R PI3K but not E545K PI3K markedly upregulated the HER3/HER4 ligand heregulin (HRG). HRG siRNA inhibited growth of H1047R but not E545K-expressing cells and synergized with the HER2 inhibitors trastuzumab and lapatinib. The PI3K inhibitor BEZ235 markedly inhibited HRG and pAKT levels and, in combination with lapatinib, completely inhibited growth of cells expressing H1047R PI3K. These observations suggest that PI3K mutants enhance HER2-mediated transformation by amplifying the ligand-induced signaling output of the ErbB network. This also counteracts the full effect of therapeutic inhibitors of HER2. These data also suggest that mammary tumors that contain both HER2 gene amplification and PIK3CA mutations should be treated with a combination of HER2 and PI3K inhibitors.
Keywords: PIK3CA mutations, HER2 overexpression, HER3, Heregulin, Breast cancer
Introduction
HER2 (ErbB2) is a member of the ErbB family of transmembrane receptor tyrosine kinases, which also includes the epidermal growth factor receptor (EGFR), HER3, and HER4. Binding of ligands to the extracellular domain of EGFR, HER3 and HER4 induces the formation of kinase active homo- and heterodimers to which activated HER2 is recruited as a preferred partner (Yarden and Sliwkowski, 2001). Amplification of the HER2 gene occurs in 25% of invasive breast cancers where it is associated with poor patient prognosis (Nahta et al., 2006). Activation of the ErbB receptor network engages multiple effectors and signaling pathways including the phosphatidylinositol-3 kinase (PI3K) survival pathway (Engelman and Cantley, 2006) In HER2-overexpressing cells, a major mechanism of PI3K activation is heterodimerization of HER2 with kinase-deficient HER3 which, when phosphorylated, can directly couple to the p85 subunit of PI3K (Holbro et al., 2003). Treatment with HER2 inhibitors such as trastuzumab or lapatinib uncouples HER3 from p85, thus inhibiting PI3K (Engelman et al., 2007; Junttila et al., 2009; Ritter et al., 2007; Yakes et al., 2002). Trastuzumab, a monoclonal antibody that binds to the extracellular domain of HER2 is approved as therapy for HER2 gene-amplified breast cancers (Slamon et al., 2001) A large proportion of patients who initially respond to trastuzumab eventually develop resistance (Nahta et al., 2006). It is speculated that sustained inhibition of PI3K is essential for the antitumor effect of HER2 inhibitors. Indeed, presence of detectable PIK3CA mutations and/or loss or low levels of PTEN measured by IHC have been associated a lower response to trastuzumab and chemotherapy in patients with HER2+ tumors (Berns et al., 2007; Nagata et al., 2004) and to lapatinib in HER2+ cells (Eichhorn et al., 2008; Junttila et al., 2009; Serra et al., 2008).
PI3K, a lipid kinase consisting of a regulatory p85 and a catalytic p110 subunit, plays a central role in normal cellular growth and metabolism (Cantley, 2002). p110 phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3) at the plasma membrane. Pleckstrin homology (PH) domain-containing proteins involved in cell survival such as the serine/threonine kinase AKT are recruited by PIP3 to the plasma membrane where they become activated (Cantley, 2002). The PI3K-AKT pathway is commonly altered in human cancers (Vivanco and Sawyers, 2002). Mutations in PIK3CA are single nucleotide substitutions occurring in about 30% of several common cancers, including carcinoma of the breast, colon, endometrium, and prostate (Bachman et al., 2004; Campbell et al., 2004; Karakas et al., 2006; Samuels et al., 2004). About 80% of these mutations occur primarily at three “hotspots” (E542K, E545K in exon 9 and H1047R in exon 20) (Samuels et al., 2004), resulting in elevated catalytic activity of p110α (Carson et al., 2008) and cell transformation (Bader et al., 2006; Isakoff et al., 2005; Kang et al., 2005; Samuels et al., 2005). In breast cancers, PIK3CA mutations are associated with HER2 overexpression (Saal et al., 2005; Stemke-Hale et al., 2008) and correlate with lymph node metastases (Saal et al., 2005) and poor patient outcome (Lai et al., 2008; Lerma et al., 2008; Li et al., 2006). In this study, we have examined potential mechanisms by which the acquisition of PIK3CA mutations enhances HER2-mediated transformation in mammary epithelial cells and confer resistance to anti-HER2 therapies.
Results
E545K and H1047R PIK3CA mutants confer a gain of function to HER2-overexpressing cells
We stably transduced hemagglutinin (HA)-tagged wild-type (WT), E545K (EK) and H1047R (HR) PIK3CA retroviral vectors in HER2-overexpressing MCF10A human mammary epithelial cells. Since p110 requires p85 for its stability (Geering et al., 2007), p85 levels limit the overexpression of transfected p110. Consistent with this, total p110 levels were overall similar in control and transfected cells (Figure 1a). We next evaluated the effects of EK and HR mutants on PI3K signaling in medium supplemented with serum, EGF, and insulin. Cells harboring the HR mutant, but not WT or EK mutated PI3K, exhibited elevated levels of activated AKT (pAKTS473 and pAKTT308). There was also an increase in phosphorylation of glycogen synthase kinase 3 (GSK3) and ribosomal protein S6, targets downstream AKT and TORC1, respectively (Engelman and Cantley, 2006) in both mutants compared to WT PI3K (Figure 1b). PI3K activation leads to cell cycle entry via stabilization of Cyclins (Cantley, 2002). Accordingly, we detected higher levels of Cyclins D1 and D2 in mutants, especially HR, but not WT PI3K expressing cells (Figure 1b).
Figure 1.
PI3K mutants increase transformation of MCF10A/HER2 cells. (a) Immunoblot (IB) comparing levels of HA, p110α, p85, in MCF10A/HER2 (HER2), MCF10A/HER2/WT PIK3CA (WT), MCF10A/HER2/E545K PIK3CA (EK) and MCF10A/HER2/H1047R PIK3CA (HR) cells. The HA tag was detected in cells expressing WT and mutant PI3K but not in parental MCF10A/HER2 cells. (b) IB comparing levels of total and phosphorylated AKT, S6, GSK3, total Cyclins D1 and D2 in WT, EK and HR cells. (c) 3D acinar structures of HER2, WT, EK and HR cells grown for 18 days on Matrigel ± 250f nM BEZ235 (BEZ). (d) Anchorage-independent growth of HER2, WT, EK and HR cells in soft agarose for 7 days. (e) Indirect immunofluorescence staining of cleaved caspase-3 on day 7 WT, EK and HR acini. Blue, nuclei (DAPI); green, cleaved caspase-3. (f) Transwell motility assay with WT, EK and HR cells for 24 h. (g) Invasion assay with Matrigel-coated transwell filters for 42 h.
MCF10A cells form polarized, quiescent acini in 3D basement membrane. Activation of HER2 in these cells reinitiates proliferation, disrupts tight junction polarity, and induces acinar expansion without invading into the surrounding matrix (Muthuswamy et al., 2001). Cells expressing PI3K mutants formed larger multiacinar structures whereas cells expressing WT PI3K or vector alone formed smaller, less complex acini with central necrosis (Figure 1c). In all cases, growth was prevented by BEZ235 (Figure 1c), a small molecule inhibitor of p110α with an IC50 against WT, HR, and EK PI3K of 4-5.7 nM (Maira et al., 2008). By immunofluorescence, WT cells exhibited staining with the apoptosis marker cleaved caspase-3, whereas in acini expressing EK and HR PI3K, caspase staining was almost undetectable (Figure 1e). Further, MCF10A/HER2 cells expressing the mutants formed large colonies in soft agarose (Figure 1d). Additionally, MCF10A/HER2 cells expressing mutant PI3K were more motile and invasive compared to cells expressing WT PI3K as measured by their ability to migrate to the underside of transwell filters without or with Matrigel coating after 24 and 42 h, respectively (Figures 1f, g).
For optimal growth, MCF10A cells require addition of EGF and insulin to the cell medium. When maintained in presence of serum and growth factors, WT and mutants proliferated at about the same rate, however, under serum and growth factor-deprivation, cells expressing the mutants (HR>EK) proliferated faster than cells expressing WT PI3K (Figure S1). Collectively, these results suggest that 1) stable expression of EK and HR PI3K enhances HER2-mediated transformation; and 2) the H1047R mutant is more transforming than E545K PI3K in HER2-overexpressing cells.
PI3K mutants upregulate pHER3 and enhance the association of p85 with HER3
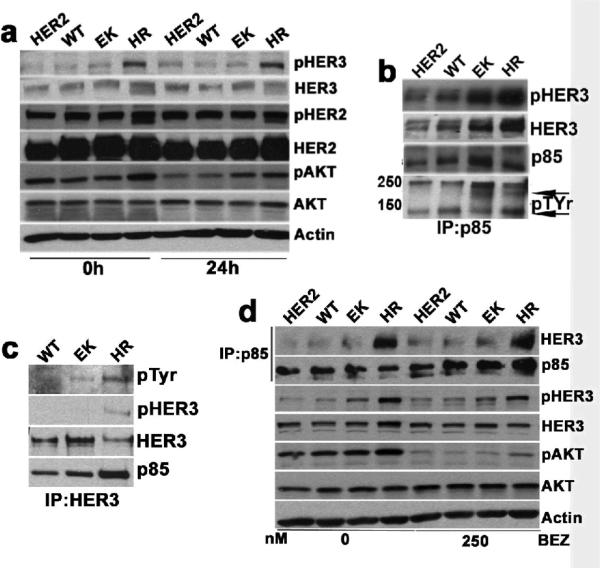
Consistent with their ability to grow better in low serum, cells expressing EK and HR PI3K maintained pAKTS473 in 0.5% serum. Cells expressing the mutants, especially HR PI3K, maintained activated HER3 as measured by pHER3Y1289 (Figure 2a). Because of the tight physical association between PI3K and its upstream activator(s), it is possible to identify PI3K-activating receptor tyrosine kinase(s) or tyrosine phosphorylated adaptor(s) by immunoprecipitation with p85 antibodies. This approach has been used to identify upstream activators of PI3K in drug-resistant cancer cells (Engelman et al., 2007; Guix et al., 2008). Therefore, p85 was precipitated from cells and the antibody pulldowns were subjected to phosphotyrosine (pTyr) immunoblotting. A pTyr band of ~200-kDa co-precipitated with p85 only in cells expressing EK and HR mutants (Figure 2b, bottom row, upper arrow). This band was confirmed to be HER3 by immunoblot (Figure 2b). The lower arrow in this blot indicates a non-specific band. Consistent with this result, precipitation with a HER3 antibody recovered a ~200-kDa pTyr band (which likely represents the p85-associated pHER3 as shown by others (Engelman et al., 2007; Ritter et al., 2007)) in both EK and HR-expressing cells. Although the HER3 immunoprecipitation recovered less HER3 in HR cells, the amount of p85 associated with HER3 was more in these cells than that in the WT or EK mutant expressing cells (Figure 2c).
Figure 2.
PI3K mutants maintain pAKT and pHER3 under serum and growth factor deprivation. (a) IB comparing pHER2Y1248, pHER3Y1289 and pAKTS473 levels in HER2, WT, EK and HR cells. Cells were harvested at 0 and 24 h following serum and growth-factor deprivation. (b and c) p85-HER3 association in WT, EK and HR cells. Cell lysates were immunoprecipitated (IP) with p85 (b) and HER3 (c) antibodies, followed by IB with pTyr, HER3, pHER3Y1289 and p85 antibodies. (d) Association of pAKT, pHER3 and p85 with HER3 in cells treated with 250 nM BEZ for 6 h in growth media. IP was performed with p85 antibody. IB were performed with the antibodies indicated to the right of the panel.
To determine whether AKT activation was dependent on mutant PI3K, we treated cells with a PI3K inhibitor. Treatment with BEZ235 for 6 h markedly reduced pAKTS473 without affecting pHER3 or the association of p85 with HER3 (Figure 2d).
RNAi of HER3 partially inhibits MCF10A/HER2 cells expressing mutant PI3K
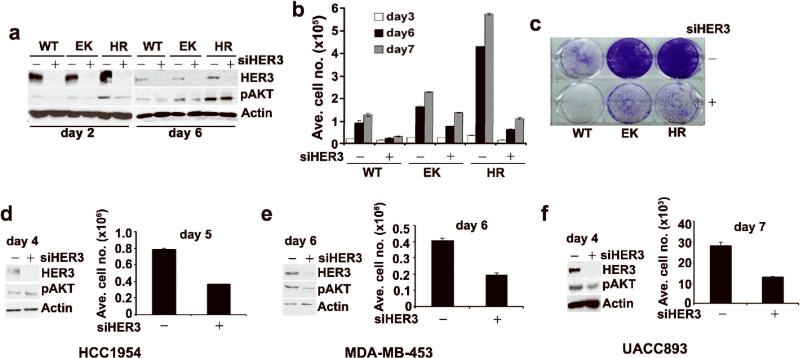
In HER2-overexpressing cells, the co-receptor HER3 directly couples to p85 and activates PI3K-AKT (Holbro et al., 2003; Lee-Hoeflich et al., 2008; Yakes et al., 2002). Thus, to determine if the PI3K mutants still depend on binding to HER3, we transfected MCF10A/HER2 cells expressing WT or mutant PI3K with HER3 or mismatch siRNA. HER3 levels were undetectable within 2 and 6 days post-transfection (Figure 3a). Growth of cells expressing WT PI3K was completely arrested upon HER3 knockdown whereas this inhibition was only partial although more obvious in H1047R than in E545K cells (Figures 3b, c). Consistent with the results on cell growth, pAKTS473 was partially downregulated upon HER3 knockdown in EK and HR cells at day 2 but recovered at day 6 particularly in cells expressing H1047R PI3K (Figure 3a). To confirm that the results were not due to off-target effect of siRNA, we used two different siRNAs and obtained similar results. Both siRNAs decreased the HER3 protein levels up to similar extent (Figure S2). We next extended this observation to HER2-overexpressing breast cancer cells expressing H1047R PI3K (HCC1954, MDA-MB-453 and UACC893). Transfection with the HER3 siRNA partially inhibited growth of all 3 cell lines but had minimal or no effect on pAKTS473 levels (Figures 3d-f). These data suggest that the PI3K mutants can both activate AKT and maintain some growth with partial autonomy from HER3.
Figure 3.
RNAi of HER3 partially inhibits growth of PI3K mutant cells. (a) IB comparing HER3 and pAKT levels in cells transfected with either control or HER3 siRNA duplexes. Cells were harvested on day 2 and 6 post-transfection. (b) Determination of cell numbers on day 3, 6 and 7. Each bar represents the mean ± SE of six replicates. (c) Crystal violet staining of transfected cells on day 7. (d) HCC-1954; (e) MDA-MB-453; and (f) UACC893 cells were transfected with control or HER3 siRNA duplexes as indicated Materials and Methods. HER3 and pAKT levels were determined by IB (day 4: HCC1954; day 6: MDA-MB-453; day 4: UACC893). Differences in cell number between control and HER3 siRNA-transfected cells on day 4 for HCC1954; day 5 for MDA-MB-453 and day 7 for UACC893 were determined in a Coulter counter.
In HER2 gene amplified cells, HER3 phosphorylation depends on the HER2 kinase activity (Holbro et al., 2003). To document this, we treated cells with the HER2 TKI lapatinib and followed HER3 phosphorylation. Lapatinib reduced pHER2Y1248 levels in cells expressing WT and mutant PI3K. Initially, lapatinib also inhibited pHER3Y1289 and pAKTS473 and the coupling of p85 with HER3 in cells with WT and EK PI3K. However, cells expressing HR PI3K maintained pHER3, pAKT, pERK, and the association of p85 with HER3 (Figure S3). This result implied that HR mutant expression was associated with cellular factors that maintained HER3 phosphorylation independent of tyrosine kinase function of HER2.
Cells expressing H1047R PI3K overexpress HER3/HER4 ligands
Breast cancer cells often express high levels of ErbB receptor-activating ligands (Revillion et al., 2008). We performed real-time qPCR analysis to compare relative levels of ErbB ligands (epidermal growth factor/EGF, transforming growth factor-α/TGFα, heparin binding-epidermal growth factor/HBEGF, epiregulin/EREG, amphiregulin/AREG, betacellulin/BTC and heregulin/HRG). Cells expressing HR mutant, but not EK or WT PI3K produced high levels of HRG and EREG mRNA. The HB-EGF mRNA levels were similar in both EK and HR-expressing cells. The most significant of these was HRG mRNA, which was up regulated by >160-fold in HR cells (Figure 4a); this was confirmed by immunoblot analysis of whole cell lysates (Figure 4b). All three ligands bind HER4, while HRG is the primary ligand for HER3 (Yarden and Sliwkowski, 2001). Consistent with HER4 activation, precipitation of HER4, followed by immunoblot with p85 and pTyr antibodies, revealed a constitutive association of HER4 with p85 and HER4 phosphorylation (Figure 4c).
Figure 4.
H1047R PI3K mutant cells overexpresses HER3/HER4 ligands. (a) Real-time qPCR comparing HB-EGF, EREG and HRG mRNA levels relative to housekeeping control FLJ22101 in MCF10A/HER2/WT, EK and HR cells. Each data point represents the mean ± SE of six readings. (b) IB comparing HRG levels in MCF10A/HER2/WT, EK and HR cells. (c) Comparison of pHER4 levels in MCF10A/HER2/WT, EK and HR cell lysates by IP with a HER4 antibody followed by IB with HER4, p85 and pTyr antibodies. (d) Real-time qPCR comparing HRG RNA levels in SKBR3 cells stably expressing the WT, E545K and H1047R PI3K (SKWT, SKEK and SKHR, respectively). (e) IB comparing the pHER2Y1248, pHER3Y1289, pHER3Y1222, pHER3Y1197, HER3, pAKTS473 and pERKT202 levels in SKWT, SKEK and SKHR cells treated with 1 μM Lapatinib for 0, 6 and 24 h. (f) IB comparing endogenous HRG protein levels in HER2-overexpressing cells SKBR3: WT; BT474: K111N (weakly oncogenic); HCC1954, UACC893: H1047R. MCF10A/HER2/HR cells: all harbor H1047R PI3K.
HRG RNA overexpression was also observed in SKBR3 cells (endogenous WT PI3K) stably transduced with H1047R but not with E545K and WT PI3K (Figure 4d). Similar to the results with MCF10A/HER2/HR cells (Figure S3), inhibition of pAKT and pERK with lapatinib was dampened in SKBR3/HR cells compared to the other two cell lines (Figure 4e). However, we did not observe a better recovery of pHER3 (Y1289, Y1222 and Y1197) in the SKBR3/HR cells than in the SKBR3/WT and EK cells following treatment with lapatinib over a 24-h time course (Figure 4e). This was probably due to much lower expression level of HRG in SKBR3/HR cells than that in the MCF10A/HER2/HR cells (4-fold versus 161-fold). Immunoblot analysis with lysates collected from human breast cancer lines harboring both amplified HER2 and mutant PI3K revealed higher levels of HRG protein in HCC1954 and UACC893 compared to BT-474 and SKBR3 cells (Figure 4f). HCC1954 and UACC893 cells have endogenous H1047R mutation whereas SKBR3 and BT474 cells express WT and a poorly oncogenic K111N mutant PI3K, respectively (Gymnopoulos et al., 2007; Saal et al., 2005). These differences between the EK and HR mutants are not inconsistent with different structural and biochemical properties of these mutants which we will discuss below.
RNAi of HRG inhibits growth of cell expressing H1047R but not E545K PI3K
To determine if autocrine HRG was important for MCF10A/HER2/HR cell growth, we transfected cells with control or HRG siRNA duplexes. Four days post-transfection, there was sustained reduction (~70%) of HRG mRNA (Figure 5a), which correlated well with HRG protein levels and with basal HER3 and HER4 activation, as measured with pHER3Y1289 and pHER4Y1284 antibodies (Figures 5b,c). HRG knockdown also resulted in uncoupling of p85 from HER3 and HER4 (Figure 5d). Further, cells transfected with HRG siRNA grew slower than control siRNA transfected cells (Figure 5e). In a head to head comparison, transfection of HRG siRNA oligonucleotides inhibited growth of MCF10A/HER2 cells expressing H1047R but not E545K cells (Figure 5f,g). HRG knockdown had minimal effect on growth of MDA-MB-361 cells, which contain HER2 gene amplification and E545K PI3K (Fig. 5h). We next added to MCF10A/HER2/WT cells serum-free medium that had been conditioned by HR cells transfected with control or HRG siRNA duplexes. Conditioned medium (CM) from control siRNA, but not HRG siRNA transfected cells upregulated pAKTS473 and pHER3Y1289 in WT cells (Figure S4a). In line with these results, WT cells incubated with CM from control siRNA transfected HR cells proliferated faster than cells incubated with CM from cells where HRG had been downregulated (Figure S4b). These data suggest that cells that contain H1047R PI3K and high levels of HER2 overexpress HRG which, in turn, can activate HER3 and HER4 in autocrine and paracrine fashion to promote cell growth.
Figure 5.
RNAi of HRG inhibits growth of H1047R but not E545K PI3K mutant cells. (a) Real-time qPCR analysis of HRG mRNA in control and HRG siRNA transfected MCF10A/HER2/HR cells. (b and c) IB comparing pHER3Y1289 (b) and pHER4Y1284 (c) levels in control and HRG siRNA transfected MCF10A/HER2/HR cells. (d) Association of p85 with HER3 and HER4 in control versus MCF10A/HER2/HR cells transfected with HRG siRNA (day 4 post-transfection). Cell lysates were precipitated with p85 antibody followed by IB with p85, HER3 and HER4 antibodies. (e) MCF10A/HER2/HR cell numbers on day 4, 5 and 6 following transfection with HRG siRNA. Each data point represents the mean ± SE of six replicates. (f) IB comparing pHER3Y1289 and HRG levels in control and HRG siRNA transfected MCF10A/HER2/EK and HR cells on day 4 post-transfection. (g) Determination of MCF10A/HER2/EK and HR cell numbers on day 7 after transfection with HRG siRNA. Each data point represents the mean ± SE of six replicates. NS, not significant; *, p<0.05, paired t-test. (h) MDA-MB-361 cells were transiently transfected with control or HRG siRNA. Cell numbers were measured in a Coulter counter on day 7 after transfection. Each data point represents the mean ± SE of six replicates. (i) IB comparing pAKTS473 levels in MCF10A/HER2/HR cells treated with vehicle (Ctrl), 20 μM LY294002 (LY) and 250 nM BEZ235 (BEZ) for 24 h. (j and k) Real-time qPCR comparing HRG mRNA levels in MCF10A/HER2/HR (j) and HCC1954 (k) cells treated or not (Ctrl) with LY and BEZ.
To determine whether HRG expression depends on the catalytic activity of mutant PI3K, we performed qPCR analysis on RNA collected from MCF10A/HER2/HR cells treated with either LY294002 or BEZ235. Treatment with each of the PI3K inhibitors almost completely eliminated pAKTS473 in MCF10A/HER2/HR cells (Figure 5i) and markedly reduced HRG mRNA levels in these and HCC1954 cells which express endogenous H1047R PI3K (Figure 5j,k). These data suggest that, in cells harboring H1047R PI3K, HRG expression is at least partially dependent on mutant PI3K.
HRG knockdown sensitizes HER2-overexpressing cells to HER2 inhibitors
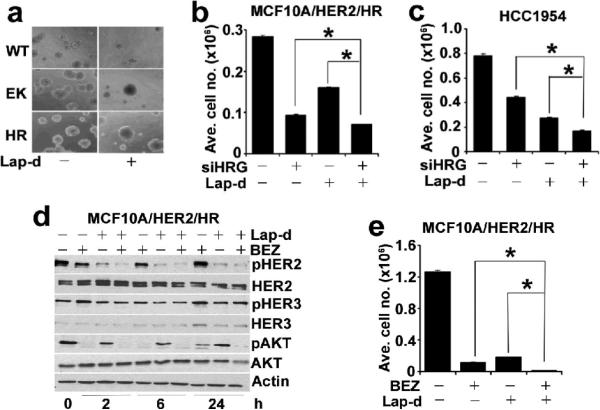
Finally, we examined whether interference of the HRG/HER3/PI3K pathway in cells expressing the exon 20 mutant would enhance the attenuated response to inhibitors of HER2. In 3D Matrigel, growth of MCF10A/HER2 acini expressing WT PI3K but not EK or HR PI3K was markedly inhibited by 1 μM lapatinib (Figure 6a). HER2-dependent breast cancer cells that are sensitive to lapatinib exhibit an IC50 of <0.1 μM (Konecny et al., 2006). The IC50 of lapatinib for HCC1954 cell growth is ≥0.3 μM (not shown). Although treatment with lapatinib inhibited pHER2Y1248 and pHER3Y1289 in HCC1954 cells, it did not inhibit pAKTS473, suggesting HER2-independent activation of PI3K (Figure S5). In both MCF10A/HER2/HR and HCC1954 cells, both lapatinib and HRG knockdown inhibited cell growth but the combination was significantly more inhibitory than each alone (Figures 6b,c). Similar experiments were performed to determine whether HRG knockdown would abrogate resistance to trastuzumab. Treatment with trastuzumab modestly inhibited growth of both cells. However, transfection of HRG siRNA oligonucleotides resulted in a more significant growth inhibition compared to trastuzumab and the combination was more effective than each intervention alone (Figure S6).
Figure 6.
RNAi of HRG synergizes with HER2 TKI lapatinib. (a) 3D structures of MCF10A/HER2 cells expressing WT, EK or HR PI3K grown for 14 days on Matrigel in absence and presence of 1 μM lapatinib. b and c, MCF10A/HER2/HR (b) and HCC1954 (c) cells were transfected with control or HRG siRNAs and treated with 0.03 and 0.1 μM lapatinib, respectively. Cells numbers were measured on day 5. Each data point represents the mean ± SE of nine replicates. (d) pHER2Y1248, pHER3Y1289 and pAKTS473 in cells treated with 1 μM lapatinib and/or 250 nM BEZ by IB. (e) MCF10A/HER2/HR cells were treated with 1 μM lapatinib and/or 250nM BEZ for 6 days and cell numbers were counted. Each bar represents the mean ± SD of six replicates. *, p<0.05, paired t-test.
In MCF10A/HER2/HR cells, a combination of HRG knockdown with lapatinib resulted in a marked reduction in pHER2Y1248 and pHER3Y1289 with further inhibition of pAKTS473 compared to lapatinib alone (Figure S7). The addition of trastuzumab following HRG knockdown did not inhibit pHER2Y1248 or pHER3Y1289 any different than siHRG alone (Figure S7) perhaps explaining why it minimally contributed to the antitumor effect of HRG knockdown in these cells (Figure S6). This result further suggests that upregulation of HRG in cells expressing H1047R PI3K amplifies signaling by the ErbB receptor network and reduces the antitumor action of HER2 inhibitors. However, it does not rule out the possibility that H1047R PI3K may have HER3- and HER2-independent functions contributing to AKT activation and escape from lapatinib or trastuzumab. In fact, lapatinib treatment did not inhibit pAKT and pHER3 despite inhibition of pHER2 in HR cells (Figure S3). This result also implies that the combination of lapatinib with an inhibitor of mutant PI3K would be more effective than the combination of lapatinib and HRG knockdown. To test this possibility, we treated MCF10A/HER2/HR cells with BEZ235, lapatinib, or the combination over a time course. Treatment with BEZ235 and lapatinib alone initially inhibited pAKTS473 but this was partially restored at 24 h. At this time, only the combination of lapatinib and BEZ235 inhibited pAKT completely (Figure 6d). In line with these data, only the combination was completely inhibitory for MCF10A/HER2/HR cell growth after 6 days (Figure 6e).
Discussion
Hyperactivation of the PI3K-AKT pathway is considered critical for the initiation and maintenance of human tumors (Garcia-Echeverria and Sellers, 2008). In breast cancer, constitutive PI3K-AKT activation occurs through HER2 amplification, somatic mutations in PIK3CA and AKT1, AKT2 amplification, and loss of the tumor suppressor PTEN (Bachman et al., 2004; Bellacosa et al., 1995; Carpten et al., 2007; Karakas et al., 2006; Li et al., 1997; Samuels et al., 2004). HER2 amplification has been shown to coexist with PIK3CA activating mutations (Saal et al., 2005; Stemke-Hale et al., 2008), suggesting that both molecular alterations may exert additive mechanisms of oncogenicity (Cantley and Yuan, 2008). In order to test these hypotheses, we ectopically expressed the E545K and H1047R “hotspot” mutations in HER2-overexpressing MCF10A human mammary epithelial cells.
HER2 overexpression in MCF10A cells resulted in elevated PI3K activity and pAKT levels (Wang et al., 2008 ). Stable expression of H1047R PI3K in MCF10A/HER2 cells induced a further increase in pAKT, consistent with the increased lipid kinase activity of this mutant compared to WT PI3K (Isakoff et al., 2005). The lack of detectable AKT activation by cells expressing the E545K mutant is consistent with a recent study which showed that in some breast cancer cells with PIK3CA mutations, AKT is minimally activated whereas phosphoinositide-dependent kinase 1 (PDK-1), another direct target of PI3K, is highly active (Vasudevan et al., 2009). Under serum deprivation, however, pAKT was activated in both EK and HR cells but not in cells expressing WT PI3K. Compared to control cells, mutant-expressing cells formed larger invasive acini in 3D with less apoptosis and larger colonies in soft agarose. In addition, the PI3K mutant cells were more motile and invasive and less dependent on added serum and growth factors for proliferation (Figures 1 and S1). These data indicate that expression of PI3K mutants enhanced the transformed phenotype of human mammary epithelial cells above that conferred by HER2 overexpression alone.
PI3K activation requires direct interaction of its regulatory subunit p85 with phosphotyrosine residues in activated growth factor receptors or adaptors (Cantley, 2002). In HER2-overexpressing cells, the HER2 co-receptor HER3 is the main adaptor that, upon phosphorylation of tyrosine residues in its cytoplasmic tail, engages and activates PI3K (Holbro et al., 2003). RNAi of HER3 partially inhibited growth of cells expressing mutant PI3K (H1047R>E545K) (Figure 3), suggesting that mutant kinases still associate with HER3 in order to be fully active. However, pAKT was partially maintained in cells where HER3 had been knocked down, implying that the PI3K mutants activated AKT and partially maintained cell growth in a HER3-independent manner. Addition of the HER2 TKI lapatinib inhibited HER2 phosphorylation, yet pHER3 and pAKT were maintained particularly in cells expressing H1047R PI3K (Figure S3), suggesting the presence of compensatory factors aimed at maintaining signaling output by the ErbB network.
ErbB ligands HB-EGF, EREG, and HRG were overexpressed mainly in cells expressing HR PI3K. Overexpression of HER3/HER4 ligand HRG was >160-fold in HR-expressing cells where more robust HER3 and HER4 phosphorylation had been observed (Figures 4 and 2). Autocrine HRG secretion is common in breast cancer cells where it confers a more malignant phenotype (Tsai et al., 2003). HRG Knockdown inhibited pHER3 and pHER4, the association of both HER3 and HER4 with p85, and growth of cells expressing H1047R but not E545K PI3K (Figures 5e-h, 6b,c). Interestingly, this growth inhibition was superior to that induced by trastuzumab (Figure S6). Consistent with this result, previous reports have shown that EGFR overexpression and HER3/4 ligands confers resistance to trastuzumab likely because of the reported inability of the antibody to block ligand-induced HER2-containing heterodimers (Ritter et al., 2007). These results also suggest that HRG is an important growth factor in cells driven by the H1047R PI3K mutant and that therapeutic antibodies against HER3 may have a role in tumors harboring H1047R PI3K.
Although both mutants enhanced HER2-mediated transformation, there were differences between both. Cells expressing H1047R PI3K appeared more dependent on HER3 (Figures 2 and 3) and maintained higher levels of pAKT, pS6, Cyclins D1, D2 and HRG (Figures 1 and 4). Differences between both mutants are not unprecedented. For example, in chick embryo fibroblasts, the transforming effect of the E545K mutant is independent of binding to p85 but requires interaction with RAS-GTP (Zhao and Vogt, 2008). In contrast, H1047R is transforming in absence of RAS-GTP binding but is highly dependent on its interaction with p85 (Zhao and Vogt, 2008). IRS-1 has been shown to activate WT and H1047R but not E542K PI3K (Carson et al., 2008), suggesting differences in growth factor dependence between both mutants. Further, crystallographic data showed that E542K disrupts the inhibitory interaction of p110α with the amino-terminal SH2 domain of p85, thus enhancing its catalytic activity (Miled et al., 2007). Related to this, Lee et al. (Lee et al., 2007) speculated that the p85 nSH2 domain, unencumbered by its interaction with the helical domain of p110α, remains more tightly associated with tyrosine phosphorylated receptors and adaptors at the cell membrane, thus protecting critical tyrosine residues from dephosphorylation and thereby prolonging production of PIP3. Similar data for H1047R PI3K have not been reported. However, structural analysis led to the speculation that this mutation most likely has a direct effect on the conformation of the p110α activation loop, changing its interaction with phosphoinositide substrates (Huang et al., 2007).
Cells expressing H1047R PI3K were partially resistant to the HER2 TKI lapatinib (Figure 6). MCF10A/HER2/HR and HCC1954 cells maintained pAKT despite inhibition of the HER2 kinase (Figures 6, S3 and S5). In MCF10A/HER2/HR cells, maintenance of pHER3 cannot be explained by persistent EGFR and HER4 phosphorylation as lapatinib also inhibits both receptors [(Medina and Goodin, 2008) and data not shown]. This suggests the possibility of HER3 transactivation by another kinase when EGFR, HER2, and HER4 are inhibited. Nonetheless, in MCF10A/HER2/HR and HCC1954 cells, the combination of lapatinib with HRG siRNA inhibited growth significantly better than each alone (Figure 6), thus implying that HRG upregulation shifts the dose response to lapatinib.
In summary, the data herein indicate that PI3K mutations enhance HER2-mediated transformation. Expression of H1047R p110α amplified signaling output of the ErbB receptor network by upregulating ErbB ligand expression, thus counteracting the response to the HER2 inhibitors trastuzumab and lapatinib. Although the PI3K mutants still coupled to HER3, cell growth and AKT activation upon HER3 knockdown were partially maintained in HER2-overexpressing cells with mutant PI3K. This suggests that p110α mutants conferred PI3K activation and viability in HER2-overexpressing cells without complete dependence on HER3. Taken together, these data imply two not mutually exclusive possibilities: 1) there is a kinase or adaptor other than HER3 responsible for activation of the mutants, and 2) the PI3K mutants are constitutively active in absence of binding to any kinase or adaptor. Finally, only combined inhibition of PI3K and HER2 with BEZ235 and lapatinib completely inhibited growth of MCF10A/HER2/HR cells (Figure 6). Based on these data, we propose that combined inhibition of PI3K and HER2 should be a preferred approach against cancers that contain both HER2 gene amplification and PIK3CA activating mutations.
Materials and Methods
Cell lines, plasmids, viruses and inhibitors
All cell lines were purchased from American Type Culture Collection. MCF10A/HER2 cells were generated and maintained as described previously (Ueda et al., 2004; Wang et al., 2006). Improved Minimal Essential Medium (IMEM) and fetal calf serum (FCS) were purchased from Invitrogen. HA–tagged WT or EK and HR PI3K variants cloned into JP1520 retroviral vector were described previously (Isakoff et al., 2005). Retrovirus production and generation of stable clones were described previously (Wang et al., 2006). LY294002, trastuzumab and lapatinib ditosylate (Lap-d) were purchased from Calbiochem, Vanderbilt University Medical Center Pharmacy and LC Laboratories, respectively. NVP-BEZ235 was provided by Carlos Garcia-Echeverria (Novartis, Basel).
Cell growth and viability assays
For growth in monolayer, 2.5×104 cells were seeded in 12-well plates and allowed to grow in presence or absence of inhibitors. Cells were harvested by trypsinization and cell number was determined in a Beckman Coulter counter. For crystal violet assay 5×104 cells were seeded in 6-well plates and grown for indicated time ± inhibitors, fixed in methanol, stained with crystal violet, and photographed. Anchorage independent growth was measured in a soft agarose colony formation assay as described previously (Wang et al., 2006).
Three-dimensional (3D) morphogenesis and indirect immunofluorescence
3D morphogenesis, indirect immunofluorescence and confocal analyses were performed as described previously (Wang et al., 2006). The cleaved caspase-3 (Asp175) antibody was from Cell Signaling Technology. Fluorescent secondary antibodies were from Molecular Probes.
Motility and invasion assays
Motility was measured by using 8-μm pore polycarbonate transwell filters (Corning Costar) as described (Wang et al., 2006). Similar protocol was followed for invasion assay with Matrigel-coated transwell filters from BD Biosciences.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed using published protocols (Isakoff et al., 2005; Wang et al., 2006; Wang et al., 2008 ). Primary antibodies included: AKT, pAKTS473, pAKT308, pHER2Y1248, pHER3Y1289, pHER3Y1222, pHER3Y1197, pHER4Y1284, S6, pS6S240/244, GSK-3β, pGSK-3α/βS21/9, HRG (Cell Signaling Technology); pTyr (BD Transduction Laboratories); p85 (Millipore); β-Actin (Sigma); HER2 (NeoMarkers); HER3, Cyclin D1, Cyclin D2 (Santa Cruz Biotechnology). The rabbit HER4 polyclonal antibody was a gift from H. Shelton Earp (University of North Carolina).
Real-time qPCR analysis
RNA was isolated using TRIzol (Invitrogen). Protocols for reverse transcription, qPCR and subsequent data analysis were described previously (Wang et al., 2008 ). Sequences for primer sets for EGF, TGFα, HB-EGF, EREG, AREG, BTC, HRG, and Human cDNA FLJ22101 (housekeeping gene) were described previously (Wang et al., 2008 ).
RNAi studies
siRNA duplexes for mismatch control and human HER3 were described in ref (Wang et al., 2008 ). ON-TARGETplus SMARTpool siRNAs against HRG were obtained from Thermo Scientific. Transfections were performed with Lipofectamine RNAiMAX (Invitrogen).
Supplementary Material
Acknowledgments
We thank Drs. H. Shelton Earp III and Carlos Garcia-Echeverria for providing the HER4 antibody and NVP-BEZ235, respectively. This work was supported by R01 CA80195 (CLA), ACS Clinical Research Professorship Grant CRP-07-234 (CLA), Breast Cancer Specialized Program of Research Excellence (SPORE) P50 CA98131, and Vanderbilt-Ingram Comprehensive Cancer Center Support Grant P30 CA68485.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated in high frequency in human breast cancers. Cancer Biol. Therapy. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Feo DD, Godwin AK, Bell DW, Cheng JQ, Altomare DA, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennnessy BT, Mardiredjo M, Hiijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzuamb resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DYH, Montgomery KG, Ciavarella ML, Hooi CSF, et al. Mutation in the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Yuan TL. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem. J. 2008;409:519–524. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/AKT pathway in cancer. Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- Geering B, Cutillas PR, Vanhaesebroeck B. Regulation of class IA PI3Ks: is there a role for monomeric PI3K subunits? Biochem. Soc. Trans. 2007;35:199–203. doi: 10.1042/BST0350199. [DOI] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J. Clin. Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymnopoulos M, Elsliger M-A, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, III, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast-cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, Fields C, Phillips GDL, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br. J. Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- Lai Y-L, Mau B-L, Cheng W-H, Chen H-M, Chiu H-H, Tzen C-Y. PIK3CA exon 20 mutation is independently associated with a poor porognosis in breast cancer pateints. Annals Surgical Oncol. 2008;15:1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, et al. A central role of HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- Lee JY, Engelman JA, Cantley LC. Biochemistry. PI3K charges ahead. Science. 2007;317:206–207. doi: 10.1126/science.1146073. [DOI] [PubMed] [Google Scholar]
- Lerma E, Catasus L, Gallardo A, Peiro G, Gallardo A, Peiro G, et al. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–139. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res. Treatment. 2006;96:91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- Maira SM, Stuffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- Medina PJ, Goodin S. Lapatinib: a dual inhibitor of human epidermal growth factor receptor tyrosine kinases. Clin. Ther. 2008;30:1426–1447. doi: 10.1016/j.clinthera.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Miled N, Yan Y, Hon W-C, Perisic O, Zvelebil M, Inbar Y, et al. Mechanisms of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Lan K-H, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trstuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Nahta R, Yu D, Huang M-C, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat. clin. Practice Oncol. 2006;3:269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- Revillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat J-P. ErbB/HER ligands in human breast cancer, and realationships with the receptors, the biopathological features and prognosis. Annals Oncol. 2008;19:73–80. doi: 10.1093/annonc/mdm431. [DOI] [PubMed] [Google Scholar]
- Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, et al. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin. Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- Slamon D, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpress HER2. New Eng. J. Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-S, Shanmon-Taylor LA, Mehmi I, Tang CK, Lupu R. Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene. 2003;22:761–768. doi: 10.1038/sj.onc.1206130. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J. Biol.Chem. 2004;279:24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Wang SE, Xiang B, Guix M, Olivares MG, Parker J, Chung CH, et al. Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol. Cell. Biol. 2008;28:5605–5620. doi: 10.1128/MCB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and AKT is required for antibody-mediated effects on p27, Cyclin D1, and antitumor action. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untanglling the ErbB signaliling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.