Abstract
Alzheimer’s disease (AD) is a common cause of dementia with a strong genetic component and risk sharply increasing with age. We performed two parallel microarray experiments to independently identify genes involved in normal aging and genes involved in AD using RNA extracted from the temporal lobe of 22 late onset AD and 23 control brain donors. We found that AD is accompanied by significant changes in the expression of many genes with up-regulation of genes involved in inflammation and in transcription regulation and down-regulation of genes involved in neuronal functions. The changes with healthy aging involved multiple genes but were not as strong. Replicating and strengthening previous reports we find a highly significant overlap between genes changing expression with age and those changing in AD and we observe that those changes are most often in the same direction. This result supports an overlap between the biological processes of normal aging and susceptibility to AD and suggests that age related genes expression changes might increase the risk to develop AD.
Keywords: Dementia, gene expression, aging, microarray, brain, temporal lobe
1. Introduction
Alzheimer’s disease is the most common cause of dementia in the U.S. affecting an estimated 5.2 million (Alzheimer’s association report 2008). With few exceptions of familial cases due to mutations in one of three known genes, APP (Goate, et al., 1991), PSEN1 (Sherrington, et al., 1995) and PSEN2 (Levy-Lahad, et al., 1995), AD has a late age of onset most often after the age of 65. It presents with progressive loss of multiple cognitive abilities leading within an average of 8 years to severe dementia and death. Although it is a complex genetic disorder, late onset AD is in large extent due to genetic predisposition with a heritability calculated between 0.6 and 0.74 (Bergem, et al., 1997,Gatz, et al., 1997). Despite this major genetic influence only one gene, APOE encoding for Apolipoprotein E, has been consistently shown to be involved in the risk for late onset AD (Strittmatter, et al., 1993), while a few more genes with variants contributing significantly less to the risk are now emerging through recent large genome wide association studies (Harold, et al., 2009,Lambert, et al., 2009).
Like most cell types, neurons respond to normal or abnormal stimuli – such as those involved in a disease process – by setting in motion signaling cascades and modifying their internal and external microenvironment. Among the responses are changes in the priorities of the protein synthesis machinery, observed as changes in gene transcription and the levels of the relevant mRNAs. Measuring these changes can provide information on the nature of the genes that play a role in the disease process and allow comparisons with other physiological or pathological states. In some cases an observed difference in the levels of a particular mRNA between diseased and control tissue might reflect the primary defect that contributes to the risk. A challenge in this type of analysis is to distinguish between the primary and secondary alterations. Nevertheless, knowledge of gene expression changes involved in a disease may prove useful in understanding the disease process and possibly exploring preventions and treatments. Further, gene expression profiling can itself be a means to test and develop new drugs (Gerhold, et al., 2002). Today there are multiple array-based choices for surveying genome wide gene expression that differ in their content, the probe preparation methods and the chemistry of the array surface. The most commonly used include laboratory developed cDNA arrays and commercial gene chip products from Affymetrix®, Illumina®, Amersham® Agilent® NimbleGen® and other biotechnology companies. Such chips examine thousands of genes often covering much more than the well characterized genes in the genome and in some cases interrogating individual exons.
There have been many studies investigating gene expression changes in AD (Blalock, et al., 2004,Colangelo, et al., 2002,Dunckley, et al., 2006,Emilsson, et al., 2006,Ginsberg, et al., 2000,Haroutunian, et al., 2009,Kong, et al., 2009,Liang, et al., 2007,Liang, et al., 2008,Loring, et al., 2001,Parachikova, et al., 2007,Ray, et al., 2008,Ricciarelli, et al., 2004). Among the multiple variables that can influence the results of such studies are the selection of tissue type or brain region, the expression analysis platform and the analytical methods. Differences in these variables between different studies together with small sample sizes, stochastic and other variation, have often led to inconsistent observations. Most studies use one of two main approaches to the interpretation of their results. Some focus on the individual dysregulated genes and make hypotheses on the possible roles of the gene products in the disease process. Others identify groups of genes either by setting a significance threshold or through gene co-expression network analyses (Zhang and Horvath, 2005) and then examine the groups for excess representation of specific functional classes. Although such groups likely contain false positives overall they are highly enriched for true positives and their composition can provide significant and reliable results. Although different platforms and analytical methods can lead to different results at the individual gene level, gene class enrichment is robust across platforms (Maouche, et al., 2008) and while this approach does not identify specific target genes it provides important insights into the possible disease mechanisms and consequences of the disease at the molecular level.
Our motivation for this study was two-fold. First, we wanted to provide new insights and add support to conclusions from previous gene expression studies of AD. Second, we wanted to test for an overlap between gene expression changes in AD and in normal aging as suggested by a previous report (Miller, et al., 2008), a phenomenon that we think could of great importance to our understanding of the genetics of AD. In two parallel studies we examined the gene expression profile of Broadman area 22 (superior temporal lobe), an area strongly affected by AD pathology, in 22 AD cases and 23 controls without brain pathology at death. The samples were split in two independent sets, one focusing in AD and using a subset of 9 controls matched to the cases and another focusing on changes with age using the remaining 14 controls with a relatively wide spectrum of ages and no AD cases. We used the Illumina Sentrix HumanRef-8 Expression BeadChips that interogate 24,000 genes recognized by the National Center for Biotechnology Information (NCBI). We report on genes showing significant changes in AD, on functional enrichments among genes changing expression and on a highly significant overlap between the two groups, an observation that we replicated using a third, public dataset.
2. Materials and methods
2.1 Samples
We obtained 3mm punch biopsies from the superior temporal lobe (Brodman area 22) of 22 deceased patients with confirmed AD pathology and 23 controls with no brain pathology. All cases and controls were of European descent and their ages, sex and the time between death flash freezing of the brain slices (Post Mortem Delay; PMD) are shown on supplementary Table 1 together with Braak staging and CERAD scores for cases.
Samples were split in two sets,(i) the AD sample-set of 22 cases and 9 controls with no significant differences in age, sex, PMD, or positioning on the Illumina Beadchips and (ii) the AGE sample-set with a wider age range (35–93) consisting of samples free of pathology and with no significant correlations between age and PMD, sex or placement on Illumina Beadchips. All the details pertaining to the sets are shown on supplementary table 1. All samples were from brains collected by the Johns Hopkins Brain resource center (courtesy of the director Dr. Juan Troncoso).
2.2 Transcript measurements
To measure transcript abundance we used the Illumina Sentrix HumanRef-8 Expression BeadChips (Illumina, San Diego, CA 92121-1975, cat. no. 11201828) containing 24,000 genes recognized by NCBI at the time of production. We extracted total RNA using Trizol (Invitrogen, Carlsbad, California 92008, cat. no. 15596-026) with additional purification on RNAeasy columns (Qiagen, Valencia, CA 913555, cat. no. 74104). We assessed the quality of total RNA on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and 0.5 µg of total RNA from each sample was labeled by using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX 78744-1832, cat. no. IL1791) in a process of cDNA synthesis and in vitro transcription. We generated and labeled single-stranded RNA (cRNA) by incorporating biotin-16-UTP (Roche Diagnosics GmbH, Mannheim, Germany, cat. no. 11388908910) and hybridized (16 hours) a total of 0.85 µg of biotin-labeled cRNA to the BeadChips. The hybridized biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using Illumina's BeadStation 500GX Genetic Analysis Systems scanner. The primary Illumina data was returned from the scanner in the form of an “.idat” file which contains single intensity data values/gene following the computation of a trimmed mean average for each probe type represented on the array by a variable number of bead probes. We performed preliminary analyses of the scanned data using Illumina BeadStudio software which returns a detection call D based on a comparison between the intensity of a single probe and the intensities of a large number of negative control beads built-in to the BeadChip arrays (D = % above negative/100, 1 = perfect, i.e. the intensity value of a gene is greater than all the intensities for every negative control tested). Any gene consistently below D=0.98 was eliminated from further analysis, leaving data for 11,326 named genes expressed in temporal lobe for analysis. Normalization of the expression values to account for differences in input RNA, processing, labeling etc. was performed by Z-transformation on each sample/array on a stand-alone basis (Cheadle, et al., 2003).
Replications using SYBR-green real time detection with Applied Biosystems reagents (Foster City, CA, cat#4312704) and an ABI 7900 sequence detection system (Applied Biosystems) were performed on newly extracted RNA from the same tissue sample. After the examination of melting curves excluding non specific PCR products, relative quantification of each sample was performed using a standard curve from standardized dilutions of a reference RNA. Measurements were normalized against the average of two housekeeping genes (M-RIP and POLR2) selected from a study that identified AD – appropriate reference genes using a neuroblastoma cell line that models aspects of Alzheimer's disease in culture (Hoerndli, et al., 2004). Ratios to the reference were log transformed resulting in normally distributed values.
2.3 Data analysis
We used Z-normalized expression values of each transcript as the dependent variable in a generalized linear model that included disease status, age, sex and PMD for the AD sample-set and age, sex and PMD for the AGE sample-set. The analyses were performed in R (version 2.4.1, http://cran.r-project.org) using the “glm” function for generalized linear models. Expression change was investigated in the AGE sample-set while changes with disease were investigated separately in the independent AD sample-set. We also used a set of public data, those from a study by Myers et al (Myers, et al., 2007), as a third independent set for the effects of age. We analyzed the temporal lobe data of that dataset using the rank invariant normalized data supplied by the authors as an outcome and including age, PMD, sex and sample source in the generalized linear model.
The results were then parsed on excel spreadsheets, where quantile quantile (Q-Q) plots were generated, false discovery rate (FDR) was calculated from the p-values following the Benjamini and Hochberg procedure (Benjamini and Hochberg, 1995), results of the AGE sample-set and AD sample-set were matched by gene and compared in parallel and gene lists were generated for functional enrichment analyses.
We used the expression data analysis tools provided by the Panther classification system website (http://www.pantherdb.org/tools/genexAnalysis.jsp) (Mi, et al., 2007,Thomas, et al., 2003) for enrichment analyses for specific gene functions. We used lists of genes showing changes in the generalized linear models described above at the chosen FRD thresholds against the reference set of all genes whose transcript was positively detected by the array (thus all genes that could possibly be included in the list of genes with expression changes). The Panther website performs a modified Bonferroni correction which accounts for the nesting of child gene ontology terms below parent terms. The p-values shown on Table 1 are Bonferroni corrected through this method. The significance of overlaps between lists of genes with expression changes was assessed using standard 2×2 tables of counts of genes present or absent in each list and were tested for independence by chi-square tests.
Table 1. Enrichments in genes expressed higher in AD.
Functional enrichment for genes expressed at higher or lower levels in AD cases. The category column refers to the ontology categories used for the analysis.
| Higher expression in AD | category | number of genes |
fold enrichment |
corrected significance |
|---|---|---|---|---|
| FDR < 0.05 – 504 genes | ||||
| Interleukin signaling pathway | Pathway | 36 | 2.3 | ** |
| mRNA transcription | Biol. Proc. | 222 | 1.4 | *** |
| Oncogenesis | Biol. Proc. | 68 | 1.8 | *** |
| Nucleoside, nucleotide and nucleic acid metabolism | Biol. Proc. | 353 | 1.2 | ** |
| Hematopoiesis | Biol. Proc. | 18 | 3.3 | ** |
| mRNA transcription regulation | Biol. Proc. | 167 | 1.4 | ** |
| Cell structure and motility | Biol. Proc. | 132 | 1.4 | ** |
| Immunity and defense | Biol. Proc. | 137 | 1.4 | ** |
| Macrophage-mediated immunity | Biol. Proc. | 21 | 2.5 | * |
| Cell proliferation and differentiation | Biol. Proc. | 119 | 1.3 | * |
| Developmental processes | Biol. Proc. | 211 | 1.2 | * |
| Transcription factor | Mol. Funct. | 221 | 1.3 | *** |
| Nucleic acid binding | Mol. Funct. | 282 | 1.3 | *** |
| Lower expression in AD | category |
number of genes |
fold enrichment |
corrected significance |
| FDR<0.05 – 526 genes | ||||
| Neuronal activities | Biol. Proc. | 117 | 1.9 | *** |
| Synaptic transmission | Biol. Proc. | 63 | 1.9 | *** |
| Nerve-nerve synaptic transmission | Biol. Proc. | 23 | 2.7 | ** |
| Ion channel | Mol. Funct. | 57 | 1.7 | ** |
| Voltage-gated ion channel | Mol. Funct. | 31 | 2.1 | * |
| Neuropeptide | Mol. Funct. | 11 | 3.6 | + |
p<0.1,
p<0.05,
p<0.01,
p<0.001. All p values are Bonferroni corrected.
3. Results
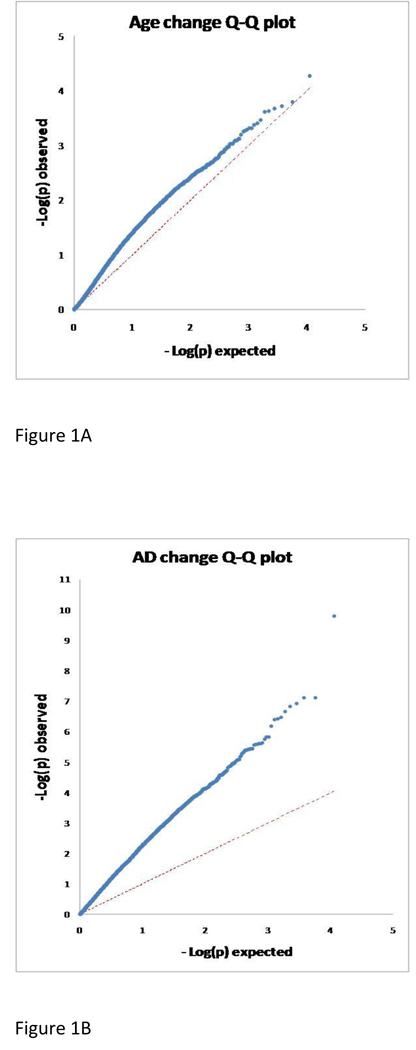
All 45 cases and controls provided good quality RNA without significant degradation, as shown by gel electrophoresis and analysis on the Agilent 2100 Bioanalyzer, and were successfully processed and analyzed by the Illumina BeadStudio software. Expression of a total of 11,326 named genes was positively detected by Beadstudio and their data were processed as described in the materials and methods. Figure 1 shows Q-Q plots of the distribution of p-values for expression changes with AD and with age and supplementary Figure 1 shows volcano plots for the two datasets. There is clear inflation of low p-values in the AD sample-set and 1,031 genes were found dysregulated at an FDR <0.05. In the AGE sample-set there is also an inflation of signals at p-values between 0.05 and 0.001 which however does not continue at lower p-values, suggesting multiple true signals yet small effects that cannot reach strong statistical significance. In agreement with this, the FDR is 0.4 at p< 0.0415 but does not improve much thereafter (lowest FDR is 0.345 at p<0.0086). For this reason we chose this relatively relaxed FDR (0.4) to define genes regulated with aging. Although this is expected to include a significant number of false positives it also has the highest enrichment for true positives we can achieve for such a large group, consisting of 1,174 genes more than half of which are expected to be true positives. Of these genes 604 (51.4%) showed decreased expression with age. Sex and PMD did not show strong effects with the exception of four genes, all located on the Y chromosome that showed a strong sex effect on both sample-sets, reaching an FDR of less than 0.05 in the larger AD sample-set. One of them also reached FDR<0.05 for sex regulation in the smaller AGE sample-set analyses. Among the genes that were downregulated with age there was significant enrichment for those whose products are involved in pre-mRNA processing (p=0.0025), splicing (p=0.004) and for ribosomal protein genes (p=0.00001). No significant enrichment for functional classes was observed among genes up-regulated with age.
Figure 1.
Figure 1A: Quantile Quantile plot for the observed p-value distribution for the effect of age on gene expression compared with the expected null distribution
Figure 1B: As in figure 1A for the effect of AD on gene expression.
The analysis of the AD sample set revealed many highly significant differences with 1,031 genes showing change in expression at FDR <0.05, 51% of these showing reduced expression in AD cases (see supplementary Table 2 for the complete list). The up-regulated genes were enriched for transcription factors (p=2.3×10−3) and genes involved in nucleic acid metabolism(p=0.023). Down-regulated genes were enriched for genes involved in neuronal activities (p=7.5×10−3), specifically for voltage-gated ion channels (p=0.022). Using a more relaxed FDR threshold of 0.2 which identified 1,804 down-regulated and 1,602 up-regulated genes we were able to achieve much stronger statistical evidence of enrichment shown on Table 1.
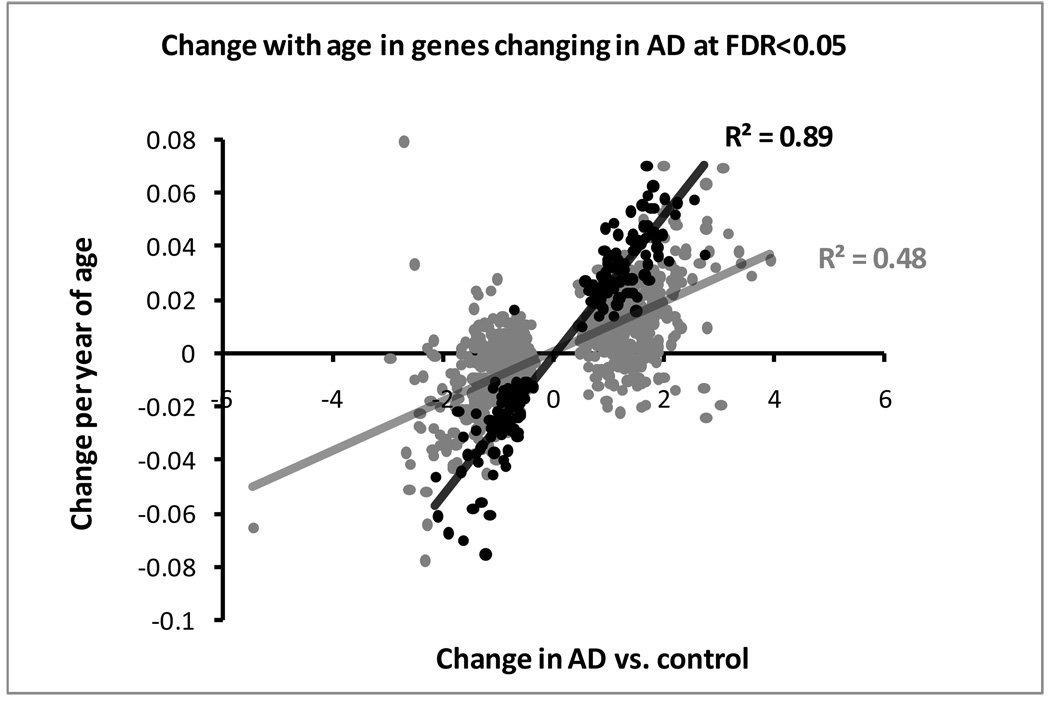
We then examined the hypothesis that the set of genes changing with age might be enriched for genes involved in AD. Using the same FDR levels above (0.2 for AD and 0.4 for age) 3,406 of the 11,326 genes change expression levels in AD and 1,174 change with age. The two groups shared 451 genes, significantly more than expected overlap by chance (p=4.5×10−11). The significance remained strong if we used the stringent FDR of 0.05 for AD with 166 overlapping genes (p=2.1×10−10). Strikingly, in all but one of the 166 (and in 95% of the 451) overlapping genes the change with increasing age was in the same direction with the change in AD. This is illustrated on the scatter plot in Figure 2. In order to exclude any systematic error in our two parallel experiments we further examined a public dataset that was informative for gene regulation with age, that of Myers at al (Myers, et al., 2007). We downloaded the data and analyzed them as described in our materials and methods for the effect of age. There were in total 9,743 transcripts with present calls in at least two thirds of individuals. This dataset provided somewhat more significant results than ours, presumably because of the larger sample size (131 samples from the temporal lobe) with 10 transcripts reaching an FDR<0.1, yet none an FDR <0.05. The Myers et al study was performed on an Affymetrix array and among the transcripts called present we could match 6,368 to those with present calls in our sample-sets. Of the 86 transcripts overlapping at FDR<0.4 between our AGE sample set and the Myers et al set 76 (88%) showed change in the same direction for both sample sets, supporting the validity of the results. The Myers et all dataset included 690 genes at FDR <0.4, 415 decreasing with age and enriched in peptide hormone genes (p=0.033) and 275 increasing with age with no significant enrichments. We compared the genes changing in AD from our AD sample set (FDR<0.2) to the genes changing with age from the Myers dataset (FDR <0.4) and again we found a significantly high overlap between the two sets. Of the 6,368 genes in common 1304 (our AD sample set) changed in AD, 503 with age (Myers dataset) and 150 were in common (p=6×10−8). Once again the vast majority (133, 89%) were changing in the same direction with age and AD.
Figure 2.
Scatter plot of effects on gene expression (linear model parameter estimates) of genes that change significantly in AD at FDR <0.05. Black and grey dots correspond to genes that change significantly with age at FDR <0.4 or not. A strong directional correlation is observed, stronger for genes with a significant age effect.
We compared our results with the list of genes reported by Miller et al to be overlapping in AD and aging with at least one significant probe set (Miller, et al., 2008) (found on their suppl. Table 6). In their results, strong excess of genes with same the directionality of change was also evident, yet not as striking as we observe. The overlap of their list of 558 genes changing both with age and AD with our list of 451 (FDR <0.2 and 0.4 for AD and age respectively) was 31 genes. For 23 of those (74%) the results of both studies were in the same direction both for aging and AD. Only one of the remaining genes showed completely discordant results between studies (see suppl. Table 3). Finally we compared our list of age regulated genes with those reported by Lu et al (Lu, et al., 2004) in their Supplementary Table 2. All 19 genes that we could match by RefSeq name from their table and were present in our list of age regulated genes at FDR <0.4 showed the same direction of effect, further validating our results.
We experimentally validated our microarray results by extracting new RNA from the AD sample-set and measuring expression by real time PCR. We tested eight genes, three with FDR <0.05 (VGF, RBP4, ADCYAP1) and five with FDR 0.2 (SOX10, NPTXR, VMD2, SP1, BDNF). In all eight cases the new measurement showed an effect in the same direction as the microarray. In five cases including all three at FDR <0.05 the result replicated with at least nominal significance (see suppl. Table 4) . The remaining three showing the same direction but without nominal significance are likely due to the platform differences, noise introduced by harvesting new tissue and extracting new RNA and likely also include false positives.
4. Discussion
We have performed a large microarray based gene expression study exploring the effects of age on gene expression and comparing it to gene expression changes in AD. We found that the effects of aging on gene expression is relatively subtle yet it involves multiple genes. Our separate study of transcript abundance differences between AD affected brains and unaffected controls showed multiple significant differences with high statistical confidence. Most importantly, when comparing the results of the expression study on AD with that on aging we found that more genes than expected were affected by both and almost always in the same direction, i.e. genes whose expression goes down with age are often found to be lower in AD affected brains and vice versa. This result had highly significant statistical support and was observed almost as strong when we analyzed data on the effect of aging from a completely independent publicly available data set using a different platform. In fact the overlap was stronger when age effects were calculated using the Myers et al data (~50% higher than expected by chance) than when using our own (~30% higher), reflecting perhaps the higher number of control brains sampled.
We are providing a list of 1030 genes in our supplementary material (suppl. Table 2) that show differences in expression in AD at FDR<0.05. This is a high confidence list likely to include mostly true positives and it includes multiple genes that have been previously reported. Some of these genes could reflect primary changes, i.e. they could be responsible for the development of the disease, but most are likely secondary changes, in response to the disease process. Our functional enrichment results are consistent with previous literature and provide additional support for specific functions while expanding the observations to Brodmann area 22. We found an enrichment for genes in the Interleukin signaling pathway, immunity and defense and specifically macrophage mediated immunity among the genes with higher expression in AD (Table 1) which supports links between inflammation and AD (DeLegge and Smoke, 2008,McGeer and McGeer, 1998,Wyss-Coray, 2006). We also found an enrichment in transcription factors and other genes involved in transcription, likely reflecting the induction of cellular responses by the disease process. Among the genes with lower expression in the AD brain we found enrichments in genes involved in synaptic transmission, ion channels and generally genes involved in neuronal activities. Our results replicate strengthen and expand previously published similar findings (Katsel, et al., 2005,Papassotiropoulos, et al., 2006) elaborating on important aspects of the AD process.
The result that we found most striking and also carried the strongest statistical support was the overlap between genes dysregulated with AD and genes that change expression with age. Such an overlap was first described by Miller et al (Miller, et al., 2008) who performed a systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. Here we provide strong replication using multiple independent datasets and we show a strong directionality of this phenomenon. This significantly increased overlap could result from many different underlying links between aging and AD related genes, and is of particular interest because advanced age is the most significant AD risk factor. It is possible that changes in some genes’ expression with age, although part of the normal aging process, can also lead to increased vulnerability to AD. It is possible that for a subset of such genes changes might happen faster for some individuals – perhaps due in part to genetic variation – leading to increased vulnerability. As these individuals would end up in our case group they could produce the observed results. The overlap might also reflect a globally accelerated aging process in the people that are vulnerable to AD, which could be due to genes, environment or both. It must be noted that the small effects on gene expression observed with age forced us to adapt an FDR of 0.4, meaning that a significant number of false positives are included in the results. This might reduce the confidence in the validity of individual gene results however it does not reduce the importance of the highly significant overlap which would be expected even stronger if we could clear false positives off our lists.
The genetic overlap of aging and AD has important implications for aging research. It would be useful to perform more and larger studies covering more brain regions and more patients and controls to confidently identify this set of overlapping genes. These genes are likely to be important to healthy aging and possibly primary culprits for vulnerability to AD, either of which would make them important targets for pharmacological intervention. The observed low effect of normal aging on gene expression which currently translates to low statistical confidence for individual genes underscores the importance of further research on expanded datasets, as defining the exact overlap between normal aging and AD could lead to significant breakthroughs in our understanding and our therapeutic approach to the disease.
Supplementary Material
Acknowledgements
This work was supported by N.I.A. grants to DA and SSB (RO1AG022099 and RO1AG021804) and an award from the Neurosciences Education and Research Foundation to D.A. We thank the Johns Hopkins Brain Resource Center and the director Dr. Juan Troncoso for contributing the tissue samples studied here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors have no actual or potential conflicts of interest to disclose.
REFERENCES
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54(3):264–270. doi: 10.1001/archpsyc.1997.01830150090013. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101(7):2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Cho-Chung YS, Becker KG, Vawter MP. Application of z-score transformation to Affymetrix data. Appl Bioinformatics. 2003;2(4):209–217. [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70(3):462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- DeLegge MH, Smoke A. Neurodegeneration and inflammation. Nutr Clin Pract. 2008;23(1):35–41. doi: 10.1177/011542650802300135. doi:23/1/35 [pii]. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG, LaFleur BJ, Coon KD, Brown KM, Caselli R, Kukull W, Higdon R, McKeel D, Morris JC, Hulette C, Schmechel D, Reiman EM, Rogers J, Stephan DA. Gene expression correlates of neurofibrillary tangles in Alzheimer's disease. Neurobiol Aging. 2006;27(10):1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson L, Saetre P, Jazin E. Alzheimer's disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiol Dis. 2006;21(3):618–625. doi: 10.1016/j.nbd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52(2):M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- Gerhold DL, Jensen RV, Gullans SR. Better therapeutics through microarrays. Nat Genet. 2002;32 Suppl:547–551. doi: 10.1038/ng1042. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer's disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48(1):77–87. [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009 doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V, Katsel P, Schmeidler J. Transcriptional vulnerability of brain regions in Alzheimer's disease and dementia. Neurobiol Aging. 2009;30(4):561–573. doi: 10.1016/j.neurobiolaging.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Toigo M, Schild A, Gotz J, Day PJ. Reference genes identified in SH-SY5Y cells using custom-made gene arrays with validation by quantitative polymerase chain reaction. Anal Biochem. 2004;335(1):30–41. doi: 10.1016/j.ab.2004.08.028. doi:S0003-2697(04)00691-8 [pii] [DOI] [PubMed] [Google Scholar]
- Katsel PL, Davis KL, Haroutunian V. Large-scale microarray studies of gene expression in multiple regions of the brain in schizophrenia and Alzheimer's disease. Int Rev Neurobiol. 2005;63:41–82. doi: 10.1016/S0074-7742(05)63003-6. [DOI] [PubMed] [Google Scholar]
- Kong W, Mou X, Liu Q, Chen Z, Vanderburg CR, Rogers JT, Huang X. Independent component analysis of Alzheimer's DNA microarray gene expression data. Mol Neurodegener. 2009;4(1):5. doi: 10.1186/1750-1326-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009 doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KA, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer's disease locus on chromosome 1. Science. 1995;269(5226):970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette C, Schmechel D, Alexander GE, Reiman EM, Rogers J, Stephan DA. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer's disease. DNA Cell Biol. 2001;20(11):683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. doi:10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Maouche S, Poirier O, Godefroy T, Olaso R, Gut I, Collet JP, Montalescot G, Cambien F. Performance comparison of two microarray platforms to assess differential gene expression in human monocyte and macrophage cells. BMC Genomics. 2008;9:302. doi: 10.1186/1471-2164-9-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL. The importance of inflammatory mechanisms in Alzheimer disease. Exp Gerontol. 1998;33(5):371–378. doi: 10.1016/s0531-5565(98)00013-8. doi:S0531-5565(98)00013-8 [pii]. [DOI] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35(Database issue):D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28(6):1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–1499. doi: 10.1038/ng.2007.16. doi:ng.2007.16 [pii] [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Fountoulakis M, Dunckley T, Stephan DA, Reiman EM. Genetics, transcriptomics, and proteomics of Alzheimer's disease. J Clin Psychiatry. 2006;67(4):652–670. doi: 10.4088/jcp.v67n0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachikova A, Agadjanyan MG, Cribbs DH, Blurton-Jones M, Perreau V, Rogers J, Beach TG, Cotman CW. Inflammatory changes parallel the early stages of Alzheimer disease. Neurobiol Aging. 2007;28(12):1821–1833. doi: 10.1016/j.neurobiolaging.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, Ruan J, Zhang W. Variations in the transcriptome of Alzheimer's disease reveal molecular networks involved in cardiovascular diseases. Genome Biol. 2008;9(10):R148. doi: 10.1186/gb-2008-9-10-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciarelli R, d'Abramo C, Massone S, Marinari U, Pronzato M, Tabaton M. Microarray analysis in Alzheimer's disease and normal aging. IUBMB Life. 2004;56(6):349–354. doi: 10.1080/15216540412331286002. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12(9):1005–1015. doi: 10.1038/nm1484. doi:nm1484 [pii] [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.